Abstract
Coupled expression of the M1 and BM2 open-reading frames (ORFs) of influenza B from the dicistronic segment 7 mRNA occurs by a process of termination-dependent reinitiation. The AUG start codon of the BM2 ORF overlaps the stop codon of the upstream M1 ORF in the pentanucleotide UAAUG, and BM2 synthesis is dependent upon translation of the M1 ORF and termination at the stop codon. Here, we have investigated the mRNA sequence requirements for BM2 expression. Termination–reinitiation is dependent upon 45 nucleotide (nt) of RNA immediately upstream of the UAAUG pentanucleotide, which includes an essential stretch complementary to 18S rRNA helix 26. Thus, similar to the caliciviruses, base-pairing between mRNA and rRNA is likely to play a role in tethering the 40S subunit to the mRNA following termination at the M1 stop codon. Consistent with this, repositioning of the M1 stop codon more than 24 nt downstream from the BM2 start codon inhibited BM2 expression. RNA structure probing revealed that the RNA upstream of the UAAUG overlap is not highly structured, but upon encountering the M1 stop codon by the ribosome, a stem–loop may form immediately 5′ of the ribosome, with the 18S rRNA complementary region in the apical loop and in close proximity to helix 26. Mutational analysis reveals that the normal requirements for start site selection in BM2 expression are suspended, with little effect of initiation codon context and efficient use of noncanonical initiation codons. This suggests that the full complement of initiation factors is not required for the reinitiation process.
Keywords: ribosome, translation, influenza B virus, termination–reinitiation, 18S rRNA, RNA structure
INTRODUCTION
The expression of viral proteins is frequently subject to regulation at the level of protein synthesis (Gale et al. 2000). The dominant target of such control is the initiation step, but regulation can also occur at the elongation and termination steps. For those eukaryotic viruses that express polycistronic mRNAs, an additional requirement is to facilitate access of ribosomes to downstream open-reading frames (ORFs). A variety of mechanisms have evolved to achieve this at the translational level, including leaky scanning of 40S subunits past the start codon of the first ORF (Pavlakis and Felber 1990), the possession of intercistronic internal ribosome entry signals (Wilson et al. 2000), programmed ribosomal frameshifting during elongation (Baranov et al. 2006; Brierley and Dos Ramos 2006), and stop codon suppression at the termination step (Bertram et al. 2001; Dreher and Miller 2006). These mechanisms also afford a level of control over the relative quantities of the proteins encoded by upstream and downstream ORFs. Another strategy that has evolved to allow regulated expression of a downstream ORF is termination–reinitiation (also referred to here as “stop–start”). Under certain circumstances, ribosomes are able to translate an upstream ORF, but 40S subunits may remain tethered following termination and go on to reinitiate at the start codon of a downstream ORF. In most cellular examples of this phenomenon, efficient termination–reinitiation is dependent upon the first ORF being short (Luukkonen et al. 1995; Kozak 2001), probably because the ribosome does not retain initiation factors for long after elongation begins (Pöyry et al. 2004). The process is best exemplified in the regulated expression of GCN4 in yeast, whose translation is repressed by the presence of four short ORFs (uORFs) in its 5′ UTR (Hinnebusch 2005). Similar regulatory elements are present in mRNAs of higher eukaryotes and are often associated with repression of expression of oncogenes and proteins that are only synthesized under stress conditions (Harding et al. 2000). Another feature of these systems is that the uORF translation product does not encode a functional protein.
Viral termination–reinitiation was first described in the synthesis of the BM2 protein of influenza B (Horvath et al. 1990) and has since been documented in the expression of the M2-2 protein of respiratory syncytial virus (RSV; Ahmadian et al. 2000; Gould and Easton 2005) the M2-2 protein of pneumovirus of mice (PVM; Gould and Easton 2007), the VP2 protein of feline calicivirus (FCV), and the VP10 protein of the calicivirus rabbit hemorrhagic disease virus (RHDV; Meyers 2003, 2007; Luttermann and Meyers 2007). The viral examples are distinct from cellular examples of reinitiation in that the upstream ORFs are comparatively long and encode functional proteins. The mechanism by which the ribosome is able to initiate on the downstream ORF is not fully understood and models must take into account the fact that the ribosome is likely to have lost some or all of the initiation factors that one would expect to be required for reinitiation (Pöyry et al. 2004). The process is clearly distinct from ribosomal frameshifting and termination codon suppression in that the two ORFs are not translationally fused, and termination of the first ORF is known to be an absolute requirement for synthesis of the downstream protein (Horvath et al. 1990; Ahmadian et al. 2000; Meyers 2003, 2007; Luttermann and Meyers 2007; Pöyry et al. 2007). Recent work has shown that termination–reinitiation is dependent on sequences upstream of the closely spaced termination and initiation codons (the stop–start window; Meyers 2003, 2007; Gould and Easton 2005; Luttermann and Meyers 2007) and that it is the primary sequence of the RNA and not the protein encoded by the first ORF that is important (Meyers 2003). This region of the mRNA is almost certainly involved in the retention of post-termination 40S subunits, and there is evidence that it is involved in the recruitment of eukaryotic initiation factor 3 (eIF3) and eIF3/40S complexes (Pöyry et al. 2007). It has also been suggested that a sequence within this region that is complementary to part of helix 26 of 18S rRNA may act to tether the 40S ribosomal subunit to the mRNA post-termination, allowing time for the ribosome to acquire the factors necessary to initiate on the downstream ORF (Luttermann and Meyers 2007; Meyers 2007). However, the region upstream of the putative stop–start window in RSV does not appear to contain sequences complementary to 18S rRNA, and so this mechanism may not apply to all viruses that employ termination–reinitiation to regulate the expression of viral proteins.
The influenza A, B, and C viruses are a group of enveloped negative-strand RNA viruses with a segmented genome. The M1 (encoding matrix protein 1) and BM2 ORFs are located on segment 7 of the influenza B genome (Briedis et al. 1982; Hiebert et al. 1986; Horvath et al. 1990), with the termination codon of M1 in close proximity to the start codon of BM2 (UAA UG; stop codon of M1 in bold, start codon of BM2 underlined) consistent with the proposed expression of BM2 by termination–reinitiation (Horvath et al. 1990). The BM2 protein is a functional homolog of the M2 protein of influenza A and encodes a small proton channel (Mould et al. 2003), which is thought to be involved in RNP acidification and consequent M1 release during entry. The relative levels of BM2 expression are important to virus viability, as reduction of its synthesis (to one-eighth of the wild-type level) results in decreased levels of infectivity (Jackson et al. 2004). Termination-dependent reinitiation is thus likely to be highly important for influenza B virus viability, although little is known of the mechanism.
In this report we describe an analysis of the mRNA signals involved in the expression of BM2. We confirm that the BM2 polypeptide is expressed by termination–reinitiation and provide evidence consistent with a functional interaction between the coding region of the M1 mRNA and 18S rRNA. The potential involvement of mRNA secondary structure in the process is also investigated. Overall, our data suggest that the mechanism of BM2 expression is similar to that of the caliciviruses, and this is the first example of such a mechanism operating outside of the positive-stranded RNA viruses.
