Abstract
Production of aberrant messenger ribonucleoprotein particles (mRNPs) is subject to quality control (QC). In yeast strains carrying mutations of the THO complex, transcription induction triggers a number of interconnected QC phenotypes: (1) rapid degradation of several mRNAs; (2) retention of a fraction of THO-dependent mRNAs in transcription site-associated foci; and (3) formation of a high molecular weight DNA/protein complex in the 3′-ends of THO target genes. Here, we demonstrate that the 3′–5′ exonucleolytic domain of the nuclear exosome factor Rrp6p is necessary for establishing all QC phenotypes associated with THO mutations. The N terminus of Rrp6p is also important presumably through its binding to the Rrp6p co-factor Rrp47p. Interestingly, the 3′–5′ exonucleolytic activity of Dis3p, the only other active exonuclease of the nuclear exosome, can also contribute to RNA QC in THO mutants, while other nuclear 3′–5′ exonucleases cannot. Our data show that exonucleolytic attack by the nuclear exosome is needed both for provoking mRNP QC and for its ensuing elimination of faulty RNA.
Keywords: mRNP quality control, 3′–5′, exoncleolysis, nuclear exosome, THO complex mutants
INTRODUCTION
Production of functional messenger ribonucleoprotein particles (mRNPs) requires correct RNA processing and RNA/protein packaging. These events are monitored by cellular surveillance systems, which detect aberrant mRNPs and rid the cell of faulty transcripts. Such control occurs throughout the mRNP lifecycle, but is effective already during cotranscriptional RNA processing and mRNP assembly (for reviews, see Jensen et al. 2003; Saguez et al. 2005; Doma and Parker 2007). In theory, quality control (QC) of mRNPs can be separated into an initial leg where the RNP aberrancy in question is detected, and an ensuing leg where the faulty mRNA is degraded. In reality, however, either these events rapidly follow one another, hampering experimental discrimination, or the two activities are mechanistically coupled or directly overlapping. In addition, production of aberrant mRNPs provokes a range of interconnected phenotypes; thus, it is a challenge to interrelate these different QC phenotypes as well as to evaluate to what extent surveillance factors and their different enzymatic activities impact the mRNP QC process.
The best-studied nuclear mRNA surveillance factor is the 3′–5′ exonucleolytic RNA exosome. This highly conserved, multisubunit complex is present in both the nucleus and the cytoplasm of eukaryotic cells, with accessory factors determining compartmental specificity. In Saccharomyces cerevisiae, the nuclear exosome contains two active 3′–5′ exonucleases, called Rrp6p and Dis3p (Rrp44p) (Liu et al. 2006; Dziembowski et al. 2007; Schneider et al. 2007). The RNase D-like enzyme Rrp6p is nucleus-restricted, while Dis3p, a member of the RNase II family, is associated with both nuclear and cytoplasmic exosomes. Another nucleus-restricted component in yeast is Rrp47p (Lrp1p), which directly contacts the N terminus of Rrp6p and promotes its catalytic activity (Mitchell et al. 2003; Stead et al. 2007). It is not yet clear whether Rrp6p/Rrp47p and Dis3p always require the core exosome for in vivo function, or whether these enzymes can also operate independently.
The study of mRNP QC requires model systems. A well studied example is provided by S. cerevisiae strains carrying mutations of the tetrameric THO complex, which is required for cotranscriptional mRNP assembly, presumably via its interaction with the mRNA export factors Sub2p and Yra1p (Strasser et al. 2002; Zenklusen et al. 2002). Individual deletion of THO components elicits a dramatic drop in the level of several mRNAs, a defect that is restored upon deletion of Rrp6p or Rrp47p (Libri et al. 2002; Rougemaille et al. 2007). Another prominent QC-related phenotype in THO/Sub2p mutants is fluorescent in situ hybridization (FISH) detection of mRNAs retained in transcription site-associated foci (Jensen et al. 2001a; Libri et al. 2002; Thomsen et al. 2003, 2008; Rougemaille et al. 2007). These RNA “dots” are post-transcriptional, as FISH signals persist after transcriptional shutoff (Rougemaille et al. 2007). Thus, at least two different pools of RNAs exist in THO mutant strains: those that are rapidly degraded shortly after their synthesis, and those that escape decay and accumulate in foci. It is not yet clear how these phenotypes relate to yet another recently discovered THO mutant phenomenon, i.e., the formation in the 3′-ends of THO target genes of a high molecular weight DNA/protein complex. Formation of this complex leads to depletion of THO complex target genes from chromatin extracts, a phenomenon that has been dubbed differential chromatin fractionation (DCF) or “heavy chromatin” (Rougemaille et al. 2008). DCF complexes harbor pre-mRNA 3′-end formation factors as well as nucleoporins, suggesting that they represent mRNPs that are docked to nuclear pore complexes but have not been released from their transcription sites. RNP retention as defined both by RNA-FISH and DCF again requires Rrp6p (Jensen et al. 2001a; Libri et al. 2002; Vinciguerra et al. 2005, Rougemaille et al. 2008).
