Abstract
Highly conserved sequences at the 5′ splice site and branch site of U12-dependent introns are important determinants for splicing by U12-dependent spliceosomes. This study investigates the in vivo splicing phenotypes of mutations in the branch site consensus sequence of the U12-dependent intron F from a human NOL1 (P120) minigene. Intron F contains a fully consensus branch site sequence (UUCCUUAAC). Mutations at each position were analyzed for their effects on U12-dependent splicing in vivo. Mutations at most positions resulted in a significant reduction of correct U12-dependent splicing. Defects observed included increased unspliced RNA levels, the activation of cryptic U2-dependent 5′ and 3′ splice sites, and the activation of cryptic U12-dependent branch/3′ splice sites. A strong correlation was observed between the predicted thermodynamic stability of the branch site: U12 snRNA interaction and correct U12-dependent splicing. The lack of a polypyrimidine tract between the branch site and 3′ splice site of U12-dependent introns and the observed reliance on base-pairing interactions for correct U12-dependent splicing emphasize the importance of RNA/RNA interactions during U12-dependent intron recognition and proper splice site selection.
Keywords: splicing, U12-dependent spliceosome, minor class intron
INTRODUCTION
The removal by splicing of introns from primary transcripts and subsequent ligation of exons is essential to the expression of most genes in higher eukaryotes. The splicing of eukaryotic genes is performed by either of two spliceosomes of differing composition. The U2-dependent spliceosome catalyzes the removal of the major U2-dependent class of introns, while the U12-dependent spliceosome removes the less-abundant U12-dependent introns. Despite their low abundance, U12-dependent introns are found in both tissue-specific and ubiquitously expressed genes, some of which carry out essential cellular functions (e.g., DNA replication and repair, transcription, RNA processing, and translation). Therefore, proper selection and precise removal of minor class introns by the U12-dependent spliceosome is critical.
Assembly of the different spliceosomes on their respective introns involves similar mechanisms, with a major difference occurring at the step of initial intron recognition (for review, see Will and Lührmann 2005). In the initial stages of U2-dependent splicing, the U1 small nuclear ribonucleoprotein complex (snRNP) interacts with the 5′ splice site followed by subsequent interaction of the U2 snRNP with the U2-dependent branch site. In contrast, splicing of U12-dependent introns begins with the cooperative recognition of the 5′ splice site and branch site by the preformed U11/U12 di-snRNP (Montzka Wassarman and Steitz 1992; Frilander and Steitz 1999). Consensus sequences at the intron splice sites and branch site (for review, see Burge et al. 1999), which interact with small nuclear ribonucleic acids (snRNAs) mainly through base pairing (for review, see Nilsen 1998), specify which splicing system will be used. The 5′ splice site of U12-dependent introns is longer and more highly conserved than U2-dependent 5′ splice sites and serves as a major determinant of splice site specificity. Several reports analyzing U12-dependent splice site dinucleotides have revealed that, in contrast to U2-type introns, there is no strict 3′ splice site sequence requirement for U12-type introns (Dietrich et al. 1997, 2001a, 2005; Levine and Durbin 2001; Hastings et al. 2005). Instead, 3′ splice site selection is driven by the sequence at the 5′ splice site junction and the distance from the highly conserved branch site.
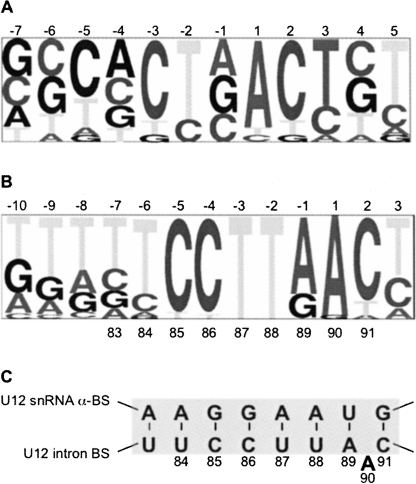
In addition to splice site sequences, the branch site consensus sequence is an important determinant of which spliceosome class (i.e., U2-dependent versus U12-dependent) is used in splicing a particular intron. In contrast to the relatively weak consensus of U2-dependent branch sequences (Fig. 1A), the U12-dependent intron branch site (Fig. 1B) is highly conserved, suggesting that it serves as a primary recognition element for the U12-dependent spliceosome (Padgett and Burge 2003).
FIGURE 1.
Pictograms of the U2-dependent (A) and U12-dependent (B) intron branch site consensus sequences using the data set described by Padgett and Burge (2003). In these pictograms, the size of the letter corresponds to the frequency in which that base is present at each position. The position labeled 1 (A,B) at the top is the predicted branch site residue. The numbers below the U12-dependent branch site pictogram (B) indicate the position of the residues in the P120 U12-dependent intron F. (C) Complementary base-pairing interactions between the U12-dependent branch site and the U12 snRNA anti-branch site. Numbered residues correspond to positions used in the mutational analysis. Adenosine 90 is the branch site residue used in this intron (Tarn and Steitz 1996).