RESULTS
The influenza BM2 protein is translated via termination-dependent reinitiation
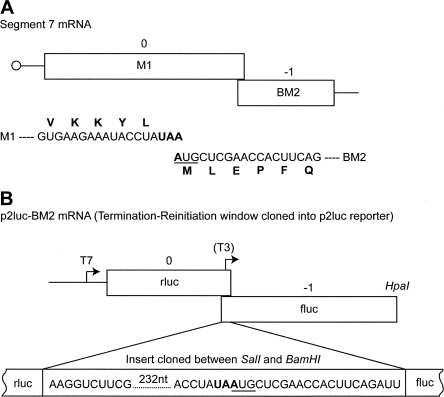
To investigate termination–reinitiation in the synthesis of the influenza B virus BM2 protein, a 270-base-pair (bp) fragment of segment 7 cDNA was cloned between the SalI and BamHI sites of the dual-luciferase reporter vector p2luc (Grentzmann et al. 1998). The cloned fragment, which contained 250 bp of sequence information upstream of the UAA UG stop–start window and some 20 bp downstream, was suspected, on the basis of work with other viruses (see the Introduction), to contain all of the required sequences for termination–reinitiation. The cDNA fragment was cloned in such a way that the Renilla and Firefly luciferase ORFs were in frame with the stop and start codons, respectively, of the termination–reinitiation motif to give an ORF configuration 5′ rlucM1-BM2fluc 3′ (Fig. 1). This vector, named p2luc-BM2wt, contains a T7 RNA polymerase promoter allowing synthetic mRNAs to be generated to investigate the stop–start process in in vitro translation reactions.
FIGURE 1.
Structure of influenza B segment 7 and p2luc-BM2wt mRNAs. (A) Schematic of the segment 7 mRNA showing the encoded ORFs M1 and BM2 and, below, the mRNA and amino acid sequence of the overlap region. The stop codon of M1 is highlighted in bold and the start codon of BM2 is underlined. (B) Schematic of the p2luc-BM2 reporter construct. The termination–reinitiation cassette (250 nt upstream of and 20 nt downstream from the UAA UG motif) was cloned into the SalI and BamHI sites of the p2luc reporter plasmid. HpaI run-off transcripts for in vitro translation were generated using T7 RNA polymerase. The position of the T3 promoter present in the structure mapping construct p2luc-BM2-T3 is indicated.
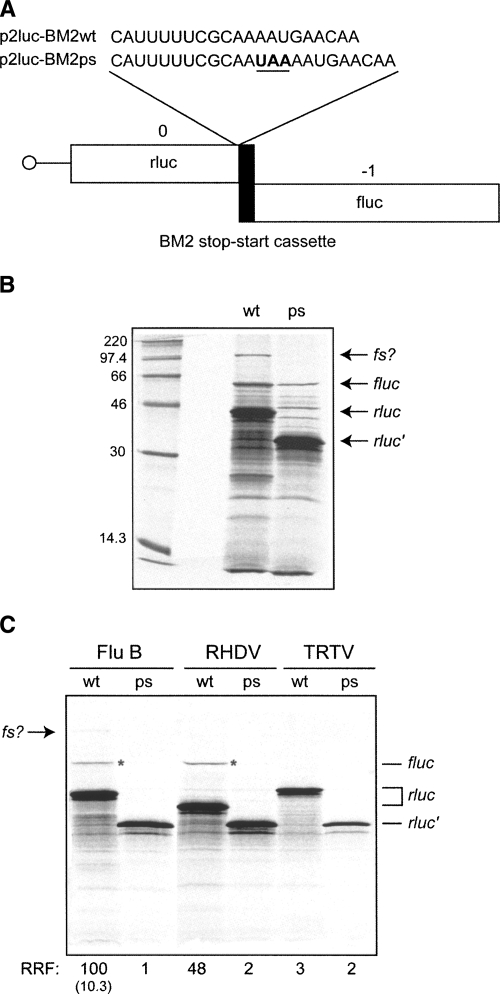
Termination–reinitiation is distinct from IRES-mediated expression of downstream ORFs, as translation through the upstream ORF is an absolute requirement (Meyers 2003; Gould and Easton 2005; Luttermann and Meyers 2007). In order to establish whether this is also the case for BM2 expression, a premature in-frame stop codon was inserted close to the end of the rluc ORF but upstream of M1 sequence information (267 bp upstream of the authentic rlucM1 termination codon) (Fig. 2A). If the expression of BM2fluc is a result of termination–reinitiation, translating ribosomes would be unable to reach the AUG start codon of BM2fluc in the mutant mRNA and the ORF could not be translated. The translation of in vitro synthesized wild-type (wt) and premature stop (ps) mutant mRNAs from p2luc-BM2wt and p2luc-BM2ps in standard rabbit reticulocyte lysate gave products of the expected sizes (rlucM1 ORF, ∼44 kDa; rlucM1ps ORF, 33 kDa) (see Fig. 2B). The product of the downstream ORF (BM2fluc) was also seen, migrating at ∼62 kDa as expected. That this was indeed the product of the second ORF was further confirmed by comparing the migration of RRL translation products from mRNAs derived from p2luc-BM2 that had been linearized at different points within the second ORF (data not shown). As is clear from Figure 2, the introduction of a premature stop codon into the rluc/M1 ORF reduced, but did not abolish, expression of the BM2/fluc product. A likely explanation for this observation is that in addition to termination–reinitiation, some of the BM2/fluc expression may arise by other mechanisms, perhaps leaky scanning or via internal initiation.
FIGURE 2.
Expression of BM2 is by a mechanism consistent with termination–reinitiation. (A) The primary sequence of the rluc/BM2 junction in the BM2wt and BM2ps mRNAs is given, showing the premature stop (ps) codon (UAA) upstream of the BM2 sequence in p2luc-BM2ps. The ps mutation was also introduced into the p2luc-RHDV and p2luc-TRTV reporter plasmids. (B) Wild-type (wt) and ps mRNAs were synthesized from HpaI-linearized p2lucBM2wt and p2lucBM2ps reporter plasmids, and the transcripts translated in nuclease-treated RRL (Promega) at a final RNA concentration of 50 μg/mL. Products were labeled with [35S]-methionine, separated by 12% SDS-PAGE, and detected by autoradiography. The product of the full-length rluc ORF (predicted size 44 kDa, marked rluc), the truncated rluc ORF (in the ps mutant, predicted size 33 kDa, marked rluc′), and the BM2 fluc product (predicted size, 62 kDa, marked fluc) are indicated with arrows. The putative frameshift product (fs?) is also arrowed. (C) p2luc-BM2wt/ps, p2luc-RHDVwt/ps, and p2luc-TRTVwt/ps reporter constructs were linearized with HpaI and run-off transcripts translated in Flexi RRL, supplemented with 140 mM KCl. Products were resolved by 12% SDS-PAGE and autoradiographed. The rluc product is shorter in the p2lucRHDVwt translations (expected size, 37 kDa, as compared to 44 kDa for the other two mRNAs) as the insert contains only ∼80 nt upstream of the stop–start window. The rluc products, generated as a result of termination–reinitiation, are marked with an asterisk. RRF denotes the relative reinitiation frequency in comparison to wt (set at 100). The figure in brackets represents the ratio of the intensity of the fluc and rluc products (adjusted for methionine content and expressed as a percentage) for the wt mRNA.