Studies of mRNP surveillance have so far mostly been carried out using deletion mutants, e.g., Δrrp6, as a convenient tool to specifically block nuclear 3′–5′ exonucleolytic degradation. An obvious caveat to this approach is that other domains with putative important functions are also removed. In this paper, we investigate the specific contributions of Rrp6p and Dis3p exonucleolytic activities to several QC-related phenotypes. Interestingly, we find that Rrp6p exonucleolytic activity is necessary for establishing all of the phenotypes associated with THO deletion. The interaction of Rrp6p with Rrp47p and the exonucleolytic activity of Dis3p can also contribute. Thus, surprisingly, exonucleolysis is involved in both triggering and responding to QC phenotypes.
RESULTS AND DISCUSSION
In vitro characterization of Rrp6p mutants
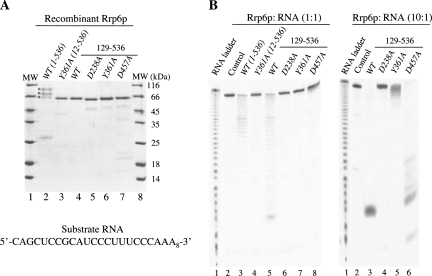
To assay the specific influence of Rrp6p functional domains on mRNP QC, we focused our attention on mutations previously reported to impact Rrp6p in vivo function. The D238A single amino acid substitution disrupts the coordination of metal ions in the active site whereas the Y361A mutation prevents the activation of the hydrolytic water and thereby presumably abolishes exonucleolytic activity of the enzyme, while the D457A mutation in the conserved helicase and RNase D C-terminal (HRDC) domain alters protein interdomain contacts, possibly affecting coordination of the RNA substrate (Phillips and Butler 2003; Midtgaard et al. 2006). In addition, deletion of the first 128 amino acids of Rrp6p has been shown to disrupt its interaction with Rrp47p (Stead et al. 2007). First, Rrp6p variants consisting of the core exonuclease and HRDC domains, either containing (1–536) or lacking (129–536) the N terminus, were expressed as 6×His-tagged proteins in Escherichia coli and purified to homogeneity using a well established three-step protocol (Fig. 1A, lanes 2,4; Midtgaard et al. 2006). The enzymatic activity of these variants was tested by incubation in a 1:1 molar ratio with a defined 30-nucleotide (nt) RNA substrate with little or no secondary structure (Fig. 1A, bottom) for 1 h at 30°C. The experiment showed that the two constructs have comparable substrate degradation activities (Fig. 1B, left panel, cf. lanes 3 and 5). We, therefore, conclude that the N terminus of Rrp6p (1–128) does not contribute significantly to the exonucleolytic activity of the enzyme in vitro.
FIGURE 1.
In vitro activity of Rrp6p mutant variants. (A) Commassie-stained gel of purified recombinant Rrp6p constructs used for RNA degradation analysis. Lengths of the individual constructs are indicated in parentheses or above the line for the amino acids 129–536 versions. (*) Full-length and presumed degradation products of the amino acids 1–536 construct. (MW) Molecular weight marker with the indicated sizes. Sequence of the RNA substrate used in degradation reactions is given below the gel. (B) 20% urea–acrylamide gel of RNA degradation reactions using a molar ratio of 1:1 (left panel) or 10:1 (right panel) between enzyme and RNA. Rrp6p constructs used are indicated above the gel as in A. Controls are RNA incubated in the absence of enzyme.
Next, we introduced the D238A, Y361A, and D457A substitutions separately into the Rrp6-129-536 construct and repeated the RNA degradation assay. When incubated in a 1:1 enzyme:substrate molar ratio, only the D457A mutant exhibited a weak exonucleolytic activity, albeit far from the activity of its wild-type (WT) counterpart (Fig. 1B, left panel, cf. lanes 5–8). However, at a 10:1 enzyme:RNA ratio, the D457A mutant was able to degrade the RNA substrate to completion (Fig. 1B, right panel, lane 6). Under these reaction conditions, the Y361A mutant exhibited only minor degradation activity, whereas the D238A mutant remained completely inactive. We conclude that the D457A mutation affects the in vitro exonucleolytic activity of Rrp6p to some extent, and that the Y361A mutation is capable of limited hydrolysis.
Characterization of single-site mutant rrp6 alleles in vivo
We next tested the in vivo activity of the Rrp6p point mutants. Due to the overlap of the centromeric region of chromosome XV with sequences in the RRP6 promoter, and the consequent instability of RRP6-containing low copy CEN plasmids, the three rrp6 variants and a RRP6 positive control were stably integrated into the RRP6 locus of the W303 test strain.