The recognition of the branch site in U2-dependent introns is through a combination of interactions. The protein SF1/BBP binds to the branch site through recognition of the sequence surrounding the branch site adenosine (Berglund et al. 1997). The large subunit of the U2AF heterodimer binds to the polypyrimidine tract of higher eukaryotic 3′ splice sites and recruits U2 snRNP (Ruskin et al. 1988; Gaur et al. 1995) while the small subunit binds to the 3′ splice site AG adjacent to the polypyrimidine tract (Guth et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). Subsequently, U2 snRNA interacts with the branch site region by base-pairing interactions (Wu and Manley 1989; Zhuang and Weiner 1989). While these interactions are conserved in U2-dependent splicing in different organisms, the relative importance of the interactions differs. For example, the introns of budding yeast have a relatively weak polypyrimidine tract at their 3′ splice sites while having highly conserved branch site sequences (Burge et al. 1999; Lopez and Seraphin 1999). Perhaps as a result, the yeast U2AF analog, MUD2, is dispensable (Abovich et al. 1994). Mammalian U2-dependent introns tend to have strong polypyrimidine tracts and weak conservation of branch site sequences (Burge et al. 1999). Alteration of the mammalian branch site is well tolerated, with many sequences able to serve as branch sites over a significant range of distances from the 3′ splice site (Padgett et al. 1985; Ruskin et al. 1985; Gao et al. 2008).
U12-dependent introns have two notable features in their 3′ splice site regions. First, in all organisms in which they have been recognized, these introns share a highly conserved branch site sequence that is complementary to U12 snRNA. Second, they conspicuously lack a polypyrimidine tract between the branch site and the 3′ splice site. These features suggest that base pairing of U12 snRNA with the branch site sequence is of primary importance in U12-dependent introns while association between the polypyrimidine tract and U2AF is of primary importance in U2-dependent introns, at least in mammalian cells. The lack of a polypyrimidine tract and the variability seen in the sequence of the 3′ splice site suggest that neither subunit of U2AF is involved. Recent biochemical evidence also suggests that U2AF is not a required factor for U12-dependent splicing (Shen and Green 2007). However, U2AF is also thought to be a major target of recruitment by SR proteins bound to enhancer sequences (Wang et al. 1995). Since U12-dependent introns have been shown to respond to exonic enhancers (Dietrich et al. 2001b; Hastings and Krainer 2001) and adjacent U2-dependent splice sites (Wu and Krainer 1996), U2AF may still function through these interactions.
Since U12-dependent introns lack a polypyrimidine tract, the high conservation and restricted distance upstream to the 3′ splice site of the U12-dependent branch sequence (i.e., 10–25 nucleotides [nt] from the branch point adenosine) (Dietrich et al. 2001a) must provide the necessary signals for U12 snRNP recruitment and 3′ splice site selection. The base-pairing interactions between the U12-dependent branch sequence and the U12 snRNA anti-branch sequence (shown for the P120 U12 branch:U12 snRNA anti-branch in Fig. 1C) are thought to specify and help activate the specific adenosine residue that initiates the chemical steps of the splicing reaction.
To investigate the importance of the conserved nucleotide composition of the U12-dependent branch site sequence, we have made and tested mutations at each position of the P120 minigene intron F branch sequence. The U12-dependent intron F branch site sequence extends from position 83 to 91 with respect to the 5′ splice site and reads -UUCCUUAAC-. Mutations at positions 84 to 91 were analyzed for their effects on in vivo U12-dependent splicing. Mutations at most positions resulted in significant defects in U12-dependent splicing. The defects observed included an increase in the amount of unspliced RNA, the activation of cryptic U2-dependent splice sites, and the activation of cryptic U12-dependent branch/3′ splice sites. The effect of each mutation on the thermostability of the U12-dependent branch site:U12 snRNA anti-branch site interaction was determined by calculating the free energy of each duplex using the nearest-neighbor model (Serra and Turner 1995). A direct correlation was observed between branch:anti-branch thermostability and accurate splice site selection, providing evidence that the U12-dependent branch site primarily facilitates intron recognition via complementarity to the U12 snRNA anti-branch sequence.
RESULTS
Splicing patterns of the P120 minigene intron F
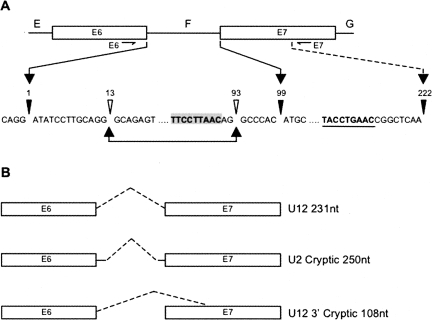
The experimental construct used in this investigation was a well-studied minigene expression construct derived from the human NOL1 (nucleolar protein P120) gene (Hall and Padgett 1996). Mutations were introduced at positions spanning the U12-dependent intron F branch site. Figure 2A shows the architecture of the region of the P120 minigene containing the U12-dependent intron F and diagrams the various splice sites that are utilized to generate the different products observed in the RT-PCR analysis. As shown in Figure 2, the PCR primers flank a region of the P120 construct that can produce three spliced isoforms. These spliced isoforms have been described previously and the spliceosomes required to produce them have been determined using in vitro methods (Dietrich et al. 1997, 2001b).
FIGURE 2.
Schematic of the U12-dependent P120 intron F diagramming the various spliced isoforms. (A) The indicated PCR primers flanking the splice sites anneal within the E6 and E7 exons surrounding intron F (the vector-specific RT primer is not indicated). The proper U12-dependent 5′ and 3′ splice sites are at positions 1 and 99, respectively. The correct U12-dependent branch site sequence is shaded, and the downstream cryptic U12-dependent branch site is underlined. The cryptic U2-dependent splice sites are at positions 13 and 93, and the cryptic U12-dependent 3′ splice site is at position 222. (B) Schematics of the three spliced isoforms observed by RT-PCR with indicated PCR product sizes after splicing.
