Abstract
Bacterial polynucleotide phosphorylase (PNPase) plays a major role in mRNA turnover by the degradation of RNA from the 3′- to 5′-ends. Here, we determined the crystal structures of the wild-type and a C-terminal KH/S1 domain-truncated mutant (ΔKH/S1) of Escherichia coli PNPase at resolutions of 2.6 Å and 2.8 Å, respectively. The six RNase PH domains of the trimeric PNPase assemble into a ring-like structure containing a central channel. The truncated mutant ΔKH/S1 bound and cleaved RNA less efficiently with an eightfold reduced binding affinity. Thermal melting and acid-induced trimer dissociation studies, analyzed by circular dichroism and dynamic light scattering, further showed that ΔKH/S1 formed a less stable trimer than the full-length PNPase. The crystal structure of ΔKH/S1 is more expanded, containing a slightly wider central channel than that of the wild-type PNPase, suggesting that the KH/S1 domain helps PNPase to assemble into a more compact trimer, and it regulates the channel size allosterically. Moreover, site-directed mutagenesis of several arginine residues in the channel neck regions produced defective PNPases that either bound and cleaved RNA less efficiently or generated longer cleaved oligonucleotide products, indicating that these arginines were involved in RNA binding and processive degradation. Taking these results together, we conclude that the constricted central channel and the basic-charged residues in the channel necks of PNPase play crucial roles in trapping RNA for processive exonucleolytic degradation.
Keywords: polynucleotide phosphorylase, mRNA turnover, RNase, RNA degradation, crystal structure
INTRODUCTION
Messenger RNA turnover plays a vital role in quality control of RNA biogenesis and modulation of protein expression, and as such, it regulates diverse physiological events (Song and Parker 2004; Carpousis 2007). Eukaryotic mRNAs are largely degraded by the protein complex, the exosome, from the 3′ to 5′ direction (Raijmakers et al. 2004; Houseley et al. 2006). In parallel, the bacterial polynucleotide phosphorylase (PNPase), which shares similar domain organization to the eukaryotic exosome core complex, plays a key role in the degradation of prokaryotic mRNAs (Marcaida et al. 2006).
PNPase is a 3′ to 5′ exonuclease, using phosphate to catalyze phosphorolysis of RNA and generating nucleoside diphosphates as the cleavage products (Sarkar and Fisher 2006). PNPase is evolutionarily conserved, present in almost all species from bacteria to plants and higher mammals, but not in yeasts, trypanosomes, or archaea (Baginsky et al. 2001; Leszczyniecka et al. 2002; Sarkar and Fisher 2006). Mammalian PNPase is intriguingly located in the mitochondrial intermembrane space where no RNA is present (Piwowarski et al. 2003). Recent studies suggest that human PNPase has a crucial role in maintaining mitochondrial homeostasis and is induced by type I interferons, likely playing a role in antiviral defense and cellular senescence (Leszczyniecka et al. 2003; Chen et al. 2006, 2007; French et al. 2007). However, the molecular basis for the mammalian PNPase regulation of these fundamental physiological events is largely unknown. Conversely, bacterial and plant chloroplast PNPases have been studied more extensively at the molecular level. Both enzymes participate in the rapid exonucleolytic degradation of polyadenylated messenger RNAs that are generated through endonucleolytic cleavage of RNA, followed by the addition of a poly(A) or poly(A)-rich tail (Schuster et al. 1999; Mohanty and Kushner 2000; Yehudai-Resheff et al. 2003).
Escherichia coli PNPase is located in the cytoplasm, and a small fraction of the enzyme is associated with RNase E in the protein complex, the degradosome (Carpousis et al. 1994; Liou et al. 2001). The enzyme is primarily involved in the processive phosphorolytic degradation of RNA, but the reverse polymerization of ribonucleoside diphosphates has also been observed in vivo (Mohanty and Kushner 2000). Bacterial PNPases are homotrimers, each monomer containing two N-terminal RNase PH domains linking by an α-helix domain, and a C-terminal RNA-binding S1 and KH domain (see Fig. 1A). The crystal structure of Streptomyces antibioticus PNPase has been determined, revealing a doughnut-shaped structure, forming by six RNase PH domains in a homotrimer (Symmons et al. 2000). This overall organization of a hexameric ring structure with the RNA-binding domain associated on the top of the ring is similar to those of archaeal (Buttner et al. 2005; Lorentzen and Conti 2005; Lorentzen et al. 2005) and human exosome (Liu et al. 2006) core complexes, indicating evolutionary links in structure and function between PNPase and exosomes in RNA degradation (Symmons et al. 2002; Lin-Chao et al. 2007).
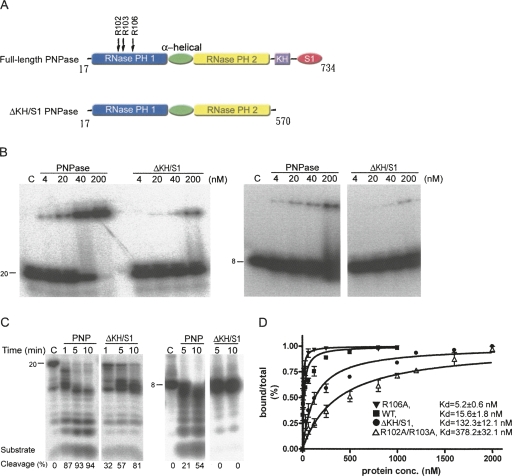
FIGURE 1.