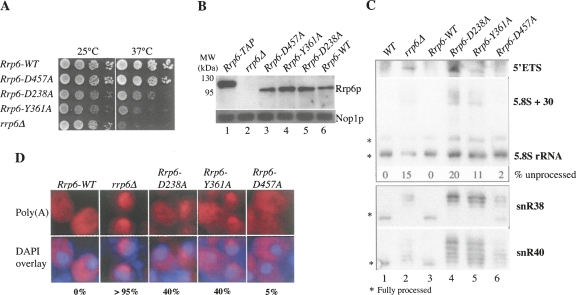
Deletion of Rrp6p is temperature sensitive at 37°C. Although strains containing the Rrp6D238A and Rrp6Y361A proteins were also growth impaired at this temperature, they grew better than rrp6Δ cells (Fig. 2A). This suggests that features other than the exonucleolytic active site of Rrp6p are important for the activity of the protein. Cells carrying the Rrp6D457A mutation grew indistinguishably from WT cells, suggesting that the (lower) exonucleolytic activity of this Rrp6p variant is sufficient for WT in vivo function. Finally, all three mutant proteins were stably expressed at levels comparable to WT Rrp6p (Fig. 2B).
FIGURE 2.
In vivo phenotypes of rrp6 mutant variants. Growth (A), Western blotting (B), Northern blotting (C), and poly(A)+-RNA-FISH (D) analyses of strains deleted for Rrp6p and harboring integrated rrp6 alleles expressing WT Rrp6p or the Rrp6D238A, Rrp6Y361A, or Rrp6D457A mutant proteins, as indicated. (A) Strains were spotted onto YM-1 plates in 10-fold dilutions at the indicated temperatures. (B) Yeast extracts were separated by SDS-PAGE, transferred to nitrocellulose membranes, and probed with Rrp6p or Nop1p antibodies. Extract prepared from a TAP-tagged Rrp6-TAP strain was used as a control (lane 1). (C) Northern blotting analysis of RNA-processing and RNA-degradation phenotypes using probes directed against 5′-ETS, 5.8S rRNA, snR38-snoRNA, or snR40-snoRNA substrates. (*) Migrations of fully processed RNA species. Individual bands were quantified and the percentage of unprocessed 5.8S + 30 pre-rRNA was calculated and denoted below the respective lanes. (WT) A bona fide W303 strain. (D) Poly(A)+-RNA-FISH on fixed samples of yeast cells after a temperature shift for 15 min to 37°C. Poly(A)+ RNA was detected using an LNA-modified dT20 probe (Thomsen et al. 2005). DNA was stained with DAPI. The ratio of cells harboring Poly(A)+ RNA signal is indicated below the images.
To further characterize the phenotypes of the rrp6 alleles, we studied the state of the 5′ ETS and 5.8S ribosomal RNA molecules as well as the snR38 and snR40 small nucleolar (sno) RNAs in these strains by Northern blotting (Fig. 2C). As previously shown, Rrp6D457A was able to degrade 5′ ETS- and pre-5.8Sr-RNAs with an efficiency comparable to WT Rrp6p, whereas snR38 and snR40 RNAs appeared as slightly extended forms (Midtgaard et al. 2006). In contrast, the Rrp6D238A and Rrp6Y361A mutants exhibited largely rrp6Δ-like phenotypes on all four RNA substrates tested.
Finally, we assayed the ability of the three Rrp6p variants to reduce the nucleolar accumulation of poly(A)+ RNA observed in rrp6Δ cells (Thomsen et al. 2005). The Rrp6D457A protein almost completely reverted this phenotype to WT conditions, whereas the Rrp6D238A and Rrp6Y361A proteins exhibited a partial reduction (Fig. 2D). Perhaps the presence of Rrp6p, albeit in a catalytic inactive form, has a positive impact on nucleolar exosome activity (i.e., Dis3p), or perhaps the affinity or coordination of cofactors on the exosome is altered.
The exonucleolytic domain of Rrp6p is required for all tested mRNP QC phenotypes
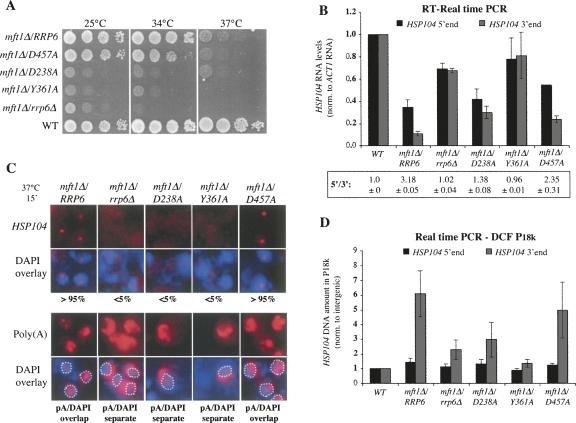
To directly assess the impact of the different Rrp6p mutants on nuclear mRNA surveillance, we crossed the respective alleles into cells lacking the THO component Mft1p. A dilution series of the resulting double mutant strains revealed that the Rrp6Y361A mutation largely had the same negative effect on growth of cells carrying the mft1Δ mutation as did a full deletion of RRP6 (Fig. 3A). The Rrp6D238A mutation also reduced the growth of mft1Δ cells, although the 37°C phenotype was not as severe as with Rrp6Y361A or rrp6Δ. Interestingly, however, in contrast to a WT environment, the exonucleolytic activity of Rrp6p was strongly rate limiting for growth in the mft1Δ context at 25°C (cf. Figs. 2A and 3A).
