Abstract
The last step in ribosome-catalyzed protein synthesis is the hydrolytic release of the newly formed polypeptide from the P-site bound tRNA. Hydrolysis of the ester link of the peptidyl-tRNA is stimulated normally by the binding of release factors (RFs). However, an unacylated tRNA or just CCA binding to the ribosomal A site can also stimulate deacylation under some nonphysiological conditions. Although the sequence of events is well described by biochemical studies, the structural basis of the mechanism underlying this process is not well understood. Two new structures of the large ribosomal subunit of Haloarcula marismortui complexed with a peptidyl-tRNA analog in the P site and two oligonucleotide mimics of unacylated tRNA, CCA and CA, in the A site show that the binding of either CA or CCA induces a very similar conformational change in the peptidyl-transferase center as induced by aminoacyl-CCA. However, only CCA positions a water molecule appropriately to attack the carbonyl carbon of the peptidyl-tRNA and stabilizes the proper orientation of the ester link for hydrolysis. We, thus, conclude that both the ability of the O3′-hydroxyl group of the A-site A76 to position the water and the A-site CCA induced conformational change of the PTC are critical for the catalysis of the deacylation of the peptidyl-tRNA by CCA, and perhaps, an analogous mechanism is used by RFs.
Keywords: ribosome, peptide release, peptidyl-tRNA hydrolysis, unacylated tRNA, CCA
INTRODUCTION
Protein synthesis catalyzed by the ribosome proceeds through the formation of consecutive peptide bonds between incoming amino acids that are ester-linked to their cognate tRNAs and the growing polypeptide chain on the peptidyl-tRNA. Pairing of the triplet codons of the mRNA with the cognate tRNAs takes place in the decoding center of the small ribosomal subunit (Ogle et al. 2001). Accommodation of correctly delivered tRNAs into the peptidyl-transferase center (PTC) results in each new amino acid attached to the A-site tRNA being added to the polypeptide chain. The peptide bond is formed by the nucleophilic attack of the αNH2-group of the aminoacyl-tRNA in the A site on the carbonyl carbon of the ester link of peptidyl-tRNA in the P site. The process continues until the ribosome encounters a stop codon, which provides a signal for the termination of protein synthesis. The stop codon is not recognized by a cognate tRNA, but by a release factor (RF) protein. Binding of RF to the ribosomal A site promotes hydrolysis of the ester link of the peptidyl-tRNA and the subsequent release of the newly synthesized peptide.
Whereas spontaneous hydrolysis of the peptidyl-tRNA on a ribosome with an empty A site occurs very slowly at a rate of 2 × 10−5 sec−1, the hydrolysis induced by RF proceeds at a rate of 1 sec−1 (Zavialov et al. 2002). Low-resolution cryo-EM and X-ray crystallography studies have revealed that RFs use both structural and functional mimicry of tRNA to promote peptide release (Rawat et al. 2003, 2006; Petry et al. 2005). Upon binding to the ribosome, their critical loops are simultaneously positioned into the decoding center and the PTC that are ∼70 Å apart (Petry et al. 2005). The 5 Å resolution of the structural studies of the RF complex with the 70S ribosome, however, did not allow understanding the structural basis of the enzymatic mechanism. Furthermore, these studies did not answer whether the RFs catalyze peptide release directly or by inducing a conformational change in the PTC that allows proper positioning of the attacking water molecule as well as deprotection and reorientation of the ester linked target. A structural study of the 70S ribosome in complex with RF1 and the unacylated tRNA bound to the P site at 3.2 Å resolution was published (Laurberg et al. 2008) while this manuscript was under revision. The structure revealed that the main-chain amide of Gln 230 of the highly conserved GGQ motif of RF1 forms a hydrogen bond with the O3′-hydroxyl of A76 of the unacylated P-site tRNA, suggesting its involvement in the termination reaction (Laurberg et al. 2008). Brunelle et al. (2008), on the other hand, proposed in their recent study that the RF-promoted peptide release depends critically on the O2′-hydroxyl group of the peptidyl tRNA substrate.