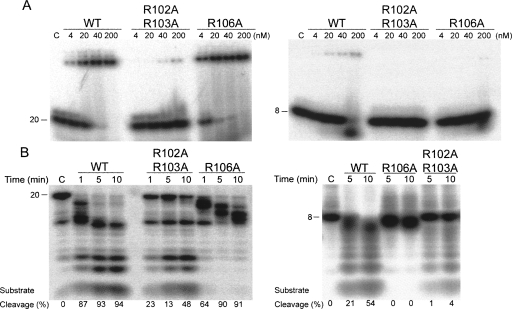
RNA-binding and digestion activities of the E. coli full-length PNPase and KH/S1-domain truncated mutant (ΔKH/S1). (A) Domain structures of the full-length PNPase and the truncated mutant ΔKH/S1. (B) RNA-binding activity of full-length PNPase and ΔKH/S1 were analyzed by gel shift assays. The 20-mer (left panel) and 8-mer (right panel) single-stranded RNA substrates (0.1 pmol) were incubated with PNPase in various concentrations from 4 to 200 nM. A control reaction without any PNPase added is shown in lane C. The ΔKH/S1 mutant bound RNA less efficiently than full-length PNPase. (C) RNase activities of full-length PNPase and ΔKH/S1 mutant were assayed by incubation of enzymes, respectively, with 8-mer and 20-mer single-stranded RNAs in time course experiments under the conditions described in Materials and Methods. The left panel shows that ΔKH/S1 had lower activities in cleaving 20-mer ssRNAs compared to the full-length PNPase. The right panel shows that ΔKH/S1 cannot degrade shorter 8-mer ssRNAs. (D) The dissociation constants between PNPase and the 20-mer ssRNA were estimated by gel shift assays.
How do PNPase and exosomes bind and cleave RNA processively from the 3′-end? The current model suggests that the conserved KH and S1 domains are involved in RNA binding, since deletion of any or both of the RNA-binding domains of E. coli PNPase reduces its RNA binding as well as its enzyme activity (Stickney et al. 2005; Briani et al. 2007; Matus-Ortega et al. 2007). The crystal structures of archaeal exosome in complex with tungstate (mimicking phosphate), and in complex with RNA, further suggest that the single-stranded RNA substrate is threaded through the central channel of the ring-like structure, as tungstates and 4-nucleotide (nt) RNAs are bound at the three active sites buried inside the channel (Buttner et al. 2005; Lorentzen and Conti 2005; Navarro et al. 2008). The threading model provides the basis for the discrimination between structured and unstructured RNAs by PNPase and exosomes since they only degrade single-stranded RNAs. Moreover, the crystal structure of the archaeal exosome bound with RNA substrates shows fragment RNA bound both at the active site and at the narrowest constriction of the central channel near the channel entrance (Lorentzen et al. 2007; Navarro et al. 2008). These results demonstrate that single-stranded RNAs are indeed threaded through the channel, particularly bound at the constricted neck region.
All the previous studies support the hypothesis that the conserved RNA-binding subunits as well as the central channel play a vital role in RNA binding and processive degradation in the exosome and in PNPase. Here, to further address the molecular basis of E. coli PNPase activity, we determined the crystal structures of the full-length and the KH/S1-truncated form of PNPase. In combination with biochemical and biophysical data, we show that the KH and S1 domains are involved not only in RNA binding but also in trimer formation. Our results also show that a properly constricted channel and the basic residues located in the channel neck regions in PNPase play critical roles in trapping RNA for processive degradation. This finding is likely a general phenomenon since only bacterial PNPase and archaeal exosomes with constricted channels are efficient enzymes in RNA degradation.
RESULTS
The KH/S1 truncated PNPase binds and cleaves RNA less efficiently
E. coli PNPase contains a C-terminal KH and an S1 domain, which have been identified in a number of nucleic acid-binding proteins (Theobald et al. 2003). Previous studies showed that the KH and S1 domains in PNPase are involved in RNA binding (Stickney et al. 2005; Briani et al. 2007; Matus-Ortega et al. 2007). To further study the interactions between PNPase and RNA, we quantitatively measured the RNA binding affinities and RNA cleavage efficiencies of wild-type PNPase and KH/S1 domain-truncated mutant. We purified full-length PNPase (Fig. 1A, residues 17–734) and the KH/S1 domain-truncated mutant (Fig. 1A, ΔKH/S1, residues 17–570), and compared their RNA binding activities by electrophoresis mobility shift assays, using a 20-mer and an 8-mer single-stranded RNA as substrates. The EMSA (Fig. 1B) showed that the full-length PNPase bound 20-mer and 8-mer RNA at concentrations >4 nM, whereas the truncated mutant ΔKH/S1 bound 20-mer and 8-mer RNA at concentrations >∼20 nM. A further quantitative gel shift study showed that wild-type PNPase bound the 20-mer RNA with a K d of 15.6 ± 1.8 nM, whereas ΔKH/S1 bound the 20-mer RNA with an eightfold reduced affinity with a K d of 132.3 ± 12.1 nM. This result is consistent with earlier studies (Stickney et al. 2005; Briani et al. 2007), suggesting that PNPase without S1/KH domains binds RNA less efficiently with an approximately one-order reduced binding affinity than full-length PNPase, and suggesting that the KH/S1 domain in PNPase is involved in RNA binding.
We then tested the exoribonucleolytic activity of full-length PNPase and ΔKH/S1 mutant in time course activity assays. The full-length PNPase digested both 20-mer and 8-mer RNA into small oligonucleotides, as shown in Figure 1C. The cleavage efficiencies of PNPase are indicated at the bottom of the gel, showing that the RNA substrates were cleaved processively with the cleavage percentage of 87%–94% in the reaction time of 1–10 min. However, ΔKH/S1 not only had less enzyme activity when cleaving 20-mer RNA, it also generated longer degradation products of ∼12-mer RNAs. The cleavage efficiencies were reduced to 32%–81% in the reaction time of 1–10 min. A 20-mer RNA and 8-mer RNA with different sequences were also analyzed for binding and cleavage assays and similar results were obtained (data not shown), verifying that all the altered properties of the truncated mutant were not sequence specific. Moreover, ΔKH/S1 could not degrade the shorter 8-mer RNA. Since ΔKH/S1 is able to bind 8-mer RNA (as shown by EMSA), it is intriguing why it cannot cleave 8-mer RNA. These results imply that the KH/S1 domain is involved not only in RNA binding, but also in modulation of enzyme activity in an unknown way.