FIGURE 3.
QC-related phenotypes of mft1Δ cells depend on Rrp6p exonucleolytic activity. Growth (A), HSP104 RNA levels (B), HSP104- and poly(A)+-RNA-FISH (C), and HSP104 gene DCF (D) analyses of mft1Δ cells deleted for Rrp6p, harboring WT Rrp6p, or carrying the Rrp6D238A, Rrp6Y361A, or Rrp6D457A mutant proteins, as indicated. (A) Strains were spotted onto YM-1 plates in 10-fold dilutions at the indicated temperatures. (B) Quantitative real-time PCR analysis of HSP104 RNA 5′- (black columns) or 3′- (gray columns) ends was performed on total RNA samples. HSP104 RNA levels were normalized to ACT1 RNA, which was unaffected by the relevant mutations. The HSP104/ACT1 RNA ratio from WT samples was set to 1. Averages and standard deviations are calculated from two experiments. Ratios of HSP104 5′-end to 3′-end levels are given below the graph. (C) HSP104- or poly(A)+-RNA-FISH on fixed samples of yeast cells after a temperature shift for 15 min to 37°C. HSP104 RNA was detected using a mixture of three Cy3-labeled oligonucleotide probes directed against the 3′-end of the transcript (Rougemaille et al. 2007). The ratio of cells harboring retained HSP104 RNA is indicated below the images. Poly(A)+ RNA and DNA were detected as in Fig. 2D. (Overlay images, white dashed line) Outskirts of the DAPI stain. Whether poly(A)+ RNA and DAPI signals largely overlapped, or were largely separate, in >95% of cells is indicated below the images. (D) Quantitative PCR analysis of HSP104 DNA 5′- (black columns) or 3′- (gray columns) ends in the pellets of ChIP-extracts after an 18,000g centrifugation step (P18k). HSP104 DNA levels were normalized to levels of intergenic DNA, which was unaffected by the relevant mutations. The HSP104/intergenic DNA ratio from WT-samples was set to 1. Averages and standard deviations are calculated from five experiments.
Next, we tested the ability of the three Rrp6p variants to restore the QC-related phenotypes of mft1Δ cells: (1) decreased levels of newly synthesized HSP104 RNA; (2) retention of HSP104- and poly(A)+-RNA in nuclear dots; and (3) formation of the DCF complex in the 3′-end of the HSP104 gene. Compared with a WT strain, HSP104 RNA levels were severely decreased in mft1Δ cells after a brief 15-min transcription induction at 37°C. Importantly, as a reliable signature for mRNA QC by the exosome, lower levels of HSP104 RNA 3′-ends than 5′-ends were readily detected (Fig. 3B, see HSP104 5′-end/3′-end ratios below the graph; Libri et al. 2002; Rougemaille et al. 2007). This 3′-end truncation bias depends on the presence of Rrp6p, as it is restored in the mft1Δ/rrp6Δ double mutant. Likewise, HSP104 5′-end/3′-end ratios were restored by introduction of the Rrp6Y361A mutant into the mft1Δ context, and to a lesser extent by introduction of Rrp6D238A, whereas the Rrp6D457A mutant behaved like WT Rrp6p. Thus, 3′-end truncation of HSP104 RNA in mft1Δ cells depends on the exonucleolytic activity of Rrp6p.
The retention of HSP104 RNA in one dot per mft1Δ cell and the accumulation of poly(A)+ RNA in granules overlapping the chromatin-stained DAPI signal were also clearly dependent on the exonucleolytic activity of Rrp6p, as signals were present in WT Rrp6p and Rrp6D457A contexts but were largely absent when the Rrp6D238A or Rrp6Y361A mutations were introduced (Fig. 3C).
To induce DCF, the exonucleolytic function of Rrp6p is also required (Fig. 3D). Formation of the DCF complex was assessed by analyzing the levels of HSP104 DNA 5′- and 3′-ends in the pellets of chromatin extracts after an 18,000g centrifugation step (P18k) (Rougemaille et al. 2008). In these fractions of extracts prepared from mft1Δ cells, HSP104 3′-ends are significantly enriched over 5′-ends (Fig. 3D). Formation of the DCF complex depends on the presence of WT Rrp6p or the Rrp6D457A protein. In contrast, the Rrp6Y361A and Rrp6D238A mutants were ineffective to promote DCF formation, the latter mutation again being less severe than the former (Fig. 3D).
It is curious that the rrp6Y361A mutation is more severe than rrp6D238A despite the fact that only the former protein product exhibits residual exonuclease activity (Fig. 1B). This is also consistent with the relative growth rates of the two mutants (Figs. 2A, 3A) and might relate to differences in their exonucleolytic activity in vivo. Alternatively, slow degradation by the defective (but not completely dead) Rrp6Y361A variant might shelter the RNA 3′-end from other exonucleases, ultimately providing some protection from degradation. Overall, however, the results demonstrate unambiguously that the exonucleolytic domain of Rrp6p is necessary for all tested mRNP QC phenotypes.