Recent mutational and kinetic studies of the ribosome suggested that the PTC residues A2637 (A2602), U2620 (U2585), A2486 (A2451), and C2104 (C2063) are important for peptide release (Polacek et al. 2003; Youngman et al. 2004); the Haloarcula marismortui ribosome numbering is used throughout with the Escherichia coli numbering shown in parentheses. The strongest effect was exerted by mutations of U2620 (U2585) and A2637 (A2602) with a 40- and 350-fold decrease of the peptide release rate, respectively (Youngman et al. 2004). Also, the most recent studies have revealed that water is indeed the favored nucleophile in the RF-promoted peptidyl-tRNA hydrolysis (Shaw and Green 2007). Peptide release has also been investigated by using either an unacylated tRNA or CCA or CA as the A-site ligand instead of the RF. Binding of an unacylated tRNA to the ribosomal A site accelerates deacylation of peptidyl-tRNA from the 70S ribosome by more than 20-fold, and remarkably the presence of 20% ethanol increases the reaction rate to the level of the RF-induced termination (Zavialov et al. 2002). Binding of an unacylated tRNA mimic of the acceptor end of tRNA, the trinucleotide CCA, in the A site promotes peptide release by two orders of magnitude in the presence of 35% acetone, whereas CA and A have no effect on the hydrolysis rate (Caskey et al. 1971).
The structure of the H. marismortui 50S ribosomal subunit (Hma50) complexed with both A-site and P-site substrates suggested that a mechanism similar to the one employed for peptide bond formation may also apply to the hydrolytic release of peptide. These structures showed that the binding of the A-site substrate induces a conformational change in the PTC, resulting both in the deprotection and reorientation of the ester linked carbonyl group and the positioning of the reactive groups. Similar models for CCA-induced as well as RF-induced peptide release were suggested by Schmeing et al. (2005a) and predicted by molecular dynamics (MD) simulations, starting with the aminoacyl-CCA induced-fit Hma50 structure (Trobro and Åqvist 2006). These MD calculations predicted that the O2′-hydroxyl group of the P-site A76 plays the same proton shuttle role in peptide release as it does in peptide bond formation (Dorner et al. 2003; Weinger et al. 2004; Schmeing et al. 2005a,b). Finally, while Schmeing et al. (2005b) proposed that the O3′-hydroxyl group of the A-site A76 of CCA orients the hydrolytic water to promote deacylation, Trobro and Åqvist (2006) suggested that both the O3′-hydroxyl of the A-site A76 and the O2′-hydroxyl group of the P-site A76 play this role.
To address the mechanism of CCA stimulated peptide deacylation, we determined two crystal structures of the Hma50 complexed with a peptidyl-tRNA analog in the P site and two oligonucleotide mimics of unacylated tRNA, CA and CCA, in the A site. Our results show that despite the ability of either CA or CCA to promote similar induced-fit conformational changes in the PTC, only CCA is capable of both orienting the hydrolytic water and stabilizing the configuration of the peptidyl-tRNA ester link that allows the attack by water. We also conclude that the apparent contradiction between the ability of CCA to stimulate peptidyl-tRNA hydrolysis and the inability of CA to do likewise is a consequence of the CA binding too remotely from the reaction site for its O3′-hydroxyl group of A76 to correctly position the attacking water molecule.
RESULTS AND DISCUSSION
CCA-pcb binds to the P site, whereas the oligonucleotide mimics of unacylated tRNA, CCA and CA, bind to the A site only
We determined two crystal structures of the Hma50 complexed with the peptidyl-tRNA analog, CCA-phenylalanyl-caproic acid-biotin (CCA-pcb), and either one of two oligonucleotide mimics of unacylated tRNA, CA or CCA. In CCA-pcb, the CCA moiety is linked to phenylalanyl-caproic acid-biotin (Phe-cap) through an amide bond rather than ester bond, providing a nonhydrolysable link to the peptidyl-tRNA analog. However, for the sake of simplicity, we shall refer to this bond as either acyl-ester or just simply ester link. The crystals containing CCA-pcb and either CCA or CA diffracted to 2.8 and 2.9 Å resolution, respectively. The difference electron density maps calculated using the observed amplitude differences between data from the complexes and the apo Hma50 data set reveal that the CCA-pcb analog is bound only to the P site in both soaks, whereas the A site is occupied by either the CA or CCA oligonucleotides (Fig. 1). The CCA-end of CCA-pcb interacts with the P loop consisting of the 23S rRNA nucleotides G2284 (G2251), G2285 (G2252), and A2486 (A2451), whereas Phe-cap moiety faces the entrance of the exit tunnel. On the other hand, both CA and CCA interact with the A loop in the A site, which is composed of the 23S rRNA nucleotides G2588 (G2553), U2589 (U2555), U2590 (U2556), and G2618 (G2583). The models of the complexes containing CCA-pcb and either CA or CCA were refined to final Rfree values of 24.5% and 25.5%, respectively (Table 1). The two 23S rRNA models were almost identical with an root mean square deviation (RMSD) value of 0.042 Å when all atoms were used for calculation.