The KH/S1 domain is involved in trimer formation
An earlier report showed that the S1 domain from PNPase was able to induce the trimerization of a RNase II-PNPase hybrid protein (Amblar et al. 2007). To test if KH/S1 domain in PNPase is involved in homotrimer formation, we measured the thermal melting points of full-length PNPase and ΔKH/S1 mutant by circular dichroism (CD) (Fig. 2A). The estimated melting point of full-length PNPase was 55°C, and that of ΔKH/S1 was 45°C, 10 degrees lower than the wild-type protein. This result showed that ΔKH/S1 was less thermal stable than the wild-type PNPase.
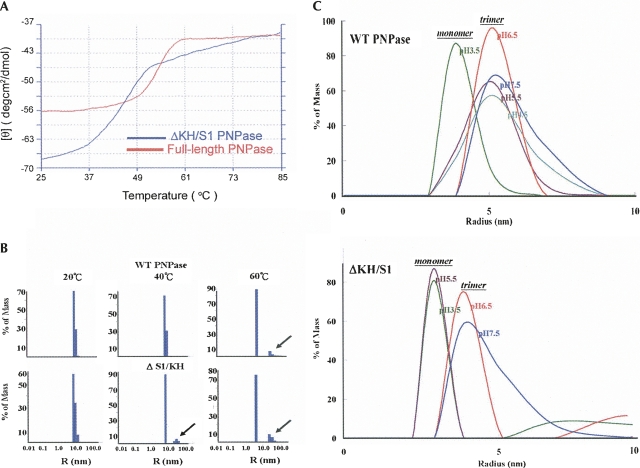
FIGURE 2.
Thermal melting and acid-induced trimer dissociation of PNPase and ΔKH/S1 assayed by circular dichroism (CD) and dynamic light scattering (DLS). (A) Thermal denaturation of PNPase was assayed by CD spectroscopy at a wavelength of 223 nm. The melting points of full-length PNPase (red) and ΔKH/S1 mutant (blue) were estimated to be 55°C and 45°C, respectively. (B) Thermal denaturation of PNPase was analyzed by DLS. Wild-type PNPase had a homogeneous particle size at 20°C and 40°C, and started to melt and formed aggregates (marked by an arrow) at 60°C, whereas ΔKH/S1 started to melt at the lower temperature of 40°C. (C) The acid-induced trimeric PNPase dissociation was measured at 20°C by DLS. The trimeric PNPase with a molecular radius of ∼5 nm (estimated molecular weight: 180–220 kDa) was dissociated into momomers (∼120 kDa) only in the highly acidic solution at pH 3.5. The truncated mutant ΔKH/S1 was less stable and dissociated into monomers at pH 5.5.
Since CD measurements reflect the thermal melting of protein secondary structures, not quaternary structures, we further analyzed the particle size of PNPase during thermal denaturation and acid-induced dissociation by dynamic light scattering (DLS). Thermal denaturation typically leads to an increase in protein particle size due to the aggregation of the unfolded proteins (Santiago et al. 2008). We observed similar phenomena that wild-type PNPase had a homogeneous trimeric particle size at the temperature of 20°C and 40°C, and it was partially denatured with an additional peak of large aggregates at 60°C (see Fig. 2B). On the other hand, ΔKH/S1 had a homogeneous trimeric size only at 20°C and it started to melt at lower temperatures of 40°C and 60°C, indicating that ΔKH/S1 was a less thermal-stable trimer than the wild-type PNPase.
The oligomeric proteins can be dissociated into monomers by acidic or basic conditions. We induced the trimer dissociation by decreasing the pH of protein solutions from 7.5 to 3.5 at 20°C (see Fig. 2C). The wild-type PNPase was a homotrimer up to pH 4.5, and it dissociated into monomers at pH 3.5, whereas ΔKH/S1 was a homotrimer at pH 7.5 and 6.5, and it dissociated into monomers at pH 5.5. This result showed that the truncated PNPase, without the KH/S1 domain, was melted at lower temperatures and dissociated into monomers in less acidic solutions. Therefore, we conclude that the KH and S1 domains contribute to the formation of a more stable trimeric PNPase.
Crystal structures of full-length and ΔKH/S1 PNPase
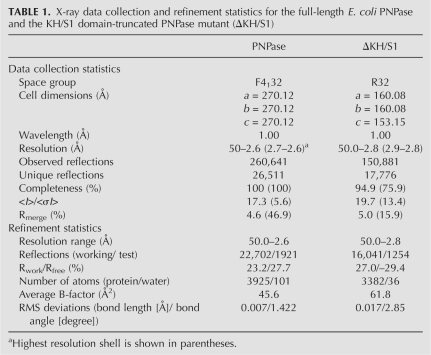
To find out why the truncated mutant ΔKH/S1 had different biochemical properties, we crystallized the two proteins for structural determination. The full-length PNPase crystallized in the cubic space group F4132, diffracting X-rays to a resolution of 2.6 Å, whereas ΔKH/S1 mutant crystallized in the rhombohedral space group R32, diffracting X-rays to a resolution of 2.8 Å. Both structures were solved by molecular replacement using the crystal structure of Streptomyces antibioticus PNPase (PDB accession code 1E3P) as the searching model. The statistics for X-ray diffraction and structural refinement are listed in Table 1.
TABLE 1.
X-ray data collection and refinement statistics for the full-length E. coli PNPase and the KH/S1 domain-truncated PNPase mutant (ΔKH/S1)
The final model of the full-length PNPase crystal structure contained two RNase PH domains and one α-helical linker domain from residues 17 to 565 (see Fig. 3). The C-terminal KH and S1 domains (Fig. 1A, residues 566–734) were not visible, containing only broken densities in the electron density maps. These KH and S1 domains were likely disordered as a whole in the crystal structure since they were connected to the second RNase PH domain by a long flexible loop. Superposition of E. coli full-length PNPase to that of Streptomyces antibioticus gave an RMSD of 1.45 Å over 443 Cα atoms, suggesting that the two bacterial structures are highly similar, whereas superposition of ΔKH/S1 to that of Streptomyces antibioticus gave a slightly higher RMSD of 1.83 Å over 384 Cα atoms.