The N-terminal domain of Rrp6p is required for QC
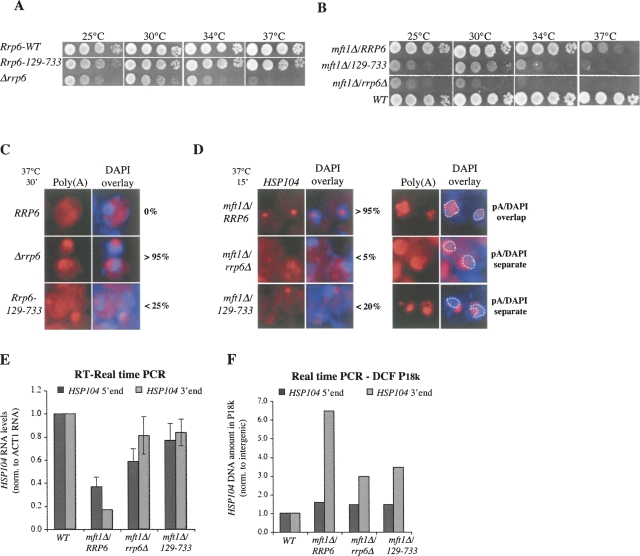
It has been shown that deletion of the Rrp6p cofactor Rrp47p elicits phenotypes similar to those observed in a rrp6Δ context (Mitchell et al. 2003; Rougemaille et al. 2007; Stead et al. 2007). Rrp47p reportedly interacts with the N terminus of Rrp6p (Stead et al. 2007), and we therefore asked what effect the deletion of the first 128 amino acids of the protein would have on QC-related activities. As seen for the Rrp6D238A and Rrp6Y361A point mutations, the Rrp6-129-733 variant had a more severe growth effect when introduced into an mft1Δ context than in a WT context (Fig. 4, cf. A and B). This trend was also manifest at the cell biological level; i.e., <25% of rrp6-129-733 cells exhibited nucleolar accumulation of p(A)+ RNA (as opposed to >95% for rrp6Δ cells) (Fig. 4C), whereas mft1Δ/rrp6-129-733 cells exhibited HSP104-and p(A)+-RNA retention phenotypes more reminiscent of mft1Δ/rrp6Δ (Fig. 4D). Moreover, HSP104 RNAs were stabilized by the Rrp6-129-733 mutation (Fig. 4E) and the level of DCF was decreased (Fig. 4F), both in a manner equivalent to the RRP6 deletion (Fig. 4E,F).
FIGURE 4.
The N-terminal domain of Rrp6p is important for its QC function. Growth (A,B), poly(A)+- and HSP104-RNA-FISH (C,D), HSP104 RNA levels (E), and HSP104 gene DCF (F) analyses of Rrp6-WT, Rrp6129-733, rrp6Δ, mft1Δ, mft1Δ/Rrp6129-73,3 and mft1Δ/rrp6Δ cells, as indicated. (A,B) Strains were spotted onto YM-1 plates in 10-fold dilutions at the indicated temperatures. (C,D) Poly(A)+- or HSP104-RNA-FISH on fixed samples of yeast cells after a temperature shift for 30 or 15 min to 37°C, as indicated. Detection of RNA and denotations are as in Figs. 2 and 3. (E) Quantitative real-time PCR analysis of HSP104 RNA 5′- (black columns) or 3′- (gray columns) ends performed as in Fig. 3B. Averages and standard deviations are calculated from two experiments. (F) Quantitative PCR analysis of HSP104 DNA 5′- (black columns) or 3′- (gray columns) ends performed as in Fig. 3D. A representative experiment is shown.
From these experiments, we conclude that the N-terminal domain of Rrp6p is essential for its QC functions. Moreover, as the Rrp6p N terminus per se does not contribute to the inherent exonucleolytic activity of the enzyme, this strongly indicates a role for the interaction between Rrp6p and Rrp47p in mRNA QC.
Dis3p exonucleolytic function impacts mRNA QC
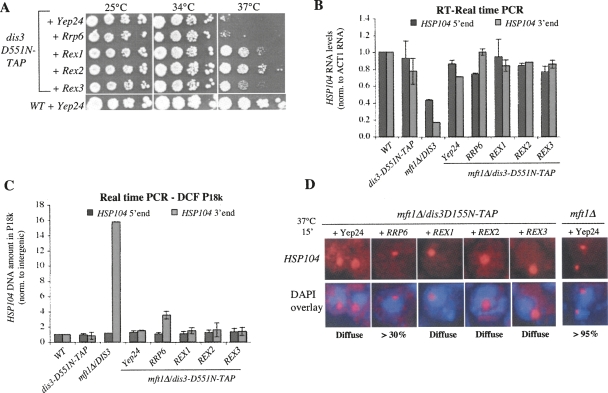
In addition to the nuclear exosome-associated Rrp6p and Dis3p proteins, the yeast nucleus harbors other predicted or proven 3′–5′ exonucleases, among these the Rex1-3p RNaseD-like proteins (van Hoof et al. 2000). Interestingly, overexpression of REX1, REX2, and REX3 from yeast 2μ plasmids partially rescued the growth of a strain harboring the catalytically dead dis3-D551N-TAP protein as its only source of Dis3p, whereas overexpression of RRP6 did not (Fig. 5A).