It is known that the proportion of an authentic translation product expressed from a monocistronic mRNA can be increased by the addition of KCl to the RRL (Dasso and Jackson 1989; Jackson 1991). To ascertain whether this would lead to increased fidelity of translation of the p2luc-BM2wt and p2luc-BM2ps mRNAs, translations were performed in the Flexi RRL system (Promega) in the presence of increasing concentrations of KCl (40 mM–200 mM added KCl; the endogenous KCl concentration of Flexi RRL was unknown but likely to be in the range of 25–40 mM; Jackson and Hunt 1983). Over this range, except at high added KCl (160–200 mM), the yield of the upstream ORF from either RNA was unaffected, but the expression of the downstream ORF from p2luc-BM2ps was greatly reduced above 130 mM added KCl (data not shown). An example of such an experiment is shown in Figure 2C, where translations were carried out in the presence of 140 mM added KCl. Translation of the p2luc-BM2wt RNA gave the expected products of ∼44 kDa (rlucM1) and ∼62 kDa correspondent with the BM2fluc product (Fig. 2C). Importantly, no BM2fluc product was observed with the mRNA containing the premature stop codon, indicating that the increased KCl concentration had suppressed the aberrant translation product. These data are thus consistent with a termination–reinitiation strategy for the expression of the BM2 protein and confirm a requirement for translation through the upstream ORF. The molar ratio of BM2fluc to rlucM1 (taking into account the methionine content of the two proteins) was typically in the region of 1:10. Thus, initiation on the downstream ORF occurred at a frequency of ∼10% of that of the upstream ORF. As controls, additional reactions were performed with wild-type and premature stop variants of p2luc reporters harboring the RHDV stop–start signal, of which VP10 is known to be expressed by termination–reinitiation (Meyers 2003, 2007), and an equivalent region of the metapneumovirus turkey rhinotracheitis virus (TRTV), whose putative M2-2 ORF was not detected in vitro (Yu et al. 1992). As can be seen in Figure 2C, a termination–reinitiation product was evident in the translation of the wild-type RHDV reporter construct and not in the case of the premature stop mutant. No termination–reinitiation product at all was observed in translations of the TRTV reporter constructs, in agreement with the view that at least in vitro, a stop–start mechanism is not employed by this virus (Yu et al. 1992). In the translation of the influenza B reporter mRNA, but not the RHDV or TRTV reporter mRNAs (Fig. 2B,C), we observed an additional species of lower abundance migrating more slowly than BM2fluc. The size of this protein is suggestive of a fusion protein of the first and second ORFs, perhaps arising from a ribosomal frameshift event. This is discussed below.
Expression of BM2 is dependent on ∼42–45 nt upstream of the UAAUG motif
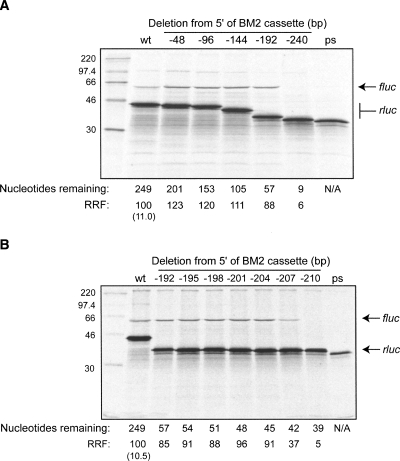
Previous work has suggested that viral termination–reinitiation events show little dependence on sequence information downstream from the stop–start window but require 69–250 nt of upstream primary sequence (Meyers 2003; Gould and Easton 2005; Luttermann and Meyers 2007). In order to determine the minimal sequence requirements for termination–reinitiation in BM2 expresssion, deletions of increasing size in 48-nt increments were made from the 5′ end of the inserted viral information (Fig. 3A). The rate of termination–reinitiation remained constant until the deletion went beyond 192 nt (p2luc-BM2-192; 57 nt remaining upstream of the stop–start window), when termination–reinitiation was abolished (Fig. 3A).
FIGURE 3.
Deletion analysis of influenza BM2 termination–reinitiation. (A) A series of p2luc-BM2 variants were prepared with stepwise, in-frame, 48-nt deletions from the 5′ end of the inserted viral sequence (BM2 cassette). The mutant plasmids were linearized with HpaI and run-off transcripts translated in Flexi RRL in the presence of [35S]-methionine and 130 mM added KCl. The products were resolved by 12% SDS-PAGE and visualized by autoradiography. The numbers above the figure correspond to the number of nucleotides deleted from the 5′ end of the insert sequence; the first set of numbers below the figure describe the number of nucleotides of viral sequence remaining up to the AUG start codon of the BM2 ORF. The ps lane is the premature stop codon control (see legend to Fig. 2). (B) Further deletion mutants were generated with stepwise 3-nt deletions using the p2luc-BM2-192 construct as a template and tested as in panel A.
In order to map more precisely the minimal sequence requirement for termination–reinitiation, the p2luc-BM2-192 plasmid was used as a template to make stepwise 3-nt deletions from the 5′ end. The stop–start product was synthesized efficiently with up to 45 nt of M1 information present upstream of the UAA UG motif, and to a lesser extent with 42 nt. However, deletion to 39 nt abolished expression of the stop–start product (Fig. 3B). These data indicate that only 42 nt of M1 primary sequence immediately upstream of the stop–start window are required for termination–reinitiation in vitro, although 45 nt are required for full activity.
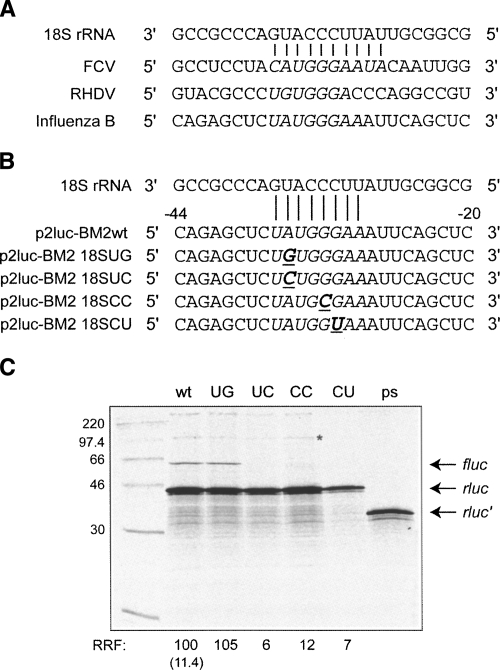
Termination–reinitiation of BM2 synthesis is dependent upon an mRNA sequence with complementarity to 18S rRNA
Two members of the calicivirus family (RHDV and FCV) that demonstrate termination–reinitiation contain primary sequence elements complementary to a region of helix 26 of 18S rRNA (Luttermann and Meyers 2007; Meyers 2007). In each case, the 5′ end of the complementary element lies 60–70 nt upstream of the stop codon of the upstream ORF, and mutational analysis has revealed that this sequence is essential for the stop–start process (Luttermann and Meyers 2007; Meyers 2007; Pöyry et al. 2007). Within the ∼45-nt minimal region of the M1 RNA required for BM2 expression, a stretch of bases with a similar level of complementarity to 18S rRNA is also found (Fig. 4A, complementary bases are shown in italics), albeit this region is much closer to the stop–start window (∼35 nt upstream of the stop codon of rlucM1) than the equivalent region of FCV or RHDV (∼60 nt). To investigate whether this region plays a role in termination–reinitiation in BM2 expression, a number of point mutations were made to disrupt potential mRNA:rRNA pairs. First, the A at −35 was mutated either to a G (18SUG, creating a presumably slightly weaker putative U-G base pair between the rRNA and mRNA) or a C (18SUC, which would be expected to disrupt the base pair between rRNA and mRNA). In separate mutations, the G at −32 was changed to a C (18SCC), and the G at −31 to a U (18SCU), both of which would act to disrupt the interaction between 18S rRNA and mRNA (Fig. 4B). With one exception, all of the mutations completely ablated expression of the BM2fluc product, supporting the idea that an interaction between the 18S rRNA and the mRNA just upstream of the termination–reinitiation site is required for stop–start (Fig. 4C). Importantly, in the one mutant where pairing was predicted to be maintained (18SUG), termination–reinitiation still occurred efficiently at approximately the same level as in the wild-type mRNA.
FIGURE 4.
Investigating the role of the 18S rRNA complementary region in termination–reinitiation. (A) Comparison of part of the sequence of helix 26 of 18S rRNA and complementary sequences present upstream of the termination–reinitiation sites of FCV, RHDV, and influenza B. Contiguous nucleotides complementary to the 18S rRNA are shown in italics. (B) Schematic of the mutants generated to address the role of the complementarity between the BM2 mRNA and the 18S rRNA. The sequence of the relevant region of 18S rRNA is shown and putative mRNA base pairing is marked, with the mRNA bases numbered relative to the stop codon of rlucM1. Mutations are shown in bold and underlined. (C) Plasmids were linearized, transcribed, translated, and analyzed according to the legend of Figure 3. Control translations (wt, ps) were performed in parallel. Lanes are labeled with the last two letters of each reporter plasmid name. The asterisked band in lane CC highlights the presence of the putative frameshift product in a translation lacking the BM2 product (see the text).