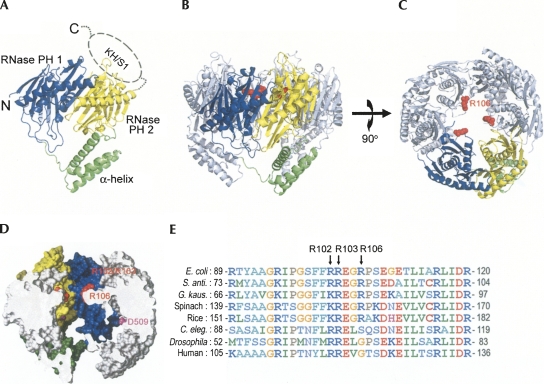
FIGURE 3.
Crystal structure of E. coli. PNPase. (A) The structure of a monomeric PNPase contains three domains: the RNase PH1 domain (blue), the α-helix linker domain (green), and the RNase PH2 domain (yellow). The C-terminal KH/S1 domain (marked by dashed circle) was disordered and thus not visible in the crystal structure. (B) The side view of the trimeric PNPase with the only one of the monomers colored in blue, yellow, and green. The other two monomers are colored in gray. The side chain of R106 is displayed as a red sphere model. (C) The top view of the trimiric PNPase. The homotrimeric PNPase is assembled into a ring-like structure with a central channel. (D) The side view of the halved PNPase trimer shows the shape of the hollow central channel. The molecular surface of RNase PH1 domain is displayed in yellow, whereas the surface of RNase PH2 domain is displayed in blue. The channel has two constricted necks formed by arginine residues in the first RNase PH domain, R102 and R103 in the upper neck closer to the channel entrance, and R106 in the lower neck closer to the active site. The D509 residue marks the putative RNase active site located in the second RNase PH domain. (D) Sequence alignment of PNPase from different species shows that the basic arginine residues, R102, R103, and R106, located at the neck regions in the central channel are highly conserved in bacteria, plants, and animals. Sequences included are from Escherichia coli, Streptomyces antibioticus, Geobacillus kaustophilus, Spinacia oleracea (spinach), Oryza sativa (rice), Caenorhabditis elegans, Drosophila melanogaster, and human. The alignment was generated by ClustalW (http://www.ch.embnet.org/software/ClustalW.html).
As expected, the homotrimer of PNPase forms a ring-like structure containing a central channel. The halved trimeric PNPase, displayed in Figure 3D, shows that the central channel contains two constricted necks with three arginine residues located in the neck regions: Arg102 and Arg103 in the upper neck closer to the channel entrance, and Arg106 in the lower neck, closer to the active site located in the second RNase PH domain. Both of the RNase PH domains, PH1 and PH2, contribute to the channel surface inside of the PNPase (surface displayed respectively in blue and yellow in Fig. 3D).
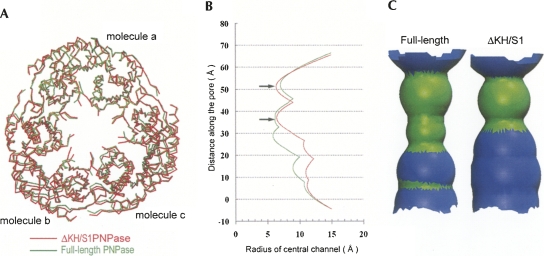
The overall structure of ΔKH/S1 was similar to that of the full-length PNPase. Superposition of only one of the monomers (Fig. 4A, molecule a) of the full-length and ΔKH/S1 PNPase gave an average RMSD of 1.1 Å for 417 Cα atoms. However, the corresponding RMSD were 1.6 Å and 1.7 Å for the molecules b and c, respectively. The top view of the superimposed trimer shows vaguely that the ΔKH/S1 truncate had a larger ring diameter (see Fig. 4A). Moreover, the surface area buried in the interface of two monomers was 1363 Å2 (for each monomer) in full-length PNPase, and 1272 Å2 in the ΔKH/S1 mutant, that was a reduction of ∼7% of buried interface without the presence of KH and S1 domains. These results suggest that the structure of ΔKH/S1 was less compact than full-length PNPase.
FIGURE 4.
Crystal structural comparison between full-length PNPase and truncated mutant ΔKH/S1. (A) One of the monomers (molecule a) in the homotrimer of full-length PNPase was superimposed on ΔKH/S1, giving an average RMSD of 1.1 Å for 417 Cα atoms. The corresponding RMSDs were 1.6 Å. and 1.7 Å for the molecules b and c, respectively. ΔKH/S1 is slightly more expanded than full-length PNPase. (B) Comparison of the radius of central channel between full-length PNPase (green line) and ΔKH/S1 mutant (red line). Two necks in the central channel are marked with arrows. The channel radius in the ΔKH/S1 mutant is larger than that in full-length PNPase at the lower region. (C) The shape of the central channel of full-length PNPase and ΔKH/S1 shows that the channel is more expanded in ΔKH/S1. The regions of the channel colored in green indicate that the diameter is 10–20 Å, and in blue is 20–30 Å. This figure and all others representing channels were generated with Hole (Smart et al. 1993).
To further confirm this observation, the sizes of central channel of full-length PNPase and ΔKH/S1 mutant were calculated by Hole (Smart et al. 1993). To ensure a fair comparison, disordered side chains or side chains with different conformations in the two structures were removed for the calculation, including the side chains of 105–107, 261–321, 394, and 409–418. We found that ΔKH/S1 indeed had a larger channel than that of full-length PNPase (see Fig. 4B,C). The most constricted region in the wild-type PNPase was about 14 Å in diameter, definitely too narrow for a double-stranded RNA. The channel size of ΔKH/S1 in the upper region was close to that of full-length PNPase, but in the lower region, the radius of the channel increased considerably, by up to ∼3 Å, or 6 Å wider in diameter. A direct measurement of the distances between a pair of Cα atoms in monomer a and b also gave increased distances in ΔKH/S1, especially for the residues located at the lower region of the channel: an increase distance of 2.77 Å for Asp509, 3.07 Å for Glu508, and 5.21 Å for Asn458. These differences were too significant to be a result of crystal packing or coordinate errors. Thus, these results support the hypothesis that the KH/S1 domain in PNPase is involved in trimeric assembly. Without this KH/S1 domain, the PNPase trimer is more loosely packed, bearing a larger central channel.