FIGURE 1.
CCA-pcb binds to the P site, whereas CCA and CA bind to the A site only. (A) A difference electron density map of the PTC calculated using the observed Fo amplitudes from the Hma50-CCA complex crystal and the Fo amplitudes of the apo Hma50 calculated at 2.8 Å resolution and contoured at 3σ. The trinucleotide CCA bound in the A site and the mimic of the peptidyl-tRNA CCA-pcb bound in the P site are shown in blue, whereas the 23S rRNA residues are shown in gold. (B) Fo-Fo difference electron density map of the same region calculated using the amplitudes from the Hma50-CA complex crystal calculated at 2.9 Å resolution and contoured at 3σ. The color coding is the same as under A.
TABLE 1.
Data collection and refinement statistics
Binding of the A-site substrates promotes conformational changes in the PTC
We observe that the binding of either CA or CCA to the A site induces nearly the same conformational changes in the RNA of the PTC as was observed previously with the aminoacylated-CCA bound to the A site (Schmeing et al. 2005b). Thus, an induced-fit mechanism similar to that proposed for peptide bond formation may be associated with peptidyl-CCA deacylation stimulated by CCA, as hypothesized earlier (Schmeing et al. 2005b).
Whereas the CCA trinucleotide binds to the A site as expected (Fig. 2A; Hansen et al. 2002; Schmeing et al. 2005a,b), somewhat surprisingly the CA dinucleotide interacts with the A loop in a similar fashion as the C74-C75 of the CCA trinucleotide (Fig. 2B). Moreover, upon binding of either CA or CCA, the PTC nucleotides G2618-U2620 (G2583-U2585) move away from the P site and adopt a similar conformation to that induced by the binding of CCPmn (Fig. 2C). Although the movement of U2620 (U2585) is not as large as it was observed in the study of peptide bond formation (Schmeing et al. 2005b), it is large enough to allow for both the necessary reorientation of the acyl ester bond of the peptidyl-tRNA and its attack by water (Fig. 2C).
FIGURE 2.
Binding of either CA or CCA to the A site promotes the same conformational change in the PTC. (A) The CCA trinucleotide (blue) binds to G2558 (G2553), U2590 (U2555), and G2618 (G2583) of the ribosomal A loop (gold). (B) The CA dinucleotide (cyan) binds to the A loop in the same place as the C74-C75 of CCA. (C) Binding of either CCA or CA (blue) to the A site causes the induced-fit conformational change in the PTC. The apo conformation of G2618-U2620 (G2583-U2585) is shown in beige, the conformation observed in Schmeing et al. (2005b) is shown in light orange (Protein Data Bank ID no. 1VQN), whereas the conformation observed upon either CA or CCA binding to the A site is shown in dark orange. CCA-pcb bound to the P site is shown in blue. Arrows show the direction of the movement of the PTC residues during the induced-fit conformational change.