RNA secondary structure analysis of the region required for termination–reinitiation in influenza BM2 synthesis
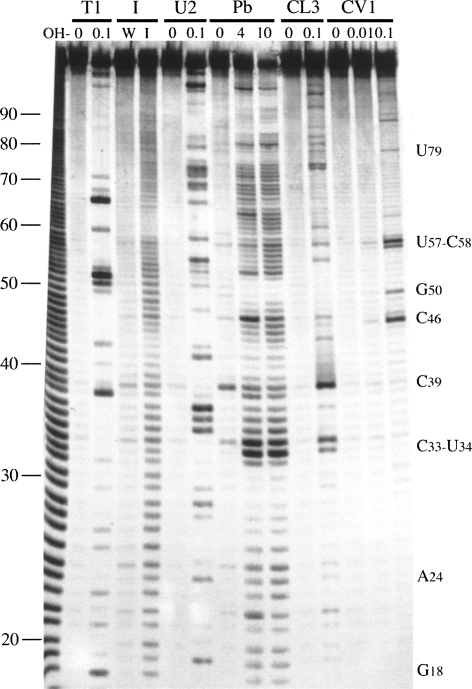
The experiments described above suggest that termination and reinitiation upon the BM2 ORF is likely to be dependent on sequence complementary to 18S rRNA. It was therefore of interest to determine the context of this complementary region within the global secondary structure of the minimal region. To achieve this, a T3 promoter was inserted ∼30 nt upstream of the viral sequence of the p2luc-BM2-204 plasmid (Fig. 1). The plasmid was linearized with BamHI, T3 run-off transcripts synthesized, and the RNA end labeled with [33P]-γATP. The labeled transcripts were subjected to limited chemical and enzymatic probing prior to analysis on denaturing polyacrylamide gels. The chemical probes used were imidazole and lead acetate, specific for cleavage of single-stranded regions. Enzymatic probes were RNases T1, U2, and CL3, which preferentially cleave single-stranded G, A, and C residues, respectively, and RNase CV1, which cuts in helical regions in double-stranded or stacked conformations. A representative stucture mapping gel is shown in Figure 5, and in Figure 6, the data are mapped onto two mfold predictions of the secondary structure of the stop–start region.
FIGURE 5.
Structure probing of the BM2 termination–reinitiation signal. RNA derived by transcription of p2luc-BM2-T3/Bam HI with T3 RNA polymerase was 5′-end labeled with [γ-33P]-ATP and subjected to limited RNase or chemical cleavage using structure-specific probes. Sites of cleavage were identified by comparison with a ladder of bands created by limited alkaline hydrolysis of the RNA (OH−) and the position of known RNase U2 and T1 cuts, determined empirically. Products were analyzed on a 10% acrylamide/7 M urea gel containing formamide. Data also were collected from 6% and 15% gels (gels not shown). Enzymatic structure probing was with RNases CL3, T1, U2, and CV1. Uniquely cleaved nucleotides were identified by their absence in untreated control lanes (0). The number of units of enzyme added to each reaction is indicated. Chemical structure probing was with imidazole (4 h, I) or lead acetate (Pb2+; millimolar concentration in reaction). The water lane (W) represents RNA that was dissolved in water, incubated for 4 h and processed in parallel to the imidazole-treated sample. The sequence of the probed RNA and the inferred secondary structure is shown in Figure 6.
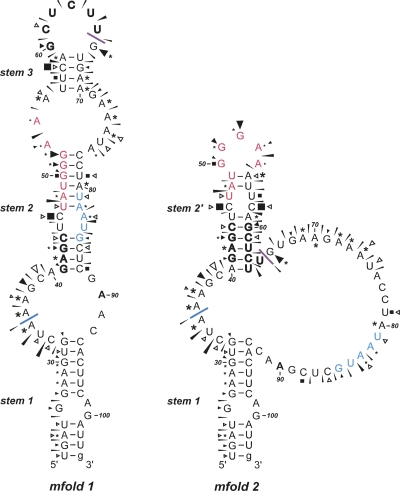
FIGURE 6.
Summary of the BM2 structure probing results. The sensitivity of bases in the BM2 termination–reinitiation region to the various probes is shown for two mfold predictions (see the text). The reactivies of the T1 (black triangle), U2 (asterisk), lead (black slender wedge), CL3 (open triangle), and CV1 (black square) probes are marked. Imidazole cleavage is not marked, but bases resistant to cleavage by this reagent are in bold/outline font. The size of the symbols is approximately proportional to the intensity of cleavage at that site. No structure mapping information was obtained beyond residue A90 (in bold). The stretch of bases in red indicate the 18S rRNA complementary region. Bases that form the stop–start overlap are in blue. The blue line indicates the start of the minimal essential region required for efficient termination–reinitiation. The purple line indicates the likely location of the 5′ edge of a ribosome poised at the termination codon (UAA, in blue). The ultimate 3′ base is in lowercase to indicate that this is a nonviral base of vector origin.
The structure probing analysis revealed that the mRNA in the region essential for termination–reinitiation is not highly structured. This was especially evident from the chemical probes, with most residues sensitive to imidazole and lead cleavage. The enzymatic probes were also active against the majority of bases in the region and consistent with this, CV1 probing identified very few double-stranded or stacked bases. We also noticed a few CL3 cuts at residues other than C, although the reason for this is uncertain. Minimal free energy mfold predictions, performed using the online server of Zuker (http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi) indicated that the most stable RNA fold was the bulged stem–loop of mfold 1 shown in Figure 6. The correspondence between mfold 1 and the mapping data was not absolute, however. While, in general, the single-stranded probes displayed more activity against regions of the model predicted to be single stranded than they did against predicted helices, there were anomalies. For example, residues 60–65 are displayed in the apical loop, yet this stretch showed resistance to imidazole probing and only weak reactivity with RNase CL3. It is also clear that the predicted duplexes showed more reactivity to single-stranded probes than one would expect for stable double-stranded stretches. Therefore, it seems likely that the RNA in this region is metastable, potentially adopting a number of coexisting structures. Examination of the imidazole probing reactions revealed only two areas of resistance to cleavage, the aforementioned region (60–65) and a second stretch from residues 41–44. Based on this, a second model was developed (Fig. 6, mfold 2) in which these regions were forced to pair. In mfold 2, stem 1 is retained, but stems 2 and 3 are replaced by an alternative second stem (Fig. 6, stem 2′) and by a longer single-stranded region connecting the second arm of stems 2′ and 1. As with mfold 1, there was a reasonable fit between the experimental mapping data and the predicted structure, with the odd anomaly. For example, residues C77 and C78, predicted to be in a single-stranded region in mfold 2 (but paired in mfold 1), were unreactive to CL3. The position of the sequence complementary to 18S rRNA varies in the two models. In mfold 1, it is sequestered mostly within a base-paired region (Fig. 6, stem 2), but in mfold 2 it forms the apical loop of the structure. Given that the termination–reinitiation process requires the ribosome to translate through the M1 ORF, it should be noted that the RNA upstream of the stop–start window would be translationally remodeled as the ribosome approaches the termination codon. Heelprinting of ribosomes paused at initiation codons has shown that the 5′ edge of the ribosome is some 12–13 nt from the first base of the AUG (Wolin and Walter 1988). This would place the 5′ edge of the terminating ribosome close to residue G66 on our mRNA. Thus a terminating ribosome would in principle prevent formation of mfold 1 but not stem 2′ of mfold 2. Therefore, we suggest that the 18S rRNA complementary region would be displayed on the apical loop of mfold 2 and immediately adjacent to the terminating ribosome.