Arginine residues in the channel neck regions are involved in RNA binding and processing
Previously published crystal structures of an archaeal exosome in complex with RNA substrates show a ribonucleotide bound at the narrowest constriction of the central channel (Lorentzen et al. 2007; Navarro et al. 2008). To find out if RNA is also bound at the neck regions in PNPase, we mutated the two arginine residues located at the upper neck (R102 and R103) closer to the channel entrance, and one arginine located at the lower neck (R106) closer to the active site, to generate the double-mutant R102A/R103A and a single-point mutant R106A. R102A/R103A had decreased binding activity to a 20-mer or an 8-mer ssRNA, compared to the wild-type PNPase, as shown in Figure 5A. The dissociation constant between R102A/R103A and the 20-mer RNA was 378.2 ± 32.1 nM, about 24-fold reduced binding affinity compared to the wild-type PNPase. On the other hand, R106A bound the 20-mer RNA substrate as efficiently as wild-type PNPase with a K d of 5.2 ± 0.6 nM, even though it bound the 8-mer RNA less efficiently. This indicates that the arginines located in the upper neck are involved in capturing RNA substrates, whereas the arginine located in the lower neck is not a determinant factor in the first step of RNA binding.
FIGURE 5.
RNA binding and degradation activities of wild-type and mutated PNPase. (A) Gel shift assays show that R102A/R103A mutant bound 20-mer ssRNAs (left panel) or 8-mer ssRNAs (right panel) less efficiently than the wild-type PNPase, whereas R106A bound 20-mer RNA as efficiently as wild-type PNPase. (B) The 20-mer RNA (left panel) and 8-mer RNA (right panel) samples were incubated, respectively, with wild-type and mutated PNPase in time course experiments described in Materials and Methods. The digested RNAs were resolved in 15% polyacryamide/7 M urea gels. R102A/R103A digested RNA less efficiently but produced similar cleaved products as those of wild-type PNPase, whereas R106A digested 20-mer RNA efficiently but produced longer digested products and it cannot digest 8-mer RNA substrates.
We then compared the RNase activities of wild-type and mutated enzymes. R102A/R103A produced similar cleaved products when it digested 20-mer and 8-mer ssRNA substrates, although less efficiently than wild-type PNPase (Fig. 5B). Conversely, R106A was almost as efficient as wild-type PNPase in digesting 20-mer ssRNA, because about 64% of RNA substrates were degraded after 1-min incubations (compared to 87% for the wild-type PNPase). However, R106A could not degrade 20-mer RNA to smaller products, and instead yielded a set of stalled intermediates even after 5-min or 10-min incubations. Moreover, R106A could not degrade shorter 8-mer ssRNA substrates at all. These biochemical properties of R106A were similar to those of the ΔKH/S1 mutant, which cleaved only longer RNA and produced longer products of digestion.
DISCUSSION
The KH/S1 domain helps PNPase to form a more stable compact trimer
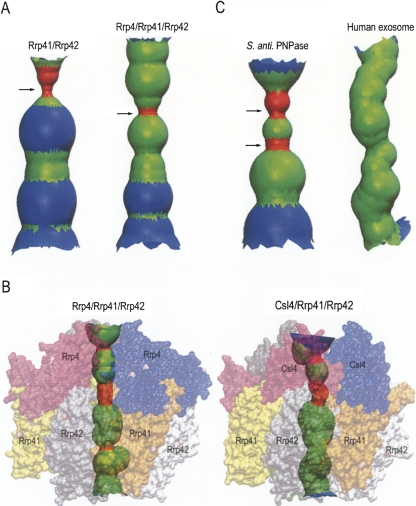
Our biochemical, circular dichroism, dynamic light scattering, and structural results consistently suggest that the KH/S1 domains are involved not only in RNA binding but also in the formation of a compact trimer with a more constricted central channel. Is it a general phenomenon that the S1 RNA-binding subunit in exosomes also modulates complex formation and channel size? Extensive structural studies have been carried out on archaeal exosome core enzymes, which contain three copies of Rrp41 and Rrp42, each bearing a RNase PH domain, to form a hexameric ring-like structure. One of the S1 homolog, Rrp4 or Csl4, is bound on the top of the ring to form a trimeric S1 pore (Lorentzen et al. 2007). The crystal structures of Sulfolobus solfataricus exosome core enzyme Rrp41/Rrp42 in the presence or absence of the RNA-binding subunit of Rrp4 have been solved (Lorentzen et al. 2005; Lorentzen et al. 2007). Interestingly, the central channel of trimeric Rrp41/Rrp42, shown in Figure 6A, appears to be wider than that of the Rrp4/Rrp41/Rrp42 complex. This suggests that Rrp4 may also contribute to complex formation, where it likely helps the nine-subunit exosome to assemble into a more compact structure. Moreover, the surface area buried in the interface of Rrp41 and Rrp42 between two heterodimers are 1210 Å2 (for each monomer) in the presence of Rrp4, and the buried surface is reduced to 1080 Å2 in the absence of Rrp4, supporting that KH and S1 domains in Rrp4 contribute to the formation of a more compact exosome.
FIGURE 6.
The shape of the central channel of two archaeal exosomes. (A) A comparison of the central channel of S. solfataricus exosome, in the absence of Rrp4 (left) and in the presence of Rrp4 (right), shows that the channel is narrower when the S1 protein Rrp4 is associated in the complex. The channel was calculated by Hole (Smart et al. 1993) based on the crystal structures of the six-subunit exosome, (Rrp41+Rrp42)3 (PDB entry: 2br2) and the nine-subunit exosome, (Rrp4+Rrp41+Rrp42)3 (PDB entry: 2jea). (B) A comparison of the channels in the A. fulgidus exosomes of (Rrp4+Rrp41+Rrp42)3 (PDB entry:2ba0) (left) and (Csl4+Rrp41+Rrp42)3 (PDB entry: 2ba1) shows that the shape of the central channel is different. The region of the channel colored in red indicates that the diameter is smaller than 10 Å, in green is 10–20 Å, and in blue is 20–30 Å. (C) The RNA-binding channel in the Streptomyces antibioticus PNPase (PDB entry: 1e3p) and human exosome (PDB entry: 2nn6).