FIGURE 5.
Dis3p is an mRNP QC factor. (A) dis3D551N-TAP and WT strains haboring 2μ/URA3 Yep24 vectors, and dis3D551N-TAP cells containing Yep24-RRP6, -REX1, -REX2, or –REX3 plasmids, were spotted onto URA drop-out plates in 10-fold dilutions at the indicated temperatures. Quantitative real-time PCR (B), quantitative PCR DCF (C), or HSP104 RNA-FISH (D) analyses were performed on the indicated strains and processed as in Fig. 3. The ratio of cells harboring HSP104 RNA signal retained in focused dots is given below the images. (Diffuse) The HSP104 RNA signal was spread out over a larger volume in >95% of the cells.
To direct our observations toward mRNP QC, we initially investigated the effect of introducing the dis3-D551N-TAP allele into a mft1Δ context. The mft1Δ/dis3-D551N-TAP strain was severely growth-impaired, suggesting that QC of aberrant mRNA in mft1Δ might be affected (data not shown). This anticipation was met as low HSP104 RNA levels in mft1Δ were almost fully restored by the introduction of dis3-D551N-TAP (Fig. 5B). Moreover, the dis3-D551N-TAP mutation completely obliterated the formation of DCF in mft1Δ cells (Fig. 5C). Finally, inactivation of Dis3p exonucleolytic activity resulted in a more diffuse HSP104 RNA-FISH signal compared with the mft1Δ mutation alone (Fig. 5D). We conclude that the exonucleolytic activity of Dis3p can impact mRNP QC in mft1Δ cells.
We next asked whether any of the dis3-D551N-TAP growth suppressors could rescue the low QC activity of the mft1Δ/dis3-D551N-TAP strain. Hence, 2μ REX1, REX2, REX3, or RRP6 plasmids were introduced into mft1Δ/dis3-D551N-TAP cells and HSP104 RNA levels, DCF, and FISH analyses were carried out. With the exception of a minor effect of high copy RRP6 on DCF and HSP104-FISH, none of the suppressors had an effect. Interestingly, a comparable experiment addressing the consequences on growth and QC phenotypes of high copy expression of REX1, REX2, REX3, or DIS3 in mft1Δ/rrp6Δ cells arrived at a similar conclusion; no prominent rescue could be detected (data not shown).
We conclude that exonucleolytic attack, specifically carried out by the nuclear exosome, is essential for establishing the mRNP QC phenotypes tested. Moreover, unlike the case in a WT strain background, the DIS3 and RRP6 genes do not readily substitute for one another when the system is sensitized toward mRNA QC by inactivation of the THO complex.
CONCLUSIONS
The present report shows that the 3′–5′ exonucleolytic activity of the nuclear exosome is a key requirement for the activation of phenotypes associated with production of aberrant mRNP in a yeast strain carrying a deletion of the THO complex component Mft1p. Notably, this is also the case for the retention of mRNP as defined by either HSP104 RNA-FISH or DCF, two phenotypes that supposedly do not directly involve RNA degradation. This observation makes it unlikely that Rrp6p and/or Dis3p, alone or together with the exosome, physically tether malformed mRNPs to a chromatin-associated locale. Rather, our data support a previous suggestion, i.e., that an export-antagonizing event, involving the nuclear exosome, negatively affects HSP104 RNA release and triggers the building up of transcripts detected by FISH or DCF (Rougemaille et al. 2007). It is tempting to speculate that the activity challenged by 3′–5′ exonucleolytic decay by the exosome is a step in mRNA 3′-end formation. Support for this suggestion comes from recent results demonstrating that 3′-end processing factors accumulate near the site of transcription in THO/sub2 mutants (Rougemaille et al. 2008), and that polyadenylation efficiency is markedly decreased (Saguez et al. 2008). The mechanistic details of how nuclear exosome-challenged mRNA 3′-end formation might signal a halt to mRNP biogenesis await further experimentation.
MATERIALS AND METHODS
Protein expression and purification
Recombinant S. cerevisiae Rrp6p protein containing the appropriate truncations and mutations was expressed and purified essentially as described previously (Midtgaard et al. 2006). Briefly, amplification of the genomic S. cerevisiae RRP6 locus was used to create two WT constructs of different lengths corresponding to residues 1–536 and 129–536 in addition to four mutants of RRP6, 12-536(Y361A), 129-536(D238), 129-536(Y361A), and 129-536(D457A). In all cases, the amplified DNA was inserted into the pET-30 Ek/LIC vector using ligation-independent cloning (Novagen). Proteins were expressed in E. coli BL21 (DE3) Rosetta cells grown at 30°C in 2× TY media until A600 = 1.0, followed by overnight induction at 18°C using 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were resuspended in 300 mM KCl, 50 mM Tris·Cl, at pH 7.5, 10% glycerol, and 5 mM β-mercaptoethanol (BME) and lysed by sonication, cleared by centrifugation, and purified on a 1-mL Ni2+-chelating agarose column (GE Healthcare). Recombinant tobacco etch virus (Tev) protease was used to cleave off the N-terminal 6× His-tag of Rrp6p 12-536(Y361), whereas the other Rrp6p proteins remained uncleaved. The proteins were further purified on a MonoQ 4.6/100 PE anion-exchange column (GE Healthcare) and eluted by a linear gradient of 0.1–1 M KCl, followed by a final separation on a Superdex 200 10/300 GL (GE Healthcare) equilibrated in 100 mM KCl, 50 mM Tris·Cl, at pH 7.5, and 5 mM β-mercaptoethanol (BME).