Effect of relocating the stop codon of rlucM1 downstream from the start codon of BM2fluc
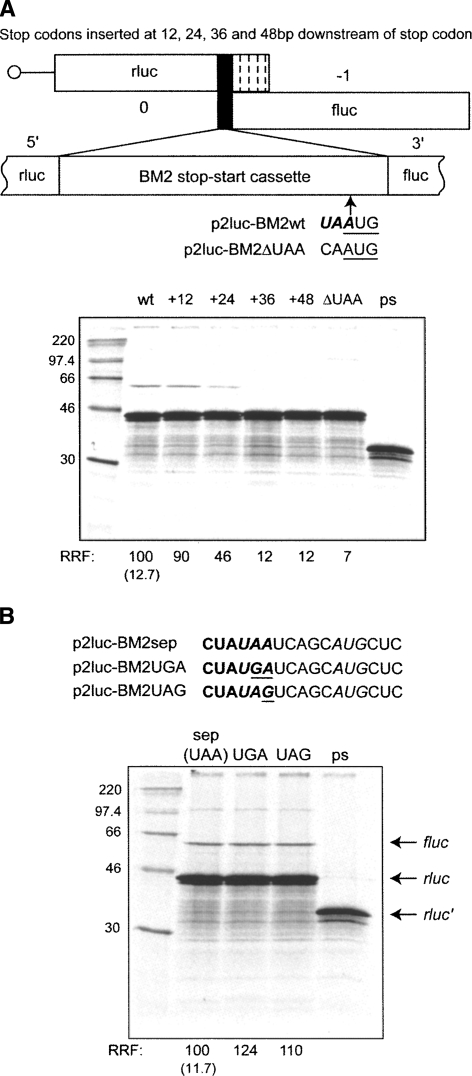
Coupled expression of the FCV VP1/VP2 and RHDV VP60/VP10 proteins is dependent on close proximity of the stop and start codons at the termination–reinitiation window (Luttermann and Meyers 2007; Meyers 2007). To investigate whether this is also true for BM2 expression, the authentic stop codon of rlucM1 was mutated from UAA to CAA such that the first ORF was extended by 21 amino acids (Fig. 7A, ΔUAA). Stop codons were then inserted (independently) such that ORF1 terminated 4, 8, 12, or 16 amino acids downstream from the natural termination site (+12, +24, +36, or +48 nt mutants, respectively). Movement of the stop codon 12 nt downstream from the normal termination site had little effect on termination–reinitiation, but at 24 nt the rate was reduced, and the process was abolished when the stop codon was moved any further downstream. Thus as with the calicivirus stop–start signals, the influenza B virus M1 stop and BM2 start codons must lie in close proximity (within eight codons) for termination–reinitiation to occur. However, in contrast to what has been described for RHDV (Meyers 2007), the identity of the stop codon does not appear to influence the rate of termination–reinitiation in BM2 expression. Variants of the reporter mRNA in which the M1 stop codon (UAA) was replaced by UGA or UAG showed unchanged levels of termination–reinitiation (Fig. 7B). These mutants were made in the background of p2luc-BM2-sep (Fig. 8A) to allow the stop and start codons to be changed without affecting the flanking sequence contexts (see below).
FIGURE 7.
The effect of M1 ORF termination codon position and identity on BM2 synthesis. (A) The stop–start window within p2luc-BM2 was modified to remove the natural rlucM1 ORF stop codon (UAA UG to CAAUG; the ΔUAA mutant), which placed the next in-frame termination codon 61 nt downstream from the AUG of the BM2fluc ORF. Subsequently, a new stop codon (UAA) was inserted at 12, 24, 36, or 48 nt downstream from the original position of the rlucM1 termination signal. The mutants were linearized, transcribed, translated, and analyzed according to the legend of Figure 3. Wild-type (wt) and premature stop (ps) controls were performed in parallel. (B) In the background of p2luc-BM2-sep (see the text and the legend to Fig. 8), the UAA termination codon at the end of the rlucM1 ORF was changed to UGA or UAG and termination–reinitiation assessed according to the legend to Figure 3. Control translations (sep, ps) were performed in parallel.
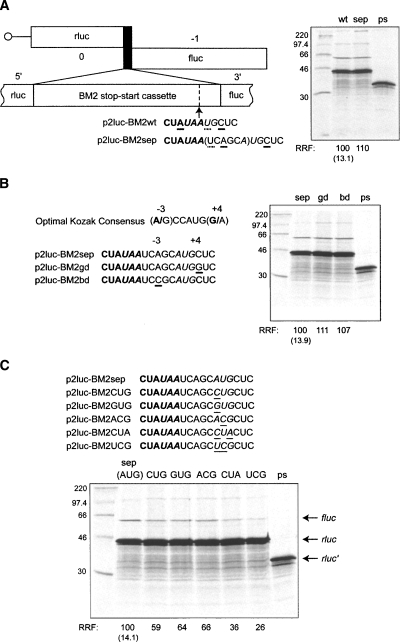
FIGURE 8.
Influence of start codon context on reinitiation of BM2 synthesis. (A) In order to modulate the start codon context of the BM2 ORF, it was first necessary to separate the rlucM1 stop codon and BM2fluc AUG. This was achieved in p2luc-BM2-sep by insertion of 6 nt (italicized in UAA UCAGCA UG), a sequence chosen to preserve the base immediately downstream from the UAA stop codon (U; dotted underline) and the A at the −3 position relative to the start codon of BM2fluc (bold underline). Translations of the p2luc-BM2-sep mRNA and relevant controls (wt, ps) are shown. (B) The optimal Kozak consensus sequence for initiation at AUG codons is shown in comparison to that of p2luc-BM2-sep and two engineered variants with good (p2luc-BM2-gd) or poor (p2luc-BM2-bd) initiation codon context (base changes are underlined). Translations of the gd and bd mRNAs are shown, alongside control mRNAs from p2luc-BM2-sep (sep) and p2luc-BM2-ps (ps). (C) The BM2 start codon in p2luc-BM2-sep was changed to alternative codons as indicated and translated, alongside sep and ps controls. In all panels, plasmids were linearized, transcribed, translated, and analyzed according to the legend of Figure 3.
The termination–reinitiation signal overcomes the requirement for good AUG context in BM2 expression and allows initiation at non-AUG codons
Translation initiation in eukaryotes typically follows the “first AUG” rule, namely, that initiation begins at the 5′ proximal AUG (Kozak 1984). However, the first AUG may be bypassed if the start codon lies in a poor sequence context. The optimal context for initiation has been defined as GCC(A/G)CCAUG(G/A), with the purines at −3 and +4 being particularly important (Kozak 1986, 1987). Termination–reinitiation requires the close proximity of the stop and start codons, and it is feasible that the ribosome is effectively “dropped” at the start site for initiation of BM2 synthesis. Under these circumstances, it is conceivable that the 40S subunit is not in the “scanning mode” adopted during conventional initiation and there may be a reduced requirement for good initiation codon context in the selection of the start site of translation. Consistent with this, the context surrounding the AUG start codon of BM2fluc is suboptimal due to the presence of a pyrimidine (cytosine) in the +4 position. To explore this more fully, it was first necessary to separate the overlapping stop–start codons to allow manipulation of context. This was achieved by insertion of six nucleotides (italicized in UAA UCAGCA UG), a sequence chosen to preserve the base immediately downstream from the UAA stop codon (U) and the A at the −3 position relative to the start codon of BM2fluc (creating p2luc-BM2sep; Fig. 8A). Importantly, this construct showed levels of termination–reinitiation similar to that of the wild-type BM2 reporter RNA (Fig. 8A). Subsequently, the context surrounding the AUG was changed to a good (−3 = A, +4 = G) or a poor (−3 = C, +4 = C) Kozak consensus. In each case, termination–reinitiation was found to occur at a level similar to that of the wild-type sequence irrespective of the context (Fig. 8B). To probe the initiation process further, we tested the effect of replacing the authentic AUG of p2luc-BM2sep with a series of non-AUG codons (Fig. 8C; CUG, GUG, ACG, CUA, UCG). All of the mutants were able to support reinitiation of the BM2/fluc ORF to some extent, including those codons with two nucleotide changes relative to the normal AUG codon. The level of reinitiation seen with the non-AUG codons paralleled the efficiency of their usage that has been described in other published work (Mehdi et al. 1990; Gupta et al. 1994). However, it should be noted that the overall rate of reinitiation at these codons was much higher than would be expected for a typical noncanonical initiation codon in a standard context (Peabody 1989; Kozak 1991). These data suggest that the start site for reinitiation is not recognized by the 40S subunit in a traditional scanning mode. Indeed, the lack of context effect and the efficient recognition of non-AUG codons support the view that the 40S subunit is placed close to or directly at the start codon. However, it is also possible that there is a relaxed requirement for the full complement of initiation factors during reinitiation, and that eIF1 and eIF1A in particular may not be needed for initiation on the BM2 ORF, a hypothesis consistent with the data presented here.