Unspliced RNA yields a RT-PCR product of 330 nt. The spliced RNA that results from utilization of the correct U12-dependent intron F splice sites (i.e., positions 1 and 99, 1 being the first nucleotide of the intron) yields a RT-PCR product of 231 nt. The construct also contains cryptic U2-dependent splice sites inside the U12-dependent intron at positions 13 and 93 that have been characterized previously (Tarn and Steitz 1996; Dietrich et al. 1997). The spliced RNA that results from utilization of the cryptic U2-dependent splice sites (i.e., positions 13 and 93) yields an RT-PCR product of 250 nt. These cryptic U2-dependent splice sites are activated by several of the branch site mutants presented here. Similar U2-dependent cryptic activation events have been observed in U12-dependent 5′ and 3′ splice site mutants (Kolossova and Padgett 1997; Incorvaia and Padgett 1998). The third spliced isoform observed here is the result of the activation of a cryptic U12-dependent 3′ splice site and utilization of an alternative branch site sequence (Fig. 2A, underlined) contained within the downstream adjacent exon. Splicing from the normal U12-dependent 5′ splice site to this cryptic U12-dependent splice site (i.e., Fig. 2A, positions 1,222) yields a RT-PCR product of 108 nt.
In vivo splicing patterns of single mutations of the branch site
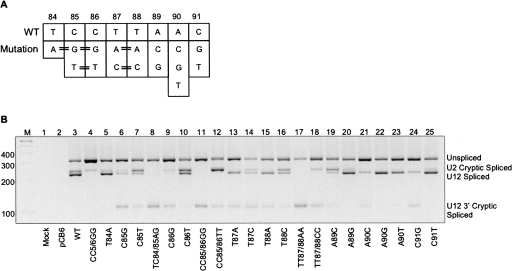
Normal U12-dependent splicing of P120 intron F utilizes the adenosine residue at position 90 as the branch site (Tarn and Steitz 1996; McConnell et al. 2002). This adenosine is embedded within a branch site consensus sequence spanning positions 83–91. The mutations introduced to the branch consensus sequence of U12-dependent intron F are presented in Figure 3A. Both single point mutations and double mutations of adjacent positions were tested. For each position normally containing a pyrimidine, a mutation was generated substituting it with a purine and vice versa. Mutations were also made at positions T84, C85, C86, A89, and C91 in which the consensus nucleotide was substituted with a nucleotide that occurs less frequently at that position (see Fig. 1B). In addition, alternate pyrimidines were tested at positions 85–88, all three nucleotide variants were investigated at the branch point adenosine (Fig. 1C, A90), and several double mutations were tested as indicated in Figure 3A.
FIGURE 3.
In vivo splicing by U12-dependent branch site mutants. (A) Diagram of constructed mutants at the indicated P120 intron F U12-dependent branch site positions. The wild-type P120 intron F branch site sequence is at the top and the nucleotide substitutions introduced at each position are indicated below. The double horizontal lines connecting adjacent positions indicate a double mutation. (B) RT-PCR analysis of in vivo RNA produced by each of the indicated P120 minigene constructs. The positions of unspliced, U2 cryptic spliced, U12 spliced, and U12 3′ cryptic spliced products are indicated to the right of the gel. (Lane M) 100 bp size marker; (lane 1) mock transfection control lane; (lane 2) CHO cells transfected with the host pCB6 vector lacking the P120 minigene; (lanes 3–25) CHO cells transfected with pCB6 constructs containing either wild-type sequence or the indicated mutations.
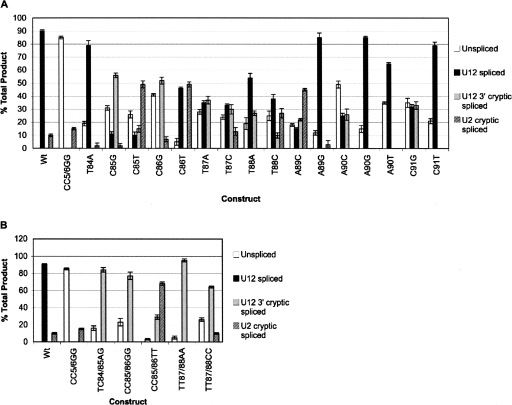
Each mutation was introduced separately into the P120 minigene plasmid and these were transfected individually into CHO cells. Total RNA was isolated from the cells 48 h after transfection and normalized for concentration prior to conducting RT-PCR. An agarose gel analysis of the spliced products is shown in Figure 3B, where the unspliced, U12 spliced, U2 cryptic spliced, and U12 3′ cryptic spliced products are visible. Band intensities of each product were determined from digitized images of gels, corrected for length and expressed as a percentage of total products (Fig. 4). In order to accurately compare results among three independent assays, the wild-type unspliced product for each was set to 0% and quantification of products in each subsequent lane was adjusted accordingly.
FIGURE 4.
Graphical representation of spliced isoforms quantified from RT-PCR analysis. Band intensities of each product (unspliced, U12 spliced, U12 3′ cryptic spliced, U2 cryptic spliced) were determined from digitized images of gels, corrected for length, and expressed as a percentage of total products for each indicated single mutant (A) and double mutant (B). Results reflect averages taken from three independent assays. Error bars represent one standard deviation from the mean.
The in vivo splicing pattern generated by each mutant varied based on position within the branch site sequence and the type of mutation, but some obvious trends were revealed when comparing mutants to the wild type (Fig. 3B, lane 3). A previously studied mutant containing the U12-dependent 5′ splice site CC5/6GG mutation is shown in lane 4 of Figure 3B. This mutant blocks U12-dependent splicing and activates the U2 cryptic splice sites, providing a marker for this product (Incorvaia and Padgett 1998).