In vitro RNA degradation assays
In vitro RNA degradation reactions were carried out with a constant amount of RNA substrate (20 nM) but two different molar ratios of enzyme (1:1 or 10:1) in a solution containing 10 mM Tris-Cl, at pH 8.0, 50 mM KCl, 7.1 mM MgCl, and 75 μM MnCl. The RNA substrate (5′-CAGCUCCGCAUCCCUUUCCCAAAAAAAAAA-3′) was synthesized with a 5′ fluorescein label and gel-purified (Invitrogen). Degradation reactions were allowed to proceed for 1 h at 30°C, after which they were quenched by addition of RNA loading buffer and separated by denaturing gel electrophoresis in 20% urea–acrylamide gels and visualized using a Typhoon Trio imager (GE Healthcare). The RNA ladder was constructed by limited alkaline hydrolysis of the RNA substrate by incubation in 50 mM NaCO3, at pH 8.9, for 10 min at 96°C followed by neutralization with 300 mM Na(CH3COO), at pH 4.5.
Yeast strain manipulations and plasmids
Yeast strains used in this study are all derived from W303, and crosses were carried out using standard laboratory procedures. RRP6 point mutants were constructed by site-directed mutagenesis of a plasmid-borne RRP6 gene, and subsequently integrated by homologous recombination. The Rrp6-129-733 allele was constructed using standard PCR methods. Western blotting analysis was performed with a rabbit polyclonal Rrp6p antibody produced by immunizing rabbits with a recombinant 129-536 fragment of Rrp6p (Cocalico Biologicals). High-copy Yep24-background plasmids expressing RRP6, REX1, REX2, and REX3 were previously described (Abruzzi et al. 2007). Presumably due to the general toxicity of DIS3 overexpression (Abruzzi et al. 2007), the Yep24-DIS3 plasmid did not complement any dis3-D551N-TAP related phenotypes well. Thus, at least two different spores from relevant crosses were tested for similar phenotypes when the dis3-D551N-TAP allele was involved.
RNA preparations, RT-qPCR, and FISH analyses
All temperature shifts were performed by the addition of an equal volume of pre-warmed medium to a 25°C culture, followed by incubation at 37°C for the appropriate times. RNA was prepared by the hot phenol method as previously described (Libri et al. 2002). Reverse transcription, quantitative PCR analyses, Northern blotting, and poly(A)- and HSP104 RNA-FISH analyses were done as previously described (Jensen et al. 2001b; Thomsen et al. 2003, 2005; Midtgaard et al. 2006; Rougemaille et al. 2007). HSP104 RNAs were detected using a mixture of Cy3 body labeled THJ203, THJ204, and THJ206 oligonucleotide probes (Jensen et al. 2001b).
DCF analysis
Differential chromatin fractionation (DCF) analyses were performed as described (Rougemaille et al. 2008). Briefly, cells were cross-linked for 10 min in 1% formaldehyde and quenched in 500 mM glycine. After washes in 20 mM Tris-HCl, at pH 8.0, the cell pellet was resuspended in 500 μL of FA-lysis buffer (50 mM HEPES-KOH, at pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, and 0.1% NonidetP-40, SDS 0.1%) and chromatin extracts were prepared. The cell lysis step was achieved with a MagnaLyser (Roche), and the chromatin was sheared with a VibraCell sonifier (Bioblock Scientific). Following sonication, chromatin extracts were spun for 10 min at 2000g and subsequently at 18,000g for 20 min. In THO mutants, the pellet of this centrifugation step (P18k) contains DNA–protein cross-linked adducts that sequester THO complex target genes from the chromatin extract, giving rise to DCF. The P18k pellet was deproteinized and decrosslinked by overnight incubation at 65°C in the presence of pronase (1 mg/mL). DNA was extracted using a QIAGEN kit, and the P18k DNA content was analyzed with specific pairs of primers by real-time PCR using a LightCycler 480 (Roche). Amplification reactions were done in duplicate and their quantification was achieved according to the efficiency of each pair of primer, which was determined by serial dilutions of total genomic DNA. The HSP104 DNA amounts were normalized to an intergenic region of DNA, relative to the reference wild-type strain, which had then an arbitrary DCF ratio of 1. DCF occurrence in P18k was plotted with standard deviation bars reflecting at least two independent experiments.