Comparison of the binding interactions exhibited by CCA, CChPmn, and ChPmn with the A-site 23S rRNA suggests an explanation for why the CA dinucleotide binds to the A site as the C74-C75 does rather than as the C75-A76 of the CCA trinucleotide. When CCA, CChPmn, or ChPmn interact with the A loop, the following common interactions are observed: (1) base pairing between C75 and G2588 (G2553); (2) the A-minor interaction between the base of A76 or Pmn and the base of G2618 (G2583) of the 23S rRNA; (3) a hydrogen bond between the O2′-hydroxyl groups of U2541 (U2506) and the A-site A76 or Pmn; and (4) the base stacking between C74 and U2590 (U2555). In addition, both CCA and CChPmn show a favorable base stacking of C74 on C75. Although the base stacking of C75 on A76 or Pmn is partially lost upon binding of all of these ligands, the aforementioned base pairing and base stacking interactions may compensate for this loss. Also, the amino acid side-chains in the CChPmn and ChPmn mimics of the aminoacylated-tRNA provide important interactions that stabilize their positioning closer to the P site. The CA dinucleotide, however, has neither a Phe amino acid side-chain nor the possibility of the favorable base stacking interactions of C74 on C75 to compensate for loss of base stacking between C75 and A76, if it were to bind in the same position as the C75-A76 does in the trinucleotide. When bound in the C74-C75 position, however, the dinucleotide assumes the conformation in which the wobble base pairing between A76 and G2588 (G2553) and base stacking C75 on A76 and C75 on U2590 (U2555) are preserved. Thus, a more stable conformation of the CA dinucleotide presumably drives its binding farther away from the reaction site.
The interactions of C74 in the A site trigger the induced fit in the PTC
The observation that the CA dinucleotide binds to the A site in the same place as C74 and C75 of CCA and is able to promote a very similar conformational change in the PTC as the CCA trinucleotide does, raises the question concerning what causes the induced-fit conformational change in the PTC. In the complexes with CA, CCA, and CChPmn (Schmeing et al. 2005b), the 5′ nucleotides of the A-site ligands, C75 and C74, form a base stack with U2590 (U2555) and the conformational change in the PTC occurs. In the case of the complex with ChPmn, on the other hand, the interaction between C75 and G2588 (G2553) is preserved while a stacking interaction with U2590 (U2555) does not occur. Since the induced fit in the PTC does not occur in this case (Schmeing et al. 2005b), the base stack of either C74 from CCA or C75 from CA on U2590 (U2555) of the 23S rRNA may indeed promote the induced fit, as proposed previously by Schmeing et al. (2005b). Finally, our structures appear not to be consistent with the proposal by Brunelle et al. (2006) that the base pairing between C75 of the A-site ligand and G2558 (G2553) of the 23S rRNA promotes the conformational change.
Because the size of the substrate-induced-conformational change in our structures is similar to the one observed in the earlier study of peptide bond formation (Schmeing et al. 2005b), we conclude that the induced-fit mechanism also plays a significant role in promoting peptide release. We also propose that interaction between either the unacylated tRNA or the RFs and the 23S rRNA residue U2590 (U2555) triggers the induced fit in the PTC.
CCA but not CA can orient the attacking water for peptidyl-tRNA hydrolysis
The apparent contradiction between the ability of CA to induce a similar conformational change as either CCA or CChPmn and its inability to promote peptide release is explained by the CCA trinucleotide, but not CA, being able to orient the hydrolytic water using the O3′-hydroxyl group of A76 and to correctly orient the ester link of the peptidyl-tRNA for the attack by water.
In the complex with CA, the carbonyl oxygen of the peptidyl-CCA is facing toward U2620 (U2585), whereas in the CCA complex, the carbonyl oxygen swings away from that base and forms a hydrogen bond with the N3 of A2486 (A2451) of the 23S rRNA (Fig. 3A). In the CCA complex, the O3′-hydroxyl of the A-site A76 forms a hydrogen bond with the water molecule Wat1, which is 3.8 Å away from the carbonyl carbon of the ester link of the peptidyl-CCA (Fig. 3A). Also, the O4 of U2620 (U2585) forms hydrogen bonds with both hydroxyl groups of the A-site A76 and is 3.6 Å away from Wat1 (Fig. 3A). This hydrogen bonding network orients a free electron pair of the oxygen atom of Wat1 toward the carbonyl carbon of the ester bond. The carbonyl oxygen of the ester link is 3.1 Å away from Wat1, but it is not oriented properly to form a hydrogen bond. Thus, Wat1 is oriented properly for a nucleophilic attack on the ester linked carbonyl carbon, with an attack angle of 104°. Our findings are in agreement with the recent study on the mechanism of the RF-promoted peptide release, which revealed that water is the preferred nucleophile during peptide release reaction (Shaw and Green 2007).
