Abstract
Chloroplast genomes in angiosperms encode ∼20 group II introns, approximately half of which are classified as subgroup IIB. The splicing of all but one of the subgroup IIB introns requires a heterodimer containing the peptidyl-tRNA hydrolase homolog CRS2 and one of two closely related proteins, CAF1 or CAF2, that harbor a recently recognized RNA binding domain called the CRM domain. Two CRS2/CAF-dependent introns require, in addition, a CRM domain protein called CFM2 that is only distantly related to CAF1 and CAF2. Here, we show that CFM3, a close relative of CFM2, associates in vivo with those CRS2/CAF-dependent introns that are not CFM2 ligands. Mutant phenotypes in rice and Arabidopsis support a role for CFM3 in the splicing of most of the introns with which it associates. These results show that either CAF1 or CAF2 and either CFM2 or CFM3 simultaneously bind most chloroplast subgroup IIB introns in vivo, and that the CAF and CFM subunits play nonredundant roles in splicing. These results suggest that the expansion of the CRM protein family in plants resulted in two subfamilies that play different roles in group II intron splicing, with further diversification within a subfamily to accommodate multiple intron ligands.
Keywords: group II intron, RIP-Chip, plastid, Arabidopsis, maize
INTRODUCTION
Group II introns are large ribozymes whose splicing involves the same chemical steps as spliceosome-catalyzed splicing in the nucleus (reviewed in Pyle and Lambowitz 2006). The mitochondrial and plastid genomes in land plants each encode ∼20 group II introns; none of these have been reported to self-splice in vitro, and some lack intron motifs known to play important roles in splicing (for review, see Schmitz-Linneweber and Barkan 2007; Bonen 2008). Thus, most or all group II introns in plant organelles appear to have lost the capacity to self-splice even under the nonphysiological conditions that promote the self-splicing of several model group II introns. Indeed, the splicing of almost all of the introns in angiosperm chloroplasts has been shown to require the participation of nucleus-encoded proteins in vivo (reviewed in Schmitz-Linneweber and Barkan 2007).
The genetic identification of nuclear genes involved in chloroplast group II intron splicing led to the recognition of a protein family defined by a novel RNA binding domain denoted the chloroplast RNA splicing and ribosome maturation (CRM) domain (also called the CRS1-YhbY domain/Pfam PF01985) (Till et al. 2001; Ostheimer et al. 2003; Barkan et al. 2007). This domain is derived from a prokaryotic pre-ribosome binding protein, and is found in a family comprising 14 orthologous groups in vascular plants (Barkan et al. 2007). Four CRM proteins have been shown to interact with group II introns in chloroplasts and to promote their splicing: CAF1, CAF2, CRS1, and CFM2. CAF1, and CAF2 are closely related paralogs, each with two CRM domains, that promote the splicing of different subsets of chloroplast subgroup IIB introns in heterodimeric complexes with the peptidyl-tRNA hydrolase homolog CRS2 (Ostheimer et al. 2003). CRS1 and CFM2 are closely related paralogs that belong to a different branch of the CRM protein family than CAF1/2 (see Fig. 1A): CRS1 is required for the splicing of the subgroup IIA atpF intron (Jenkins et al. 1997; Asakura and Barkan 2006), whereas CFM2 is required for the splicing of two subgroup IIB introns (ndhA and ycf3-int1) and for the splicing of the group I intron in trnL-UAA (Asakura and Barkan 2007). (See Fig. 11 below for summary of the intron specificities of each protein.) Coimmunoprecipitation assays showed that each of these proteins is found in stable ribonucleoprotein complexes with those introns whose splicing it promotes (Ostheimer et al. 2003; Schmitz-Linneweber et al. 2005; Asakura and Barkan 2007).
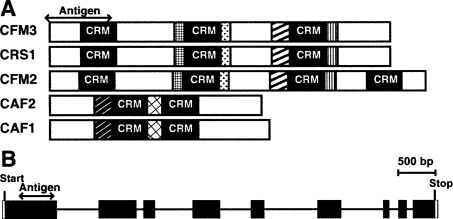
FIGURE 1.
Graphical representations of CFM3. (A) Domain organization of CFM3 and previously characterized CRM-domain splicing factors. CFM3, CRS1, and CFM2 are in the “CRS1” subfamily, whereas CAF1 and CAF2 are in the “CAF” subfamily (Barkan et al. 2007). Regions of high sequence similarity are represented by like shading. There is no discernable sequence similarity between members of the CAF and CRS1 subfamilies outside of their CRM domains. The region used for production of CFM3 antisera is indicated. (B) The ZmCfm3 gene. Exons are shown as rectangles and introns as lines.