ACKNOWLEDGMENTS
We thank Bertrand Seraphin, Michael Rosbash, and Louise V. Laursen for reagents; members of the D.L. and T.H.J. laboratories for stimulating discussions; and Martin T. Rasmussen for excellent technical assistance. This work was supported by the Danish National Research Foundation (Grundforskningsfonden) (D.L., D.E.B., and T.H.J.), the Novo Nordisk Foundation (D.E.B. and T.H.J.), and the CNRS (D.L.). This research is carried out within the scope of the Associated European Laboratory LEA “Laboratory of Nuclear RNA Metabolism,” and the ESF, RNA quality Eurocores, program NuRNASu.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1108008.
REFERENCES
- Abruzzi K., Denome S., Olesen J.R., Assenholt J., Haaning L.L., Jensen T.H., Rosbash M. A novel plasmid-based microarray screen identifies suppressors of rrp6Δ in Saccharomyces cerevisiae . Mol. Cell. Biol. 2007;27:1044–1055. doi: 10.1128/MCB.01299-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M.K., Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Dziembowski A., Lorentzen E., Conti E., Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Jensen T.H., Boulay J., Rosbash M., Libri D. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 2001a;11:1711–1715. doi: 10.1016/s0960-9822(01)00529-2. [DOI] [PubMed] [Google Scholar]
- Jensen T.H., Patricio K., McCarthy T., Rosbash M. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell. 2001b;7:887–898. doi: 10.1016/s1097-2765(01)00232-5. [DOI] [PubMed] [Google Scholar]
- Jensen T.H., Dower K., Libri D., Rosbash M. Early formation of mRNP: License for export or quality control? Mol. Cell. 2003;11:1129–1138. doi: 10.1016/s1097-2765(03)00191-6. [DOI] [PubMed] [Google Scholar]
- Libri D., Dower K., Boulay J., Thomsen R., Rosbash M., Jensen T.H. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 2002;22:8254–8266. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Greimann J.C., Lima C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Midtgaard S.F., Assenholt J., Jonstrup A.T., Van L.B., Jensen T.H., Brodersen D.E. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc. Natl. Acad. Sci. 2006;103:11898–11903. doi: 10.1073/pnas.0604731103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Houalla R., Podtelejnikov A., Mann M., Tollervey D. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell. Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S., Butler J.S. Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA. 2003;9:1098–1107. doi: 10.1261/rna.5560903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M., Dieppois G., Kisseleva-Romanova E., Gudipati R.K., Lemoine S., Blugeon C., Boulay J., Jensen T.H., Stutz F., Devaux F., et al. THO/Sub2p functions to coordinate 3′-end processing with gene–nuclear pore association. Cell. 2008 doi: 10.1016/j.cell.2008.08.005. (in press). [DOI] [PubMed] [Google Scholar]
- Rougemaille M., Gudipati R.K., Olesen J.R., Thomsen R., Seraphin B., Libri D., Jensen T.H. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J. 2007;26:2317–2326. doi: 10.1038/sj.emboj.7601669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saguez C., Olesen J.R., Jensen T.H. Formation of export-competent mRNP: Escaping nuclear destruction. Curr. Opin. Cell Biol. 2005;17:287–293. doi: 10.1016/j.ceb.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Saguez C., Schmid M., Olesen J.R., El-Hady Ghazy M.A., Qu X., Poulsen M.B., Nasser T., Moore C., Jensen T.H. Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Mol. Cell. 2008;31:91–103. doi: 10.1016/j.molcel.2008.04.030. [DOI] [PubMed] [Google Scholar]
- Schneider C., Anderson J.T., Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol. Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead J.A., Costello J.L., Livingstone M.J., Mitchell P. The PMC2NT domain of the catalytic exosome subunit Rrp6p provides the interface for binding with its cofactor Rrp47p, a nucleic acid-binding protein. Nucleic Acids Res. 2007;35:5556–5567. doi: 10.1093/nar/gkm614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondon A.G., Aguilera A., Struhl K., Reed R., et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Thomsen R., Libri D., Boulay J., Rosbash M., Jensen T.H. Localization of nuclear retained mRNAs in Saccharomyces cerevisiae . RNA. 2003;9:1049–1057. doi: 10.1261/rna.5170303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen R., Nielsen P.S., Jensen T.H. Dramatically improved RNA in situ hybridization signals using LNA-modified probes. RNA. 2005;11:1745–1748. doi: 10.1261/rna.2139705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen R., Saguez C., Nasser T., Jensen T.H. General, rapid, and transcription-dependent fragmentation of nucleolar antigens in S. cerevisiae mRNA export mutants. RNA. 2008;14:706–716. doi: 10.1261/rna.718708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Lennertz P., Parker R. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP, and RNase P RNAs in yeast. EMBO J. 2000;19:1357–1365. doi: 10.1093/emboj/19.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra P., Iglesias N., Camblong J., Zenklusen D., Stutz F. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J. 2005;24:813–823. doi: 10.1038/sj.emboj.7600527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D., Vinciguerra P., Wyss J.C., Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]