FIGURE 3.
Only CCA can orient the hydrolytic water, whereas CA binds too remotely from the reaction site. (A) In the Hma50-CCA complex A76 of the A site CCA (blue) forms a hydrogen bond with the hydrolytic water Wat1 (red sphere) through its O3′-hydroxyl group. Wat1 is 3.8 Å away from the carbonyl carbon of the ester link in CCA-pcb. The ribosomal residue U2620 (U2585) (gold) forms hydrogen bonds with both the O2′- and O3′-hydroxyl groups of the A-site A76, whereas the carbonyl oxygen of the P-site ester link forms a hydrogen bond with N3 of A2486 (A2451) (gold). (B) In the Hma50-CA complex, the dinucleotide CA (cyan) binds too remotely from the hydrolytic water Wat1, which forms a hydrogen bond with the carbonyl oxygen of the ester link in CCA-pcb (cyan). The A site is occupied with additional ordered water molecules, Wat2 and Wat3 that form hydrogen bonds with U2620 (U2585). The Fo-Fo difference electron density map covering water molecules is contoured at 4σ in A and at 3.5σ in B and is shown as green mesh.
Further, these structural results are in partial agreement with the conclusions of the MD calculations on the CCA-promoted peptide release (Trobro and Åqvist 2006). However, this MD study suggested that the O2′-hydroxyl group of the P-site A76 served to orient the hydrolytic water molecule (Trobro and Åqvist 2006), while our structure shows that it is the O3′-hydroxyl group that is positioning the water molecule and the O2′-hydroxyl of the P-site A76 is facing away from the A site and is hydrogen bonded with the O2′-hydroxyl of C2104 (C2063) of the 23S rRNA (data not shown). This suggests that the A76 O2′-hydroxyl group of the P-site substrate is not involved in the CCA-induced peptide release, but rather it stabilizes binding of the P-site ligand. However, we cannot exclude the possibility that this group indeed participates in peptide release when promoted by RF binding (Trobro and Åqvist 2006). The A-site ligand, on the other hand, is stabilized by interactions with U2541 (U2506) and U2620 (U2585). Both the O2′-hydroxyl group of U2541 (U2506) and the base atom O4 of U2620 (U2585) form hydrogen bonds with the O2′-hydroxyl group of the A-site A76 (data not shown). Further, it has also been proposed that the base of U2541 (U2506) forms a stacking-like interaction with the Gln side-chain from the RFs' GGQ motif (Trobro and Åqvist 2006; Brunelle et al. 2008). Indeed, the recent structure of the 70S-RF1-tRNA complex has revealed that U2541 (U2506) participates in forming the binding pocket for the Gln 230 side-chain (Laurberg et al. 2008). In our structure, on the other hand, the base of U2541 (U2506) does not interact with the A-site ligand, but it rather interacts with the base of the G2618 (G2583) nucleotide (data not shown), suggesting that this residue may play different roles in the CCA- and RF-promoted reactions.
In the complex that contains CA in the A site, the carbonyl oxygen of the ester link of the peptidyl-CCA forms a hydrogen bond with the water Wat1, which is not appropriately positioned for nucleophilic attack on the carbonyl carbon (Fig. 3B). Another major difference from the CCA complex is that the Wat1 molecule forms hydrogen bonds to additional water molecules, Wat2 and Wat3, rather than to A76 of CA (Fig. 3B). This is a direct consequence of the CA binding up higher in the A site. The base of A76 forms a wobble base pair with the base of G2588 (G2553) of the 23S rRNA, whereas C75 stacks with the base of U2590 (U2555) (Figs. 2B, 3B). These interactions place the A76 nucleotide of CA farther away from the reaction site. Consequently, the O3′-hydroxyl group of the A-site A76 is >6 Å away from the attacking water Wat1 and >10 Å away from the reaction site, thus rendering the CA dinucleotide incapable of catalyzing the reaction.