Examination of the in vivo splicing phenotypes of the branch site mutations shows that substituting the central pyrimidines (CCTT) with purines resulted in a significant reduction in splicing to the normal 3′ splice site and activation of the U12 cryptic 3′ splice site (Fig. 3B, lanes 6,8,9,11,13,15,17). This result is consistent with the high conservation of these positions in U12-dependent branch sites (Fig. 1B). Mutation of T84 to A (Fig. 3B, lane 5) had only a modest effect on splicing efficiency and did not activate the downstream cryptic 3′ splice site.
Inspection of the consensus sequence shows that the A89 position adjacent to the branch point A90 can frequently be substituted by G. This is also observed in U2-dependent branch sites (Fig. 1A; Query et al. 1994). Consistent with this, the A89G mutation (Fig. 3B, lane 20) had only a small effect on U12 splicing. In contrast, mutation of A89 to a nonconsensus C residue (Fig. 3B, lane 19) effectively blocked U12-dependent splicing and activated the cryptic pathways. Mutation of the C91 residue on the other side of the A90 branch point showed a similar allele sensitivity (Fig. 3B, lanes 24,25). Mutation of C91 to T had a modest effect on U12 splicing while mutation to G inhibited normal U12 splicing and activated the U12 cryptic 3′ splice site.
The consensus data shows that the branch point residue is almost always an adenosine. However, data from both the U2-dependent system and the analysis of a rare case of a U12-dependent intron with a guanosine at this position shows that splicing can still occur either by branching to the G residue or to the immediate upstream A residue (Query et al. 1994; McConnell et al. 2002). Consistent with this, the A90G mutant (Fig. 3B, lane 22) showed only a modest reduction in splicing efficiency. The two pyrimidine substitutions, A90C and A90T (Fig. 3B, lanes 21,23), in contrast, significantly reduced normal splicing with the A90C allele being the most deleterious. Interestingly, all three A90 mutants showed increased levels of unspliced RNA relative to the wild type, suggesting that the cryptic pathways were somehow blocked in these mutants, perhaps due to the formation of dead end spliceosomal complexes involving the normal U12-dependent 5′ and 3′ splice site regions.
In vivo splicing patterns of double mutations of the branch site
The effects on splicing of the double mutations largely mirror those seen with the single mutations. The TC84/85AG mutation (Fig. 3B, lane 8) completely blocks normal U12-dependent splicing and leads to activation of the U12 cryptic 3′ splice site. This was the original mutation used to show the function of U12 snRNA in vivo through reactivation of normal splicing in the presence of a compensatory U12 snRNA mutant (Hall and Padgett 1996). Comparison of this double mutant phenotype to the individual T84A and C85G mutant phenotypes (Fig. 3B, lanes 5,6) shows that the bulk of the effect is due to the mutation at C85. A similarly dramatic phenotype is observed with the CC85/86GG (Fig. 3B, lane 11) and TT87/88AA (Fig. 3B, lane 17) mutations where almost all splicing is shifted to the U12 cryptic 3′ splice site.
The CC85/86TT mutation (Fig. 3B, lane 12) as well as the individual C85T (Fig. 3B, lane 7) and C86T (Fig. 3B, lane 10) mutations led to an activation of splicing at the U2-dependent splice sites. This result can be understood by noting that these mutations disrupt the U12 branch site consensus while retaining the pyrimidine tract. Thus, the mutant U12-dependent branch site now becomes the polypyrimidine tract for the U2-dependent 3′ splice site at position 93. Interestingly, a similar result is not seen in the case of the TT87/88CC mutation (Fig. 3B, lane 18). Here the U2-dependent cryptic splice sites are poorly activated while the amount of unspliced RNA and U12 3′ cryptic splicing is increased. This is consistent with data showing that clusters of T residues are more important for U2-dependent 3′ splice site activation than clusters of C residues (Roscigno et al. 1993).
Splicing defects are related to branch site:U12 snRNA duplex thermostability
The very different extents of branch site conservation seen between mammalian U2- and U12-dependent introns suggests that base-pairing interactions with U2 and U12 snRNAs, respectively, might differ significantly in their importance for splicing. While evidence shows that mammalian branch site:U2 snRNA pairing can be important for U2-dependent 3′ splice sites (Noble et al. 1988; Wu and Manley 1989; Zhuang et al. 1989), protein–RNA interactions appear to play a large if not predominant role (Valcarcel et al. 1996).
To gauge the role of base-pairing interactions in U12-dependent branch site specification, the thermodynamic stability of the U12-dependent branch site consensus sequence and U12 snRNA anti-branch sequence interaction was calculated using the Turner Table nearest-neighbor model (Serra and Turner 1995) for the normal branch site consensus sequence and each mutated branch site sequence. Using this method, the free energy of the normal U12-dependent branch site complex is calculated to be −4.1 kcal/mol. Free energy estimates were plotted versus the relative amount of correct U12 spliced, U12 3′ cryptic spliced, or U2 cryptic spliced products (Fig. 5A,B,C, respectively). Disruption of complementarity within the branch site:anti-branch site duplex decreased the thermostability of the complex, resulting in three possible outcomes: decreased correct U12 splicing, U2 cryptic splice site activation, and/or U12 cryptic 3′ splice site activation.