DISCUSSION
In this paper, we show that expression in vitro of the influenza B proton channel BM2 occurs by a process of coupled translation termination–reinitiation, confirming the original prediction of Horvath and colleagues (1990). The process requires the close proximity of termination and reinitiation codons and a defined region of mRNA upstream of the stop–start site that includes a functionally essential stretch of bases with complementarity to helix 26 of 18S rRNA. This complementary region is potentially located within the loop of a hairpin structure adjacent to the terminating ribosome, although a role for this secondary structure in the reinitiation process remains to be confirmed.
By placing a premature stop codon upstream of the viral information in our reporter mRNA, we were able to confirm that ribosomes must translate through the M1 ORF to allow synthesis of the BM2 product (Horvath et al. 1990). The absence of reinitiation in such premature-stop mRNAs also rules out the possibility that BM2 expression occurs by ribosome recruitment to an internal ribosomal entry site or by shunting from the rlucM1 untranslated region. Other modes of expression of BM2 are also viewed as unlikely. Termination codon suppression seems to be ruled out by the simple fact that the BM2 ORF lies in a different reading frame than M1. In segment 7 of influenza B virus, the reading frame of the BM2 ORF extends a considerable distance (∼260 nt) 5′ of the AUG, and there is significant overlap between this region and the M1 ORF. Conceivably, some BM2 expression could be as a fusion with the upstream M1 protein as a result of ribosomal frameshifting. In a number of the in vitro translation reactions carried out in the present work, an ∼95 kDa species was seen that might correspond to a fusion of the rlucM1/BM2fluc ORFs and it may be that in vitro, a low-level of frameshifting can occur, although no obvious signals are evident (Brierley et al. 1991, 1992). However, there is no evidence that an M1–BM2 fusion protein is expressed in virus-infected cells or in transfected tissue culture cells (Horvath et al. 1990). Furthermore, in RRL, the ∼95-kDa product was expressed in some cases when the BM2 product was not (e.g., Fig. 4C). It seems likely, therefore, that the ∼95-kDa protein arises as a result of an in vitro frameshift and that in infected cells, BM2 is expressed solely by termination–reinitiation.
The importance of sequences flanking the stop–start window was first investigated in RHDV (Meyers 2003). Reinitiation on the VP10 ORF showed little dependence on downstream sequences and the entire VP10 ORF could be replaced with that of another protein without effect (Meyers 2003). In our analysis of BM2 expression, we did not examine specifically the effect of sequences downstream from the stop–start window, but only 18 nt of BM2 sequence are present upstream of the fluc reporter gene; thus the BM2 protein itself is not required. Taking into account the RHDV work, it also seems likely that the remaining BM2 primary sequence (excluding the AUG) will have little effect on the process in BM2. In contrast, sequences upstream of the stop–start overlap region are crucial in all the known viral examples of termination–reinitiation, although the length of the essential region varies (Meyers 2003, 2007; Gould and Easton 2005; Luttermann and Meyers 2007). In RSV, >200 nt are required for full activity, and stepwise deletions into this region reduce the termination–reinitiation efficiency incrementally (Gould and Easton 2005). In contrast, deletion analysis reveals that a shorter stretch is sufficient in RHDV (84 nt; Meyers 2003), FCV (69–87 nt; Luttermann and Meyers 2007; Pöyry et al. 2007), and BM2 (45 nt; this study), with a sharp cut-off point. Similar to the caliciviruses (Luttermann and Meyers 2007; Meyers 2007), BM2 synthesis showed strong dependence on sequences likely to base-pair to 18S rRNA, and mutation of complementary bases to noncomplementary resulted in abrogation of BM2 synthesis (Fig. 4). It has been suggested that such an interaction would serve to tether the 40S subunit to the mRNA post-termination (Luttermann and Meyers 2007). Previous evidence suggests that termination–reinitiation is dependent on the stop and start codons lying in close proximity (Horvath et al. 1990; Ahmadian et al. 2000; Meyers 2003, 2007; Luttermann and Meyers 2007). Indeed, movement of the stop codon of the M1 ORF more than 24 nt downstream from the stop–start window inhibited expression of the BM2/fluc reporter. In support of a tethering interaction between the rRNA and mRNA, it may be that at this distance, the 40S subunit cannot relocate the AUG, perhaps because it is placed too far away from a tethering site, be it the 18S rRNA complementary region, an eIF3 binding site (as discussed below), or both.
The region of 18S rRNA to which the BM2, FCV, and RHDV mRNAs could interact forms the loop and upper base pairs of the hairpin that forms helix 26 (Gutell 1993; Matassova et al. 1998). If such an interaction involves straightforward Watson–Crick base-pairing, then the top of helix 26 would need to be unwound for pairing across the whole complementary region. Recent evidence supports the view that the apical stem–loop of helix 26 is accessible for interaction with the mRNA near to the trailing edge of ribosomal 48S and 80S complexes (Pisarev et al. 2008). In RSV, by contrast, no region with obvious complementarity to helix 26 is present, and the mechanism of termination–reinitiation may thus differ. In the caliciviruses, a second essential primary sequence motif of unknown function (termed the termination upstream ribosomal binding site [TURBS] motif 2), is present some 20–30 bases upstream of the stop–start overlap (Luttermann and Meyers 2007; Meyers 2007). Whether such a motif is present in BM2, whose minimal essential region is considerably shorter than that of the caliciviruses, is not known.
Secondary structure probing revealed that the RNA upstream of the BM2 stop–start window is largely unstructured, although hairpins of modest stability predicted by mfold showed reasonable concordance with the experimental data. Whether these secondary structures play a role in termination–reinitiation remains to be determined. It is possible that during translation of this region, the RNA is remodeled and upon termination, a structure is formed that promotes reinitiation. From the models of Figure 6, we predict that stem 2′ of mfold 2 would still form in the presence of the terminating ribosome. In this model the 18S rRNA complementary region would be located in the apical loop and in close proximity to helix 26 of rRNA (Clemons et al. 1999; Chandramouli et al. 2008), consistent with its proposed role in tethering the 40S subunit. The deletion analysis of Figure 3 revealed that RNAs retaining 45 nt of BM2 sequence upstream of the stop codon retained function, but deletion of a further three bases reduced the reinitiation frequency and removal of a further three bases (to 39 nt) abolished reinitiation. In mfold 2, the deletion retaining only 39 nt of BM2 information would begin to compromise stem 2′ formation, perhaps accounting for the reduced activity of this mRNA.