We conclude that CCA but not CA bound to the A site positions its O3′-hydroxyl group within hydrogen bonding distance from the attacking water molecule, which is appropriately placed to attack a correctly oriented carbonyl group. Consequently, the O3′-deoxy derivative of CCA should be incapable of promoting peptide release. The CCA binding also stabilizes the conformation of the ester link that allows the hydrolysis reaction to occur. Thus, the CCA-promoted hydrolysis of peptidyl-tRNA depends on both its ability to induce the required conformational change in the PTC and its ability to appropriately position the attacking water. While CA achieves the former requirement, it cannot achieve the latter, thereby explaining the observations of Caskey et al. (1971) that it cannot promote hydrolysis of the peptidyl-tRNA.
The role of A2637 (A2602), U2620 (U2585), A2486 (A2451), and C2104 (C2063) in peptide release
We finally sought to explain the role PTC residues play during peptide release. Kinetic studies have shown that mutants of A2637 (A2602), U2620 (U2585), A2486 (A2451), and C2104 (C2063) of the PTC affect RF-promoted peptide release to varying degrees (Polacek et al. 2003; Youngman et al. 2004). The strongest effect was exerted by mutating either A2637 (A2602) or U2620 (U2585), which results in a decrease of the rate of peptide release by 350- and 40-fold, respectively (Polacek et al. 2003; Youngman et al. 2004).
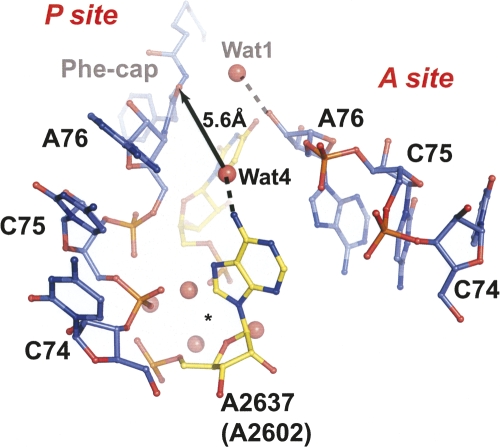
The binding of CCA-pcb induces the movement of A2637 (A2602), which is necessary for opening the P site (Schmeing et al. 2005a,b). The conformational change is reminiscent of the movement observed in the earlier study of peptide bond formation (Schmeing et al. 2005b). The base of A2637 (A2602) hydrogen bonds to a water molecule, Wat4, which is 5.6 Å away from the carbonyl oxygen of the ester link (Fig. 4). This water may play a role in stabilizing the oxyanion of the tetrahedral intermediate formed upon the attack of Wat1 on the carbonyl carbon. In addition, the backbone and base atoms of A2637 (A2602) interact with the phosphate oxygens of the P-site C75. A water oriented in part by A2637 (A2602) is seen to hydrogen bond to an analog of the oxyanion in the structure of a complex with a transition state analog of peptide bond formation as well (Schmeing et al. 2005b), yet mutating A2637 (A2602) did not affect the rate of peptide bond formation (Youngman et al. 2004). This suggests that A2637 (A2602) may play a more complex role in the RF-promoted peptide release reaction than that observed in the CCA-mediated reaction. The structure of the 70S ribosome complexed with RF1 and the unacylated tRNA bound to the P site implicates A2637 (A2602) in binding and/or positioning of domain 3 of RF1 for catalysis (Laurberg et al. 2008). On the other hand, U2620 (U2585) forms hydrogen bonds to both the O2′- and O3′-hydroxyl groups of the A-site A76. More importantly, the backbone phosphate oxygens of A2637 (A2602), U2620 (U2585), and the P-site C75 form hydrogen bonds with the common cluster of water molecules (Fig. 4). A strong positive peak in the difference electron density map using Fo-Fc as coefficients suggests that these interactions are also stabilized by a metal ion, possibly Mg2+ or Sr2+ (data not shown). These observations show that A2637 (A2602) and U2620 (U2585) stabilize both the P- and A-site ligand binding.
FIGURE 4.