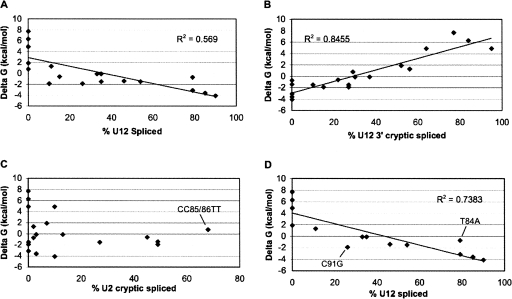
FIGURE 5.
Plots showing the relationship between U12-dependent splicing and U12-dependent branch:anti-branch stability. Free energy values were calculated using the Turner Table nearest-neighbor model (Serra and Turner 1995) to determine the stability of the U12-dependent branch site:U12 snRNA anti-branch interaction for the wild type U12 branch site sequence and each mutated branch site sequence. Free energy estimates were plotted versus the percentage of U12 spliced (A), U12 3′ cryptic spliced (B), U2 cryptic spliced (C), and U12 spliced excluding U2 activators (D). Lines are least-square best fits. R 2 values are as indicated.
Several trends can be observed in the plots shown in Figure 5. As the free energy of the branch site:U12 snRNA complex increased (delta G approaching zero), U12-dependent splicing at the correct splice sites and utilization of the normal branch site decreased (Fig. 5A) while splicing to the U12 cryptic 3′ splice site increased (Fig. 5B). The free energy of the downstream cryptic U12-dependent branch site complex has a delta G value of 1.9 kcal/mol. In most cases, mutations with a delta G >1.9 kcal/mol were associated with >50% cryptic splicing while mutations with a delta G <1.9 kcal/mol yielded >50% correct splicing (Fig. 5B).
As noted above, several mutations led to the activation of U2-dependent cryptic splicing. These mutations decreased the thermostability of the normal branch site complex and either maintained or increased the abundance of pyrimidines within the branch site. As shown in Figure 5C, this activation of the U2-dependent splicing system did not correlate with the stability of the branch site:U12 snRNA interaction. For example, the T88C mutation resulted in activation of U2 cryptic splicing whereas the T88A mutation, despite having a similar free energy, did not generate the U2 cryptic product (see Fig. 4A). In addition, the CC85/86TT double mutation activated U2-cryptic splicing (indicated in Fig. 5C) whereas the CC85/86GG double mutation did not.
Since the activation of the U2 cryptic splice sites was independent of the free energy of U12 snRNA binding, this splicing pathway would tend to mask a correlation between complex free energy and cryptic splice site usage. To test this idea, we excluded mutants that activated >25% U2 cryptic splicing from the analysis (Fig. 5D). This improved the correlation between complex free energy and splicing (Fig. 5, cf. A and D), but several outliers remained. These included mainly double mutants with high delta G values that block correct U12-dependent splicing and the two exceptional mutants T84A and C91G (indicated in Fig. 5D) that are discussed below.
DISCUSSION
The results described here extend our understanding of the role of the highly conserved branch site of U12-dependent introns. The lack of a polypyrimidine tract between the branch site and 3′ splice site of U12-dependent introns increases the importance of the U12-dependent branch site for ensuring intron recognition and accurate splice site selection. This study provides a thorough analysis of the U12-dependent branch site consensus sequence by investigating the effects on splicing in vivo that occur when mutations are introduced at each highly conserved nucleotide position.
Mutations at most positions resulted in splicing defects including reduction in intron F splicing, activation of cryptic U12-dependent branch/3′ splice sites, and activation of cryptic U2-dependent splice sites. Comparing the percent of each splice isoform (i.e., U12 spliced, U12 3′ cryptic spliced, U2 cryptic spliced) to the U12-dependent branch site:U12 snRNA anti-branch free energy value for each mutant revealed a direct correlation between branch:anti-branch thermostability and correct U12-dependent splicing with the exception of T84A and C91G. The surprisingly high level of correct U12-dependent splicing by T84A and low level by C91G reflects the frequency of these noncanonical bases at the respective positions (as seen in Fig. 2B) where an A can occur at the position corresponding to T84 but a G does not occur at C91. The downstream U12 cryptic branch site, which is used by several of the constructs studied here, also has an A at the position analogous to T84, providing additional evidence that an A at this position can effectively support splicing.
Mutants whose canonical U12 branch:anti-branch complex free energy value exceeded that of the downstream cryptic U12 branch:anti-branch complex (delta G=1.9 kcal/mol) typically resulted in the production of >50% U12-dependent 3′ cryptic spliced product (see Fig. 5B). The strong direct correlation between the stability of U12 base pairing at the wild-type branch site and the use of the U12 cryptic branch site seen in Figure 5B suggests that the competition between these two sites is largely governed by thermodynamics. While all the mutations studied here were at the wild-type branch site, a previous study in this system showed that use of the U12 3′ cryptic branch site could be activated by improving its base pairing to U12 snRNA. In this study by Dietrich et al. (2001a), when a “Cryptic-Up” mutation was created within the downstream U12 cryptic branch site at the analogous position to T84, converting it from an A to the consensus T, splicing to the 3′ cryptic site was increased from 0% to ∼30%. The Cryptic-Up mutant changed the delta G value of the U12 3′ cryptic branch:anti-branch complex from 1.9 to −1.5 kcal/mol. Figure 5B shows that when free energy values for the branch mutants were plotted versus the percent of splicing to the U12 3′ cryptic site, an increase in delta G from −1.5 to 1.9 kcal/mol resulted in an approximate 30% increase in U12 3′ cryptic spliced product, agreeing closely with the previous results. Collectively, these results emphasize the importance of the stability of the U12 branch:anti-branch complex and suggest that RNA/RNA interactions are the main determinants of U12-dependent intron recognition and proper splice site selection. The mutations that generated the U2-dependent cryptic product were generally those that disrupted the U12-dependent branch site consensus while retaining or improving the pyrimidine content of the region that leads to splicing by the U2-dependent pathway.