The crystal structures of two isoforms of Archaeoglobus fulgidus exosome, Rrp4/Rrp41/Rrp42 and Csl4/Rrp41/Rrp42, have been reported (Buttner et al. 2005). The channels of these two complexes are different in shape and size (see Fig. 6B). The channel in Csl4/Rrp41/Rrp42 is more curved and slightly larger than that in Rrp4/Rrp41/Rrp42. This result indicates that the RNA-binding subunit located on the top of the hexameric ring may modulate channel shape allosterically. Our E. coli ΔKH/S1 PNPase mutant does not bind RNA as well as full-length PNPase, likely because the KH/S1 domain pair is responsible for direct interaction with RNA. Additionally, without the KH/S1 domains, the size of the channel is also changed accordingly. A wider channel is likely to be less efficient at trapping RNA for continuous degradation, and therefore leads to a defect enzyme in processive degradation, i.e., it cannot effectively cleave shorter RNAs and produces longer cleaved products.
A constricted central channel plays a crucial role in RNA binding and processing
How does the channel grasp single-stranded RNA substrates? The structure of the archaeal exosome in complex with RNA demonstrated that a single-stranded RNA is likely threaded through the S1 pore and bound at a constricted neck, where a conserved basic arginine residue (Arg65 in Rrp41) is responsible for the interactions with RNA (Lorentzen et al. 2007). In E. coli PNPase, two necks, upper and lower, are identified in the crystal structure (marked by arrows in Figs. 3, 4). These necks are formed by arginine residues in the first RNase PH domain, Arg102/Arg103 constituting the upper neck, and Arg106 constituting the lower neck. Mutation of Arg102/Arg103 in the upper neck produced a mutant less efficient in RNA binding and cleavage. The decreased RNase activity in cleaving a 20-mer RNA is a reflection of the decreased RNA-binding activity, since both activities reduced about 10- to 20-fold. This result suggests that the constricted neck formed by basic residues close to the channel entrance is involved in capturing RNA. Both the basic charged residues and the size of the channel contribute to the binding of RNA.
The mutation of Arg106 located in the lower neck produced an interesting defective mutant in R106A, which was as efficient as wild-type PNPase, although it could not degrade a shorter 8-mer ssRNA and the degradation reaction stalled whenever the RNA was trimmed to ∼12 nt. This phenotype is similar to that of ΔKH/S1 truncated PNPase. Based on the crystal structure of ΔKH/S1, we know that the truncated mutant has a wider central channel. Mutation of Arg106 to alanine likely produced similar effects since removal of the three copies of Arg106 side chain in the channel would lead to a bigger channel at the lower region as well. A wider neck must be disadvantageous for RNA binding, as RNA cannot be properly bound here to allow a continuous feed of substrate into active sites, thus leading to longer cleaved RNA products. We, therefore, suggest that the lower neck close to the active site is involved in processive RNA degradation.
A further structural comparison to the Streptomyces antibioticus PNPase shows that the S. antibioticus PNPase also has two constricted necks in its RNA-binding channel, one closer to channel entrance, one closer to the active site, similar to that of E. coli PNPase (see Fig. 6C). On the other hand, the inactive human exosome has a channel without obvious constricted necks (see Fig. 6C). This observation supports our suggestions that necks are important in trapping RNA substrates for degradations, and therefore only the bacterial PNPases and archaeal exosomes, with high RNase activities, have obvious constricted neck regions. Moreover, the three arginine residues located in the neck regions are highly conserved in PNPase (see Fig. 3), suggesting that these necks are also conserved features in PNPase. Together with the structural data from archaeal exosomes, we conclude that the KH/S1 domains and the upper neck in the channel recruit RNA into the channel, and the lower neck keeps hold of the RNA as it guides it into the PNPase active site for processive degradation. Constrictions of the appropriate size at the necks in the central channel of exosomes and PNPases play a crucial role in RNA binding and processive degradation.
MATERIALS AND METHODS
Cloning, protein expression, and purification
The genes of the full-length PNPase (residues 17–734) and truncated ΔKH/S1 mutant (residues 17–570) were amplified by PCR and subcloned respectively into NdeI/XhoI sites of expression vectors pET-22b and pET-28c (Novagen) to generate N-terminal His-tagged constructs. All PNPase point mutants were generated by Quickchange site-directed mutangenesis kits (Invitrogen). All expression plasmids were transformed into the BL21 (DE3) strains cultured in LB medium supplemented with 100 μg/mL ampicillin to an OD600 of 0.6, and then induced with 1.0 mM IPTG at 37°C for 4 h. Crude cell extracts were first loaded onto a Ni-NTA resin affinity column (Qiagen, Germany) followed by a Q column (Pharmacia). The PNPase-containing fractions were then applied to a gel filtration chromatography column (Superdex 200, Pharmacia). Purified PNPase was dialyzed against a buffer containing 20 mM Tris buffer (pH 7.0) and 100 mM NaCl, and concentrated to a suitable concentration for crystallization and activity assay.
RNA-binding and RNase activity assays
Electrophoresis mobility shift assays (EMSA) were preformed using a single-stranded 20-mer RNA: 5′-ACUGGACAAAUACUCCGAGG-3′ and an 8-mer RNA: 5′-AAAAAAAA-3′, as substrates. The RNA substrates were labeled first at their 5′ end with [γ-32P]ATP by T4 polynucleotide kinase, and then purified by a Microspin G-25 column (GE Healthcare) to remove the nonincorporated nucleotides. The RNA substrate (0.1 pmol) was then incubated with different concentrations of PNPase (4 to 200 nM) in a buffer containing 20 mM Tris-HCl pH 7.6, 100 mM KCl, 2 mM DTT, 10 mM ZnCl2 at 37°C for 10 min. The ZnCl2 was added to ensure that PNPase can only bind but cannot cleave RNA substrates since zinc ions inhibited the enzyme activity. After incubation, the reaction mixture was applied to 20% TBE gels, which were exposed to the phosphorimaging plate (Fujifilm) for autoradiographic visualization.