The role of A2637 (A2602) in peptide release. A2637 (A2602) forms a hydrogen bond with the water molecule Wat4 that is 5.6 Å away from the carbonyl oxygen of the P-site ester link. Also, A2637 (A2602), CCA-pcb, and U2620 (U2585) form hydrogen bonds to the same cluster of water molecules (*) and possibly with the metal ion. The A- and P-site substrates are shown in blue, whereas the ribosomal residues are in gold. U2620 (U2585) is the only residue that is not annotated.
Our results confirm previous implications that A2637 (A2602) and U2620 (U2585) play an important role in peptide release. Both nucleotides undergo conformational changes upon ligand binding, and both stabilize ligands and water molecules in the PTC. Also, A2637 (A2602) may be involved in stabilizing the GGQ motif during the RF-promoted release. These observations may explain why mutating either A2637 (A2602) or U2620 (U2585) abolishes the ability of the ribosome to promote the hydrolytic deacylation of peptidyl-tRNA. Finally, our results suggest that the PTC residues are not involved directly in the catalysis of peptide release as it is thought to be the case for peptide bond formation (Sievers et al. 2004; Weinger et al. 2004; Youngman et al. 2004; Schmeing et al. 2005a,b; Bieling et al. 2006).
Concluding remarks
Our structures reveal that both the conformational change in the PTC that is induced by the binding of the CCA substrate to the A site and the position of the O3′-hydroxyl group of its A76 are critical for promoting hydrolytic release of the peptide from the peptidyl-tRNA. Interaction of either CA or CCA with the 23S rRNA residue U2620 (U2585) in the A site triggers the conformational change in the PTC. However, only CCA is able to appropriately position the attacking water molecule through a hydrogen bond by the O3′-hydroxyl group of A76. Furthermore, in this state the repositioning of G2618-U2620 (G2583-U2585) allows free rotation of the ester link of the peptidyl-tRNA as well as the water attack. Interaction of the CA dinucleotide with the A site, on the other hand, places its O3′-hydroxyl group too far from a potential attacking water to be able to position it for peptide release. Thus, these structural studies provide an explanation for the early observation of Caskey et al. (1971) that CCA, but not CA, is able to promote peptidyl-tRNA hydrolysis. Finally, we show that the ribosomal residues are not directly involved in the catalysis of peptide release; rather they stabilize the A- and P-site ligands as well as water molecules in the PTC. We propose that binding of the RFs to the A site induces a conformational change in the PTC similar to the one seen in these structures, as well as those of substrate complexes, and that RFs orient the hydrolytic water molecule by using their highly conserved GGQ motif. The recent structure of the 70S ribosome with a bound RF and a tRNA in the P site published when this manuscript was under revision appears consistent with this possibility, though the structure of a complex with peptidyl-tRNA is still required to establish the mechanism.
MATERIALS AND METHODS
Substrate analogs CA, CCA, and CCA-pcb were purchased from Dharmacon. The H. marismortui 50S ribosomal subunits were purified and crystallized as described previously (Ban et al. 2000). The crystals were stabilized in 1.6 M NaCl, 20% (v/v) ethylene glycol, 0.5 M NH4Cl, 100 mM SrCl2, 100 mM KCH3COO, and 12% (w/v) polyethylene glycol 6000 (pH 6.0), at 4°C. The CCA and CA oligonucleotides were dissolved in water, whereas CCA-pcb was dissolved in DMSO. Ligands were added to the stabilizing buffer to a final concentration of 1 mM; crystals were soaked in that solution for 1–2 h and then flash-frozen in liquid propane.
Data sets were collected at the beam line ID24 of the Advanced Photon Source (APS). Data were indexed, scaled, and merged using the program HKL2000 (Otwinowski and Minor 1997). Structures were determined by rigid-body refinement of the apo-subunit structure (Klein et al. 2004) with the program CNS (Brunger et al. 1998), followed by iterative rounds of gradient energy minimization and individual B-factor refinement. Modeling and electron density map analysis were performed with the program Coot (Emsley and Cowtan 2004), and figures were made using PyMOL (DeLano 2002).