Despite mechanistic similarities between the U2- and U12-dependent splicing pathways, there are major differences in the initial stages of spliceosome assembly. The splicing of U2-dependent introns begins with interactions involving RNA–RNA base pairing between U1 snRNA and the 5′ splice site, and by interactions mediated by protein factors (Krämer 1996). The large subunit of U2AF contains two C-terminal RNA recognition motifs (RRMs) that make sequence-specific contacts with the U2-dependent intron polypyrimidine tract. A third U2AF C-terminal RRM interacts with the splicing factor SF1/BBP, enabling cooperative recognition of the polypyrimidine tract and branch site (Berglund et al. 1998). The small subunit of U2AF interacts with the 3′ splice site (Guth et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). During the formation of the initial commitment complex, communication across exons occurs (exon definition), where the binding of U1 snRNA at the adjacent downstream 5′ splice site enhances interaction between protein factors and the 3′ splice site (Kuo et al. 1991). Interaction between U2AF and the U2 snRNP then enables the U2 snRNP to replace SF1/BBP at the branch site where RNA/RNA interactions between the U2-dependent branch site and U2 snRNA anti-branch sequence occur, establishing the pre-spliceosome (for review, see Will and Lührmann 2006). The presence of a polypyrimidine tract and the poor conservation of the U2-dependent branch site sequence indicate that the recognition of U2-dependent introns is facilitated in large part by protein/RNA interactions. In contrast to the U2-dependent pathway, the results presented here reveal a direct correlation between U12 branch:anti-branch complementarity and accurate U12 splicing, suggesting that the recognition of U12-dependent introns is more reliant on RNA/RNA interactions. This is supported by the lack of a polypyrimidine tract and the high conservation of the U12-dependent branch site sequence.
The U2AF protein does not appear to play a direct role in U12-dependent intron recognition. In addition to the RRMs of U2AF that are responsible for interaction with the U2-type intron polypyrimidine tract, U2AF has an arginine/serine-rich (RS) domain that contacts the U2-dependent branch site in the commitment complex (Gaur et al. 1995; Valcarcel et al. 1996; Shen and Green 2004). A recent study demonstrating U12-dependent splicing in U2AF-depleted nuclear extract suggests that the U2AF RS domain does not make functional interactions with the U12-dependent branch site during spliceosome assembly (Shen and Green 2007). Thus, U12-type introns are spliced in a U2AF-independent manner, consistent with the absence of a polypyrimidine tract.
The results here provide evidence that when the stability of the U12 branch:anti-branch complex is disrupted while retaining or increasing pyrimidines, the U2 system can “hijack” splicing if splice sites recognized by the U2-type spliceosome are available. This might be facilitated by U2AF successfully competing with the U12 snRNP due to a decrease in affinity of the U12 snRNA anti-branch for a compromised U12 branch site consensus sequence. However, the reported detection of U2AF in Neurospora whose introns lack the canonical polypyrimidine tract (Henscheid et al. 2008) and a possible role in interaction with exonic splicing enhancers (Wang et al. 1995; Zuo and Maniatis 1996; Guth et al. 2001) suggest that U2AF may still have some level of involvement in U12-dependent splicing.
Protein factors might also be involved in directing the utilization of proper splice sites when cryptic sites are present as with the U12-dependent intron used here, which contains internal U2-dependent splice sites. A similar architecture is contained within the prospero gene of Drosophia melanogaster, where the choice of U2- or U12-dependent splicing appears to be regulated by protein factors that bind to an intronic purine-rich element (Scamborova et al. 2004). In addition, exon-spanning interactions between factors bound to the splice sites of adjacent introns may play a key role in defining the exon–intron boundaries during U12-dependent splicing. Evidence that protein factors facilitate cooperation between U2- and U12-dependent splicing was provided by the observation that the splicing of a U12-type intron could be stimulated when immediately upstream of a U2-dependent intron (Wu and Krainer 1996). Thus, as with U2-dependent splicing, protein factor interactions with both intronic and exonic elements appear to contribute to U12-dependent exon definition and splice-site choice.
This study and others like it are necessary to increase our understanding of how and why U12-dependent splicing exists. It has been suggested that the scarcity of U12-type introns in modern organisms might reflect their less efficient splicing and the tendency to convert to the more loosely defined U2-type splice/branch sites via mutational changes during evolution (Burge et al. 1998). Results here demonstrate that point mutations within the U12 branch site can activate alternative splice sites, suggesting that marginal disruptions in important RNA/RNA interactions can severely influence U12-dependent splicing. The importance of the stringency of U12-dependent splicing signals is illustrated by the U12-type intron from the tumor suppressor gene LKB1. A 5′ splice site mutation in this gene is responsible for the autosomal dominant disorder Peutz-Jeghers syndrome associated with gastrointestinal polyposis and an increased cancer risk (Hastings et al. 2005).
In addition to their prevalence in several housekeeping genes, U12-dependent introns are also found in genes involved in specific cellular processes (e.g., Ras-Raf signaling pathway) where they might function in regulating the expression of their host genes (Chang et al. 2007). Our results reveal that the U12-dependent branch site consensus sequence is a key determinant of intron recognition and accurate splicing. Furthermore, the U12-dependent branch site:U12 snRNA anti-branch site interaction appears to provide the necessary competitive force enabling U12-type introns to persist alongside the highly abundant U2-dependent splicing system.