For RNase activity assays, 0.1 pmol of single-stranded RNA substrates were incubated with wild-type or mutated PNPase (400 nM) in a reaction buffer containing 20 mM Tris-HCl pH 7.6, 100 mM KCl, 2 mM DTT, 1 mM MgCl2, and 5 mM KH2PO4 at 37°C. The reaction was stopped at the time point indicated in the figures by adding TBE-Urea sample buffer (Bio-Rad). Reaction products were resolved in 15% polyacrylamide/7 M urea gels, which were exposed to the phosphorimaging plate (Fujifilm) and analyzed by the imaging system FLA-5000 (Fujifilm).
Circular dichroism (CD)
The thermal denaturing melting points of PNPase were measured three times by a circular dichroism spectrometer AVIV CD400. The CD spectra were scanned from 25°C to 85°C at a wavelength of 223 nm and the melting point was estimated by AVIV program. The protein concentration was 0.1 mg/mL in a buffer containing 25 mM sodium phosphate (pH 7.2).
Dynamics light scattering (DLS)
DLS measurements were carried out on a Dyna-Pro 99 MS800 instrument (Protein Solutions). The wild-type PNPase and ΔKH/S1 mutant (0.5 mg/mL) in a buffer of 50 mM potassium phosphate (pH 7.5) and 20 mM NaCl were filtered with a 0.1-μm Anodisk filter, and placed in a 12-μL cuvette (b = 1.5 nm). The protein samples were incubated for 5 min in different temperatures (20°C, 40°C, and 60°C) before data acquisition over an acquired time of 15 min.
For the analysis of the acid-induced trimer dissociation, the PNPase samples in buffers of different pH, ranging from 3.5 to 7.5 were measured at 20°C (pH 6.5 and 7.5:50 mM potassium phosphate buffer; pH 5.5, 4.5, and 3.5:50 mM glycine-HCl buffer). The size distribution plots, the x axis showing a distribution of estimated particle radius (nm) and the y axis showing the relative intensity of the scattered light (% of mass), were analyzed and prepared with the software Dynamics V5.26.60 (Protein Solutions).
Crystallization and data collection
Both full-length and ΔKH/S1 mutant were concentrated to 10 mg/mL in a buffer of 100 mM NaCl and 20 mM Tris-HCl at pH 7.0. Crystals of full-length PNPase were grown by hanging drop vapor-diffusion method at room temperature, by mixing 1 μL of protein solution with 1 μL of reservoir solution containing 15% w/v PEG 4000, 2 M MgCl2, and 0.1 M Tris-HCl at pH 8.5. Crystals of ΔKH/S1 mutant were grown by the same method using a reservoir containing 1.5 M ammonium sulfate and 0.1 M Tris-HCl at pH 8.5. Diffraction data were collected at −150°C at beamline 13C1 of the NSRRC in Hsinchu, Taiwan, and were processed and scaled by HKL2000 (Otwinowski and Minor 1997). All diffraction statistics are listed in Table 1.
Structure determination and refinement
The full-length PNPase crystallized in the F4132 cubic space group, whereas the ΔKH/S1 mutant crystallized in the rhombohedral space group of R32, with one molecule per asymmetric unit in both crystal systems. The structures of full-length PNPase and ΔKH/S1 were solved by molecular replacement by EPMR, using the crystal structure of Streptomyces antibioticus PNPase (PDB accession code: 1E3P) as the searching model. The structure model was subjected to manual rebuilding with WinCoot and then refined with the program CNS. Structural coordinates and diffraction structure factors have been deposited in the RCSB Protein Data Bank with PDB ID codes of 3CDI for full-length PNPase and 3CDJ for the ΔKH/S1 mutant.
ACKNOWLEDGMENTS
This work was supported by research grants from Academia Sinica (Summit Projects) and the National Science Council, Taiwan, Republic of China. Portions of this research were carried out at the National Synchrotron Radiation Research Center (BL-13B1 and BL-13C1), a national user facility supported by the National Science Council of Taiwan, Republic of China. The Synchrotron Radiation Protein Crystallography Facility is supported by the National Research Program for Genomic Medicine. We thank Dr Harry Wilson of Academia Sinica for manuscript editing.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1244308.