ACKNOWLEDGMENTS
We thank the staff at the beam line ID24 of the Advanced Photon Source (Argonne, IL) for help during the data collection. We also thank G. Blaha for help during structure determination, and S. Palioura and C.A. Innis for comments and suggestions.
Footnotes
Abbreviations: Pmn, puromycin; hPmn, hydro-puromycin.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1118908.
REFERENCES
- Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Bieling P., Beringer M., Adio S., Rodnina M.V. Peptide bond formation does not involve acid-base catalysis by ribosomal residues. Nat. Struct. Mol. Biol. 2006;13:423–428. doi: 10.1038/nsmb1091. [DOI] [PubMed] [Google Scholar]
- Brunelle J.L., Youngman E.M., Sharma D., Green R. The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity. RNA. 2006;12:33–39. doi: 10.1261/rna.2256706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle J.L., Shaw J.J., Youngman E.M., Green R. Peptide release on the ribosome depends critically on the 2′ OH of the peptidyl tRNA substrate. RNA. 2008;14:1526–1531. doi: 10.1261/rna.1057908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Caskey C.T., Beaudet A.L., Scolnik E.M., Rosman M. Hydrolysis of fMet-tRNA by peptidyl transferase. Proc. Natl. Acad. Sci. 1971;68:3163–3167. doi: 10.1073/pnas.68.12.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. The PyMOL user's manual. DeLano Scientific; Palo Alto, CA: 2002. [Google Scholar]
- Dorner S., Panuschka C., Schmid W., Barta A. Mononucleotide derivatives as ribosomal P-site substrates reveal an important contribution of the 2′-OH to activity. Nucleic Acids Res. 2003;31:6536–6542. doi: 10.1093/nar/gkg842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Hansen J.L., Schmeing T.M., Moore P.B., Steitz T.A. Structural insights into peptide bond formation. Proc. Natl. Acad. Sci. 2002;99:11670–11675. doi: 10.1073/pnas.172404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D.J., Moore P.B., Steitz T.A. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- Laurberg M., Asahara H., Korostelev A., Zhu J., Trakhanov S., Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- Ogle J.M., Brodersen D.E., Clemons W.M., Jr, Tarry M.J., Carter A.P., Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Petry S., Brodersen D.E., Murphy F.V.T., Dunham C.M., Selmer M., Tarry M.J., Kelley A.C., Ramakrishnan V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Polacek N., Gomez M.J., Ito K., Xiong L., Nakamura Y., Mankin A. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol. Cell. 2003;11:103–112. doi: 10.1016/s1097-2765(02)00825-0. [DOI] [PubMed] [Google Scholar]
- Rawat U.B., Zavialov A.V., Sengupta J., Valle M., Grassucci R.A., Linde J., Vestergaard B., Ehrenberg M., Frank J. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- Rawat U., Gao H., Zavialov A., Gursky R., Ehrenberg M., Frank J. Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM. J. Mol. Biol. 2006;357:1144–1153. doi: 10.1016/j.jmb.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Schmeing T.M., Huang K.S., Kitchen D.E., Strobel S.A., Steitz T.A. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell. 2005a;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Schmeing T.M., Huang K.S., Strobel S.A., Steitz T.A. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005b;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- Shaw J.J., Green R. Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol. Cell. 2007;28:458–467. doi: 10.1016/j.molcel.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers A., Beringer M., Rodnina M.V., Wolfenden R. The ribosome as an entropy trap. Proc. Natl. Acad. Sci. 2004;101:7897–7901. doi: 10.1073/pnas.0402488101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobro S., Åqvist J. Analysis of predictions for the catalytic mechanism of ribosomal peptidyl transfer. Biochemistry. 2006;45:7049–7056. doi: 10.1021/bi0605383. [DOI] [PubMed] [Google Scholar]
- Weinger J.S., Parnell K.M., Dorner S., Green R., Strobel S.A. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat. Struct. Mol. Biol. 2004;11:1101–1106. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- Youngman E.M., Brunelle J.L., Kochaniak A.B., Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- Zavialov A.V., Mora L., Buckingham R.H., Ehrenberg M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell. 2002;10:789–798. doi: 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]