MATERIALS AND METHODS
DNA constructs
The construction of the P120 minigene and the intron F mutants derived from it are as described (Kolossova and Padgett 1997). Mutations used in this study were introduced by PCR methods using pairs of oligonucleotides containing the desired mutations. All mutations were confirmed by DNA sequencing.
Analysis of in vivo splicing
Transient transfection of the P120 minigene expression plasmid into cultured CHO cells was done as described (Hall and Padgett 1996; Kolossova and Padgett 1997; Incorvaia and Padgett 1998). For these experiments, 1 μg of P120 plasmid and 9 μg of pUC19 carrier DNA were added to 1 × 106 cells. Total RNA was isolated from cells 48 h after transfection using a Roche High Pure RNA Isolation Kit and reverse transcribed using a vector-specific primer. Primary PCR amplification was performed using a P120 exon 5/6 forward primer, followed by a secondary PCR amplification using P120 exon 6 and 7 specific primers as described (Kolossova and Padgett 1997; Dietrich et al. 2001b). The products were analyzed by agarose gel electrophoresis, visualized using ethidium bromide, and digitally photographed using a UV transilluminator (BioDoc-It Imaging System). Independent transfections and analyses gave results that differed by <10%. The observed ratios of the various RNA precursors and products were insensitive to PCR cycle number or RNA input.
Quantification and thermodynamic calculation
Splice isoforms were quantified from the digital images using the Java image processing program ImageJ. Stacking energies for the branch:anti-branch duplex were calculated using Turner tables version 2.3 free energy parameters for RNA folding at 37°: http://www.bioinfo.rpi.edu/∼zukerm/cgi-bin/efiles.cgi?T=37.
ACKNOWLEDGMENTS
We thank Robert Incorvaia, Andrew Seyboldt, and John Fuller for construction of mutants, and Girish Shukla and Kwaku Dayie for helpful suggestions. This work was supported by grant GM079527 from the National Institutes of Health.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1189008.
REFERENCES
- Abovich N., Liao X.C., Rosbash M. The yeast MUD2 protein: An interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes & Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- Berglund J.A., Chua K., Abovich A., Reed R., Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- Berglund J.A., Abovitch N., Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes & Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C.B., Padgett R.A., Sharp P.A. Evolutionary fates and origins of U12-type introns. Mol. Cell. 1998;2:773–785. doi: 10.1016/s1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Tuschl T., Sharp P.A. Splicing of precursors to mRNAs by the spliceosomes. In: Gestland R.F., et al., editors. The RNA world. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. pp. 525–560. [Google Scholar]
- Chang W.C., Chen Y.C., Lee K.M., Tarn W.Y. Alternative splicing and bioinformatic analysis of human U12-type introns. Nucleic Acids Res. 2007;35:1833–1841. doi: 10.1093/nar/gkm026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich R.C., Incorvaia R., Padgett R.A. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol. Cell. 1997;1:151–160. doi: 10.1016/s1097-2765(00)80016-7. [DOI] [PubMed] [Google Scholar]
- Dietrich R.C., Peris M.J., Seyboldt A.S., Padgett R.A. Role of the 3′ splice site in U12-dependent intron splicing. Mol. Cell. Biol. 2001a;21:1942–1952. doi: 10.1128/MCB.21.6.1942-1952.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich R.C., Shukla G.C., Fuller J.D., Padgett R.A. Alternative splicing of U12-dependent introns in vivo responds to purine-rich enhancers. RNA. 2001b;7:1378–1388. [PMC free article] [PubMed] [Google Scholar]
- Dietrich R.C., Fuller J.D., Padgett R.A. A mutational analysis of U12-dependent splice site dinucleotides. RNA. 2005;11:1430–1440. doi: 10.1261/rna.7206305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frilander M.J., Steitz J.A. Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes & Dev. 1999;13:851–863. doi: 10.1101/gad.13.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K., Masuda A., Matsuura T., Ohno K. Human branch point consensus sequence is yUnAy. Nucleic Acids Res. 2008;36:2257–2267. doi: 10.1093/nar/gkn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur R.K., Valcarcel J., Green M.R. Sequential recognition of the pre-mRNA branch point by U2AF65 and a novel spliceosome-associated 28-kDa protein. RNA. 1995;1:407–417. [PMC free article] [PubMed] [Google Scholar]
- Guth S., Martinez C., Gaur R.K., Valcarcel J. Evidence for substrate-specific requirement of the splicing factor U2AF35 and for its function after polypyrimidine tract recognition by U2AF65. Mol. Cell. Biol. 1999;19:8263–8271. doi: 10.1128/mcb.19.12.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth S., Tange T.O., Kellenberger E., Valcarcel J. Dual function for U2AF(35) in AG-dependent pre-mRNA splicing. Mol. Cell. Biol. 2001;21:7673–7681. doi: 10.1128/MCB.21.22.7673-7681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S.L., Padgett R.A. Requirement of U12 snRNA for the in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science. 1996;271:1716–1718. doi: 10.1126/science.271.5256.1716. [DOI] [PubMed] [Google Scholar]
- Hastings M.L., Krainer A.R. Functions of SR proteins in the U12-dependent AT-AC pre-mRNA splicing pathway. RNA. 2001;7:471–482. doi: 10.1017/s1355838201002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings M.L., Resta N., Traum D., Stella A., Guanti G., Krainer A.R. An LKB1 AT-AC intron mutation causes Peutz-Jeghers syndrome via splicing at noncanonical cryptic splice sites. Nat. Struct. Mol. Biol. 2005;12:54–59. doi: 10.1038/nsmb873. [DOI] [PubMed] [Google Scholar]