REFERENCES
- Amblar M., Barbas A., Gomez-Puertas P., Arraiano C.M. The role of the S1 domain in exoribonucleolytic activity: Substrate specificity and multimerization. RNA. 2007;13:317–327. doi: 10.1261/rna.220407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S., Shteiman-Kotler A., Liveanu V., Yehudai-Resheff S., Bellaoui M., Settlage M., Shabanowitz J., Hunt D.F., Schuster G., Gruissem W. Chloroplast PNPase exists as a homodimer enzyme complex that is distinct from the Escherichia coli degradosome. RNA. 2001;7:1464–1475. [PMC free article] [PubMed] [Google Scholar]
- Briani F., Favero M.D., Capizzuto R., Consonni C., Zangrossi S., Greco C., Gioia L.D., Tortora P., Deho G. Genetic analysis of polynucleotide phosphorylase structure and functions. Biochimie. 2007;89:145–157. doi: 10.1016/j.biochi.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Buttner K., Wenig K., Hopfner K.-P. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol. Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Carpousis A.J. The RNA degradosome of Escherichia coli: An mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- Carpousis A.J., Van Houwe G., Ehretsmann C., Krisch H.M. Copurification of E. coli RNase E and PNPase: Evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- Chen H.-W., Rainey R.N., Balatoni C.E., Dawson D.W., Troke J.J., Wasiak S., Hong J.S., McBride H.M., Koehler C.M., Teitell M.A., et al. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitrochondrial homeostasis. Mol. Cell. Biol. 2006;26:8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-W., Koehler C.M., Teitell M.A. Human polynucleotide phosphorylase: Location matters. Trends Cell Biol. 2007;17:600–608. doi: 10.1016/j.tcb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- French S.W., Dawson D.W., Chen H.-W., Rainey R.N., Sievers S.A., Balatoni C.E., Wong L., Troke J.J., Nguyen M.T.N., Koehler C.M., et al. The TCL1 oncoprotein binds the RNase PH domains of the PNPase exoribonuclease without affecting its RNA degrading activity. Cancer Lett. 2007;248:198–210. doi: 10.1016/j.canlet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Houseley J., LaCava J., Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Leszczyniecka M., Kang D.C., Sarkar D., Su Z.Z., Holmes M., Valerie K., Fisher P.B. Identification and cloning of human polynucleotide phosphorylase, hPNPaseold-35, in the context of terminal differentiation and cellular senescence. Proc. Natl. Acad. Sci. 2002;99:16636–16641. doi: 10.1073/pnas.252643699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczyniecka M., Sua Z.-z., Kanga D.-c., Sarkara D., Fishera P.B. Expression regulation and genomic organization of human polynucleotide phosphorylase, hPNPaseold-35, a Type I interferon inducible early response gene. Gene. 2003;316:143–156. doi: 10.1016/s0378-1119(03)00752-2. [DOI] [PubMed] [Google Scholar]
- Lin-Chao S., Chiou N.-T., Schuster G. The PNPase, exosome and RNA helicases as the building components of evolutionarily conserved RNA degradation machines. J. Biomed. Sci. 2007;14:523–532. doi: 10.1007/s11373-007-9178-y. [DOI] [PubMed] [Google Scholar]
- Liou G.G., Jane W.N., Cohen S.N., Lin N.S., Lin-Chao S. RNA degradosomes exist in vivo in Escherichia coli as multicomponent complexes associated with the cytoplasmic membrane via the N-terminal region of ribonuclease E. Proc. Natl. Acad. Sci. 2001;98:63–68. doi: 10.1073/pnas.011535498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Greimann J.C., Lima C.D. Reconstitution, activities, and structure of the Eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Lorentzen E., Conti E. Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH domain. Mol. Cell. 2005;20:473–481. doi: 10.1016/j.molcel.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Lorentzen E., Dziembowski A., Lindner D., Seraphin B., Conti E. RNA channelling by the archaeal exosome. EMBO Rep. 2007;8:470–476. doi: 10.1038/sj.embor.7400945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen E., Walter P., Fribourg S., Evguenieva-Hackenberg E., Klug G., Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- Marcaida M.J., DePristo M.A., Chandran V., Carpousis A.J., Luisi B.F. The RNA degradosome: Life in the fast lane of adaptive molecular evolution. Trends Biochem. Sci. 2006;31:359–365. doi: 10.1016/j.tibs.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Matus-Ortega M.E., Regonesi M.E., Pina-Escobedo A., Tortora P., Deho G., Garcia-Mena J. The KH and S1 domains of Escherichia coli polynucleotide phosphorylase are necessary for autoregulation and growth at low temperature. Biochim. Biophys. Acta. 2007;1769:194–203. doi: 10.1016/j.bbaexp.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Mohanty B.K., Kushner S.R. Polynucleotide phosphorylase functions both as a 3′ to 5′ exonuclease and a poly(A) polymerase in Escherichia coli . Proc. Natl. Acad. Sci. 2000;97:11966–11971. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro M.V.A.S., Oliveira C.C., Zanchin N.I.T., Guimara B.G. Insights into the mechanism of progressive RNA degradation by the archaeal exosome. J. Biol. Chem. 2008;283:14120–14131. doi: 10.1074/jbc.M801005200. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Piwowarski J., Grzechnik P., Dziembowski A., Dmochowska A., Minczuk M., Stepien P.P. Human polynucleotide phosphorylase, hPNPase, is localized in mitochondria. J. Mol. Biol. 2003;329:853–857. doi: 10.1016/s0022-2836(03)00528-x. [DOI] [PubMed] [Google Scholar]
- Raijmakers R., Schilders G., Pruijn G.M. The exosome, a molecular machine for controlled RNA degradation in both nucleus and cytoplasm. Eur. J. Cell Biol. 2004;83:175–183. doi: 10.1078/0171-9335-00385. [DOI] [PubMed] [Google Scholar]
- Santiago P.S., Moura F., Moreira L.M., Domingues M.M., Santos N.C., Tabak M. Dynamic light scattering and optical absorption spectroscopy study of pH and temperature stablities of the extrocellular hemoglobin of Glossocolex paulistus . Biophys. J. 2008;94:2228–2240. doi: 10.1529/biophysj.107.116780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D., Fisher P.B. Polynucleotide phosphorylase: An evolutionary conserved gene with an expanding repertoire of functions. Pharmacol. Ther. 2006;112:243–263. doi: 10.1016/j.pharmthera.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Schuster G., Lisitsky I., Klaff P. Polyadenylation and degradation of mRNA in the chloroplast. Plant Physiol. 1999;120:937–944. doi: 10.1104/pp.120.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart O.S., Goodfellow J.M., Wallace B.A. The pore dimensions of gramicidin A. Biophys. J. 1993;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Parker R. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Stickney L.M., Hankins J.S., Miao S., Mackie G.A. Function of the conserved S1 and KH domains in polynucelotide phosphorylase. J. Bacteriol. 2005;187:7214–7221. doi: 10.1128/JB.187.21.7214-7221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons M.F., Jones G.H., Luisi B.F. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000;8:1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- Symmons M.F., Williams M.G., Luisi B.F., Jones G.H., Carpousis A.J. Running rings around RNA: A superfamily of phosphate-dependent RNases. Trends Biochem. Sci. 2002;27:11–18. doi: 10.1016/s0968-0004(01)01999-5. [DOI] [PubMed] [Google Scholar]
- Theobald D.L., Mitton-Fry R.M., Wuttke D.S. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff S., Portnoy V., Yogev S., Adir N., Schuster G. Domain analysis of the chloroplast polynucleotide phosphorylase reveals discrete functions in RNA degradation, polyadenylation, and sequence homology with exosome proteins. Plant Cell. 2003;15:2003–2019. doi: 10.1105/tpc.013326. [DOI] [PMC free article] [PubMed] [Google Scholar]