It should be noted that tethering of the 40S subunit to the viral RNA is not the only function that has been proposed for the mRNA in this region. Cross-linking analysis has shown that the FCV RNA is able to bind the multisubunit eukaryotic initiation factor 3 (eIF3; Pöyry et al. 2007). Recent work has also revealed that eIF3 plays a role in dissociating the 60S and 40S ribosomal subunits following termination, a process that is enhanced further by eIFs 1A, 1, and the loosely associated eIF3j (Pisarev et al. 2007). It has been suggested that eIF3 may bind upstream of the stop–start overlap and promote dissociation of the 60S ribosomal subunit following termination, speeding the rate of ribosome recycling and allowing reinitiation on the downstream ORF (Pöyry et al. 2007). The bound eIF3 may then play a further role in recruiting initiation factors for reinitiation on the downstream ORF, although this has yet to be demonstrated experimentally. Interestingly, mutation of 3 nt of FCV TURBS motif 1 (thought to interact with 18S rRNA) also reduces binding of the viral RNA to eIF3 (Pöyry et al. 2007). While these data challenge models that invoke simply the tethering of the 40S subunit through mRNA:rRNA interactions, it is possible that the upstream sequence plays a dual role, both in ribosome dissociation/recycling, and in 40S tethering. The essential upstream mRNA sequence required for termination–reinitiation in BM2 (∼45 nt) is shorter than that of FCV (70–87 nt), hinting at mechanistic differences. However, it is known that the region between the FCV TURBS motifs can be deleted with only a modest reduction (twofold) in reinitiation frequency (Luttermann and Meyers 2007). Thus the BM2 termination–reinitiation “module” may simply represent an evolutionarily streamlined version.
In the BM2 termination–reinitiation process, there is little requirement for good start codon context in efficient selection of the start site. Also, there is less dependence on an AUG start codon for initiation of BM2 synthesis, and a variety of noncanonical initiation codons can be used efficiently, even CUA, which differs by two bases from the normal start codon. While these observations have been made in an in vitro system, similar data have also been observed in vivo using reporter plasmids encoding VP1 and VP2 of feline calicivirus (Luttermann and Meyers 2007). However, unlike in the calicivirus RHDV, the identity of the stop codon at the BM2 stop–start window has little effect on termination–reinitiation efficiency. These data raise the possibility that there is a reduced requirement for the full complement of initiation factors in the reinitiation process, including eIF1 and eIF1A, thought to play important roles in locating and correct recognition of the AUG start codon (Cheung et al. 2007; Passmore et al. 2007).
We suggest a model for termination–reinitiation in BM2 expression whereby the 80S ribosome translates through the M1 ORF of BM2, and as it does so a structure is created 5′ of the ribosome which, after termination, tethers a proportion of 40S ribosomes to the mRNA involving a region of complementarity between the ribosome and the mRNA. The tethering of the 40S subunit, which may also involve eIF3, allows time for initiation factors to be recruited to the complex and places the ribosome in the optimal position for initiation to occur, bypassing the requirement for good initiation codon context (and also allowing the use of noncanonical initiation codons). Initiation then occurs on the BM2 ORF and translation proceeds as normal, allowing tightly regulated expression of the polypeptides encoded by the M1 and BM2 open reading frames. Future work will be aimed at testing this model.
MATERIALS AND METHODS
Extraction of viral RNAs
TRTV and RHDV RNAs were extracted from TRTV-infected DF1 cells and RHDV-infected rabbit livers, respectively, using Trizol reagent (Invitrogen) according to the manufacturer's instructions. Influenza B RNAs were obtained from a clinical sample of a patient infected with influenza B using the High Pure Viral RNA Kit (Roche) according to the manufacturer's instructions.
Construction of plasmids
All of the plasmids used in this study were based on the p2luc reporter vector (Grentzmann et al. 1998). Sequences encompassing the putative termination–reinitiation sites of RHDV (100 bp upstream of the VP60 stop codon and 19 bp downstream), TRTV (247 bp upstream of the M2 stop codon and 22 bp downstream), and influenza B (detailed in Results) were generated by RT-PCR using AMV reverse transcriptase (Promega) and Taq polymerase (Roche) according to standard protocols. The PCR products and p2luc were digested with SalI and BamHI and ligated together. Sequences were confirmed by commercial dideoxy sequencing (using the facility at the Department of Biochemistry, University of Cambridge).
Site-directed mutagenesis
Site-directed mutagenesis was performed using the Quikchange II site-directed mutagenesis kit (Stratagene) according to manufacturer's instructions. For large deletions (> 48 bp) a modification of the manufacturer's protocol was used with the primers containing ∼30 bp of complementary sequence either side of the site of deletion, as described previously (Makarova et al. 2000). Mutagenesis to introduce insertions longer than 6 bp was performed in two steps (Wang and Malcolm 1999), by first subjecting mutagenesis reactions (containing either the sense or antisense primer) to three cycles of PCR, then mixing the reactions and performing a further 18 cycles according to manufacturer's instructions.
In vitro transcription and translation
Reporter plasmids were linearized with HpaI and capped run-off transcripts generated using T7 RNA polymerase as described (Girnary et al. 2007). Messenger RNAs were recovered by a single extraction with phenol/chloroform (1:1 [v/v]) followed by ethanol precipitation. Remaining unincorporated nucleotides were removed by gel filtration through a NucAway spin column (Ambion). The eluate was concentrated by ethanol precipitation, and the mRNA was resuspended in water, checked for integrity by agarose gel electrophoresis, and quantified by spectrophotometry.
Unless otherwise stated, mRNAs were translated in Flexi rabbit reticulocyte lysate (Flexi RRL, Promega) programmed with 50 μg/mL template mRNA. Typical reactions were of 10 μL and composed of 60% (v/v) Flexi RRL, 20 μM amino acids (lacking methionine), 500 μM MgOAc, 2 mM DTT, 5 U RNAse inhibitor (RNAguard, GE Healthcare Life Sciences), 130 mM–160 mM KCl (optimized for each batch of Flexi RRL) and 0.2 MBq [35S]-methionine. Reactions were incubated for 1 h at 30°C and stopped by the addition of an equal volume of 10 mM EDTA, 100 μg/mL RNase A followed by incubation at room temperature for 20 min. Samples were prepared for SDS-PAGE by the addition of 10 volumes of 4× Laemmli's sample buffer (Laemmli 1970), boiled for 3 min and resolved on 12% SDS-PAGE gels. The relative abundance of products on the gels was determined by direct measurement of [35S]methionine incorporation using a Packard Instant Imager 2024.
RNA structure mapping
A plasmid encoding the minimal required sequence for termination–reinitiation (p2luc-BM2-204) was modified by site-directed mutagenesis to include a T3 RNA polymerase promoter 30 bp upstream of the minimal required viral sequence generating plasmid p2luc-BM2-T3. RNA for structure mapping was prepared by in vitro transcription of BamHI-digested p2luc-BM2-T3 using T3 RNA polymerase. Transcription reactions were performed on a 200-μL scale essentially as described (Girnary et al. 2007). Structure mapping was performed using a 5′ end-labeling procedure as described previously (Manktelow et al. 2005; Girnary et al. 2007). All probing reactions were performed in a final volume of 50 μL and contained ∼40,000 cpm 5′ 33P-end-labeled transcript, 10 μg Escherichia coli rRNA, and the relevant enzymatic or chemical probe. Further details are provided in the legend to Figure 5.
ACKNOWLEDGMENTS
This work was supported by a grant from the Biotechnology and Biological Sciences Research Council, United Kingdom (I.B., T.D.K.B., R.J.J.). We thank Dr. Tuija Pöyry for helpful advice and discussions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1231008.