- Henscheid K.L., Voelker R.B., Berglund J.A. Alternative modes of binding by U2AF65 at the polypyrimidine tract. Biochemistry. 2008;47:449–459. doi: 10.1021/bi701240t. [DOI] [PubMed] [Google Scholar]
- Incorvaia R., Padgett R.A. Base pairing with U6atac snRNA is required for 5′ splice site activation of U12-dependent introns in vivo. RNA. 1998;4:709–718. doi: 10.1017/s1355838298980207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolossova I., Padgett R.A. U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) introns. RNA. 1997;3:227–233. [PMC free article] [PubMed] [Google Scholar]
- Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Kuo H.-C., Nasim F.H., Grabowski P.J. Contol of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991;251:1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- Levine A., Durbin R. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res. 2001;29:4006–4013. doi: 10.1093/nar/29.19.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez P.J., Seraphin B. Genomic-scale quantitative analysis of yeast pre-mRNA splicing: Implications for splice-site recognition. RNA. 1999;5:1135–1137. doi: 10.1017/s135583829999091x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell T.S., Cho S.J., Frilander M.J., Steitz J.A. Branchpoint selection in the splicing of U12-dependent introns in vitro. RNA. 2002;8:579–586. doi: 10.1017/s1355838202028029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montzka Wassarman K., Steitz J.A. The low-abundance U11 and U12 small nuclear ribonucleoproteins (snRNPs) interact to form a two-snRNP complex. Mol. Cell. Biol. 1992;12:1276–1285. doi: 10.1128/mcb.12.3.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T.W. RNA–RNA interactions in nuclear pre-mRNA splicing. In: Simons R.W., Grunberg-Manago M., editors. RNA structure and function. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1998. pp. 279–307. [Google Scholar]
- Noble J.C.S., Prives C., Manley J.L. Alternative splicing of SV40 early pre-mRNA is determined by branch site selection. Genes & Dev. 1988;2:1460–1475. doi: 10.1101/gad.2.11.1460. [DOI] [PubMed] [Google Scholar]
- Padgett R.A., Burge C.B. Splice sites. In: Cooper D., editor. Encyclopedia of the human genome. Wiley; New York: 2003. pp. 1134–1145. [Google Scholar]
- Padgett R.A., Konarska M.M., Aebi M., Hornig H., Weissmann C., Sharp P.A. Nonconsensus branch-site sequences in the in vitro splicing of transcripts of mutant rabbit β-globin genes. Proc. Natl. Acad. Sci. 1985;82:8349–8353. doi: 10.1073/pnas.82.24.8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query C.C., Moore M.J., Sharp P.A. Branch nucleophile selection in pre-mRNA splicing: Evidence for the bulged duplex model. Genes & Dev. 1994;8:587–597. doi: 10.1101/gad.8.5.587. [DOI] [PubMed] [Google Scholar]
- Roscigno R.F., Weiner M., Garcia-Blanco M.A. A mutational analysis of the polypyrimidine tract of introns. Effects of sequence differences in pyrimidine tracts on splicing. J. Biol. Chem. 1993;268:11222–11229. [PubMed] [Google Scholar]
- Ruskin B., Green J.M., Green M.R. Cryptic branch point activation allows accurate in vitro splicing of human β-globin intron mutants. Cell. 1985;41:833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Zamore P.D., Green M.R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Scamborova P., Wong A., Steitz J.A. An intronic enhancer regulates splicing of the twintron of Drosophila melanogaster prospero pre-mRNA by two different spliceosomes. Mol. Cell. Biol. 2004;24:1855–1869. doi: 10.1128/MCB.24.5.1855-1869.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra M.J., Turner D.H. Predicting thermodynamic properties of RNA. Methods Enzymol. 1995;259:242–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- Shen H., Green M.R. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Shen H., Green M.R. RS domain-splicing signal interactions in splicing of U12-type and U2-type introns. Nat. Struct. Mol. Biol. 2007;14:597–603. doi: 10.1038/nsmb1263. [DOI] [PubMed] [Google Scholar]
- Tarn W.-Y., Steitz J.A. A novel spliceosome containing U11, U12 and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- Valcarcel J., Gaur R.K., Singh R., Green M.R. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- Wang Z., Hoffmann H.M., Grabowski P.J. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol. Chem. 2005;386:713–724. doi: 10.1515/BC.2005.084. [DOI] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. Spliceosome structure and function. In: Gestland R.F., et al., editors. The RNA world. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 369–400. [Google Scholar]
- Wu Q., Krainer A.R. U1-mediated exon definition interactions between AT-AC and GT-AG introns. Science. 1996;274:1005–1008. doi: 10.1126/science.274.5289.1005. [DOI] [PubMed] [Google Scholar]
- Wu J., Manley J.L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes & Dev. 1989;3:1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Wu S., Romfo C.M., Nilsen T.W., Green M.R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A.M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes & Dev. 1989;3:1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Goldstein A.M., Weiner A.M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc. Natl. Acad. Sci. 1989;86:2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio D.A.R., Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans . Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]
- Zuo P., Maniatis T. The splicing factor U2AF35 mediates critical protein–protein interactions in constitutive and enhancer-dependent splicing. Genes & Dev. 1996;10:1356–1358. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]