REFERENCES
- Ahmadian G., Randhawa J.S., Easton A.J. Expression of the ORF-2 protein of the human respiratory syncytial virus M2 gene is initiated by a ribosomal termination-dependent reinitiation mechanism. EMBO J. 2000;19:2681–2689. doi: 10.1093/emboj/19.11.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov P.V., Fayet O., Hendrix R.W., Atkins J.F. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22:174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Bertram G., Innes S., Minella O., Richardson J., Stansfield I. Endless possibilities: Translation termination and stop codon recognition. Microbiol. 2001;147:255–269. doi: 10.1099/00221287-147-2-255. [DOI] [PubMed] [Google Scholar]
- Briedis D.J., Lamb R.A., Choppin P.W. Sequence of RNA segment 7 of the influenza B virus genome: Partial amino acid homology between the membrane proteins (M1) of influenza A and B viruses and conservation of a second open reading frame. Virology. 1982;116:581–588. doi: 10.1016/0042-6822(82)90150-7. [DOI] [PubMed] [Google Scholar]
- Brierley I., Dos Ramos F.J. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 2006;119:29–42. doi: 10.1016/j.virusres.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Rolley N.J., Jenner A.J., Inglis S.C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Jenner A.J., Inglis S.C. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouli P., Topf M., Menetret J.F., Eswar N., Cannone J.J., Gutell R.R., Sali A., Akey C.W. Structure of the mammalian 80S ribosome at 8.7Å resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung Y.N., Maag D., Mitchell S.F., Fekete C.A., Algire M.A., Takacs J.E., Shirokikh N., Pestova T., Lorsch J.R., Hinnebusch A.G. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes & Dev. 2007;21:1217–1230. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons W.M., Jr, May J.L., Wimberly B.T., McCutcheon J.P., Capel M.S., Ramakrishnan V. Structure of a bacterial 30S ribosomal subunit at 5.5Å resolution. Nature. 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- Dasso M.C., Jackson R.J. On the fidelity of mRNA translation in the nuclease-treated rabbit reticulocyte lysate system. Nucleic Acids Res. 1989;17:3129–3144. doi: 10.1093/nar/17.8.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher T.W., Miller W.A. Translational control in positive strand RNA plant viruses. Virology. 2006;344:185–197. doi: 10.1016/j.virol.2005.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M., Jr, Tan S.L., Katze M.G. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 2000;64:239–280. doi: 10.1128/mmbr.64.2.239-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girnary R., King L., Robinson L., Elston R., Brierley I. Structure-function analysis of the ribosomal frameshifting signal of two human immunodeficiency virus type 1 isolates with increased resistance to viral protease inhibitors. J. Gen. Virol. 2007;88:226–235. doi: 10.1099/vir.0.82064-0. [DOI] [PubMed] [Google Scholar]
- Gould P.S., Easton A.J. Coupled translation of the respiratory syncytial virus M2 open reading frames requires upstream sequences. J. Biol. Chem. 2005;280:21972–21980. doi: 10.1074/jbc.M502276200. [DOI] [PubMed] [Google Scholar]
- Gould P.S., Easton A.J. Coupled translation of the second open reading frame of M2 mRNA is sequence dependent and differs significantly within the subfamily Pneumovirinae. J. Virol. 2007;81:8488–8496. doi: 10.1128/JVI.00457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G., Ingram J.A., Kelly P.J., Gesteland R.F., Atkins J.F. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- Gupta K.C., Ono E., Ariztia E.V., Inaba M. Translation initiation from non-AUG codons in COS1 cells is mRNA species dependent. Biochem. Biophys. Res. Commun. 1994;201:567–573. doi: 10.1006/bbrc.1994.1739. [DOI] [PubMed] [Google Scholar]
- Gutell R.R. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 1993;21:3051–3054. doi: 10.1093/nar/21.13.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Hiebert S.W., Williams M.A., Lamb R.A. Nucleotide sequence of RNA segment 7 of influenza B/Singapore/222/79: Maintenance of a second large open reading frame. Virology. 1986;155:747–751. doi: 10.1016/0042-6822(86)90237-0. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Horvath C.M., Williams M.A., Lamb R.A. Eukaryotic coupled translation of tandem cistrons: Identification of the influenza B virus BM2 polypeptide. EMBO J. 1990;9:2639–2647. doi: 10.1002/j.1460-2075.1990.tb07446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J. Potassium salts influence the fidelity of mRNA translation initiation in rabbit reticulocyte lysates: Unique features of encephalomyocarditis virus RNA translation. Biochim. Biophys. Acta. 1991;1088:345–358. doi: 10.1016/0167-4781(91)90124-5. [DOI] [PubMed] [Google Scholar]
- Jackson R.J., Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- Jackson D., Zurcher T., Barclay W. Reduced incorporation of the influenza B virus BM2 protein in virus particles decreases infectivity. Virology. 2004;322:276–285. doi: 10.1016/j.virol.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Kozak M. Selection of initiation sites by eucaryotic ribosomes: Effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984;12:3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Luttermann C., Meyers G. A bipartite sequence motif induces translation reinitiation in feline calicivirus RNA. J. Biol. Chem. 2007;282:7056–7065. doi: 10.1074/jbc.M608948200. [DOI] [PubMed] [Google Scholar]
- Luukkonen B.G., Tan W., Schwartz S. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J. Virol. 1995;69:4086–4094. doi: 10.1128/jvi.69.7.4086-4094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova O., Kamberov E., Margolis B. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques. 2000;29:970–972. doi: 10.2144/00295bm08. [DOI] [PubMed] [Google Scholar]
- Manktelow E., Shigemoto K., Brierley I. Characterization of the frameshift signal of Edr, a mammalian example of programmed −1 ribosomal frameshifting. Nucleic Acids Res. 2005;33:1553–1563. doi: 10.1093/nar/gki299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassova N.B., Venjaminova A.G., Karpova G.G. Nucleotides of 18S rRNA surrounding mRNA at the decoding site of translating human ribosome as revealed from the cross-linking data. Biochim. Biophys. Acta. 1998;1397:231–239. doi: 10.1016/s0167-4781(98)00015-3. [DOI] [PubMed] [Google Scholar]
- Mehdi H., Ono E., Gupta K.C. Initiation of translation at CUG, GUG, and ACG codons in mammalian cells. Gene. 1990;91:173–178. doi: 10.1016/0378-1119(90)90085-6. [DOI] [PubMed] [Google Scholar]
- Meyers G. Translation of the minor capsid protein of a calicivirus is initiated by a novel termination-dependent reinitiation mechanism. J. Biol. Chem. 2003;278:34051–34060. doi: 10.1074/jbc.M304874200. [DOI] [PubMed] [Google Scholar]
- Meyers G. Characterization of the sequence element directing translation reinitiation in RNA of the calicivirus rabbit hemorrhagic disease virus. J. Virol. 2007;81:9623–9632. doi: 10.1128/JVI.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould J.A., Paterson R.G., Takeda M., Ohigashi Y., Venkataraman P., Lamb R.A., Pinto L.H. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev. Cell. 2003;5:175–184. doi: 10.1016/s1534-5807(03)00190-4. [DOI] [PubMed] [Google Scholar]
- Passmore L.A., Schmeing T.M., Maag D., Applefield D.J., Acker M.G., Algire M.A., Lorsch J.R., Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Pavlakis G.N., Felber B.K. Regulation of expression of human immunodeficiency virus. New Biol. 1990;2:20–31. [PubMed] [Google Scholar]
- Peabody D.S. Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- Pisarev A.V., Hellen C.U., Pestova T.V. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev A.V., Kolupaeva V.G., Yusupov M.M., Hellen C.U.T., Pestova T.V. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry T.A., Kaminski A., Jackson R.J. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes & Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry T.A., Kaminski A., Connell E.J., Fraser C.S., Jackson R.J. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes & Dev. 2007;21:3149–3162. doi: 10.1101/gad.439507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Malcolm B.A. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. Biotechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- Wilson J.E., Powell M.J., Hoover S.E., Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin S.L., Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Davis P.J., Brown T.D., Cavanagh D. Sequence and in vitro expression of the M2 gene of turkey rhinotracheitis pneumovirus. J. Gen. Virol. 1992;73:1355–1363. doi: 10.1099/0022-1317-73-6-1355. [DOI] [PubMed] [Google Scholar]