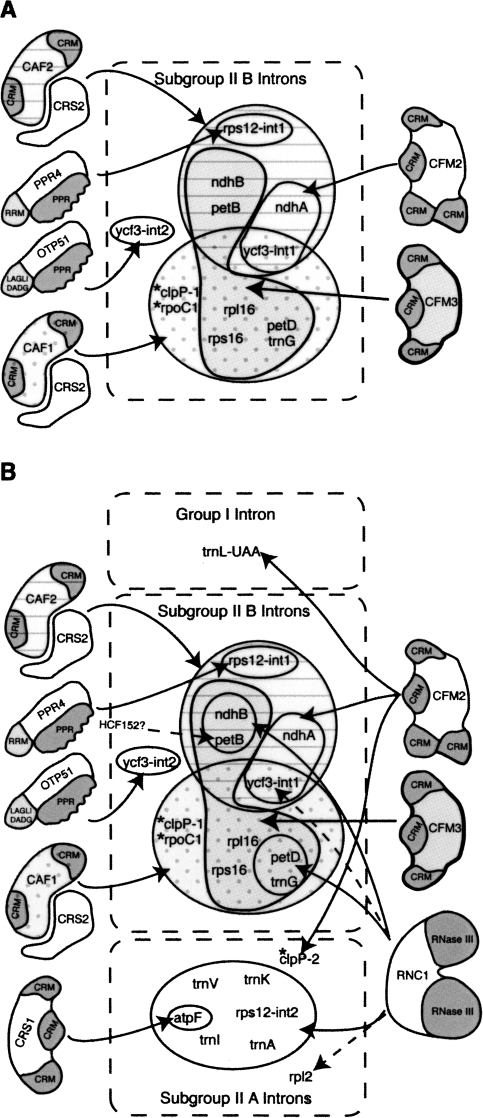
FIGURE 11.
Chloroplast splicing factors in land plants. (A) Cooperation between CFM2/3 and CAF1/2 during the splicing of Subgroup IIB introns. The intron sets that require CAF1, CAF2, CFM2, or CFM3 are marked with distinct fill patterns. These assignments for CFM3 are based on a composite of the RNA coimmunoprecipitation data from maize, and the genetic data from rice and Arabidopsis. Rps12–int1 is the sole trans-spliced intron in land plant chloroplasts. Ycf3–int2 is the sole subgroup IIB intron whose splicing does not require a CAF/CRS2 complex. Introns found in Arabidopsis but not in maize and rice are marked with asterisks. Current data are consistent with the possibility that the AtCFM3 co-orthologs play redundant roles in the splicing of the clpP-1 and rpoC1 introns, but such a role has not been demonstrated. (B) Summary of nucleus-encoded chloroplast splicing factors in land plants. The group II introns are divided into subgroup IIA and IIB according to Michel et al. (1989). Solid arrows mark interactions supported both by splicing defects in mutants and by RNA coimmunoprecipitation data. Dashed arrows mark interactions supported by either genetic or coimmunoprecipitation data, but not both. A role for the plastid-encoded protein MatK in subgroup IIA splicing has been proposed (reviewed in Bonen and Vogel 2001). HCF152 is required for the accumulation of spliced petB RNA but not for the accumulation of excised intron (Meierhoff et al. 2003). Results are summarized from this work and from Jenkins et al. (1997), Vogel et al. (1999), Ostheimer et al. (2003), Schmitz-Linneweber et al. (2005), Asakura and Barkan (2006), Schmitz-Linneweber et al. (2006), Asakura and Barkan 2007; Watkins et al. (2007), and de Longevialle et al. (2008).
To explore the functional repertoire of CRM domain proteins and to elucidate the protein machinery for plant organellar splicing, we are using reverse genetics and RNA coimmunoprecipitation assays to define the functions and RNA ligands of additional members of the CRM family. Here we present evidence that another member of the CRS1 subfamily, CFM3, acts in a nonredundant fashion with CRS2/CAF complexes to promote the splicing of a subset of group IIB introns in angiosperm chloroplasts. CFM2 and CFM3 interact with and promote the splicing of nonoverlapping subsets of CRS2/CAF-dependent introns. Thus, most chloroplast subgroup IIB introns require either CAF1 or CAF2, together with either CFM2 or CFM3. The results suggest that CFM2/CFM3 paralogs and CAF1/CAF2 paralogs play distinct biochemical roles in splicing, and that diversification within each of these CRM protein subfamilies generated proteins that promote splicing in similar ways but that recognize distinct intron subsets.
RESULTS
CFM3 localizes to chloroplasts and mitochondria
Proteins in the angiosperm CRM domain family comprise 14 orthologous groups that can be grouped into four subfamilies according to their domain organization and sequence similarity (Barkan et al. 2007). Uncharacterized members of the family were assigned the names CRM Family Member (CFM) 1–11. CFM3 is in the “CRS1 subfamily,” which includes the group II intron splicing factors CRS1 and CFM2. CAF1 and CAF2 are members of a separate subfamily. The relationships between CAF1, CAF2, CRS1, CFM2, and CFM3 are diagrammed in Figure 1A. A phylogenetic tree of the CRS1 subfamily (Supplemental Fig. 1) shows that CFM3 is represented by a single gene in rice (Os11g37990, denoted OsCFM3) and by two co-orthologs in Arabidopsis (At3g23070 and At4g14510); these genes are referred to here as AtCFM3a and AtCFM3b, respectively. Sequence for a putative maize ortholog was identified from public cDNA and genomic sequence data, and the sequence of a full-length cDNA was determined (GenBank Accession No. EU084957). Phylogenetic evidence for the orthology between the corresponding maize gene, ZmCfm3, and Os/AtCFM3 is shown in Supplemental Figure 1. An alignment between the Arabidopsis, maize, and rice CFM3 proteins is shown as Supplemental Figure 2.
AtCFM3a, AtCFM3b, ZmCFM3, and OsCFM3 are predicted to localize to chloroplasts and/or mitochondria by both the Target P (Emanuelsson and Heijne 2001) and Predotar (Small et al. 2004) algorithms. GFP fused to the N-terminal 113 amino acids of ZmCFM3 localized to both chloroplasts and mitochondria when transiently expressed in onion epidermal cells (Fig. 2A). The analogous experiment with OsCFM3 and AtCFM3a gave similar results, whereas the AtCFM3b transit peptide targeted GFP solely to chloroplasts (Fig. 2A).
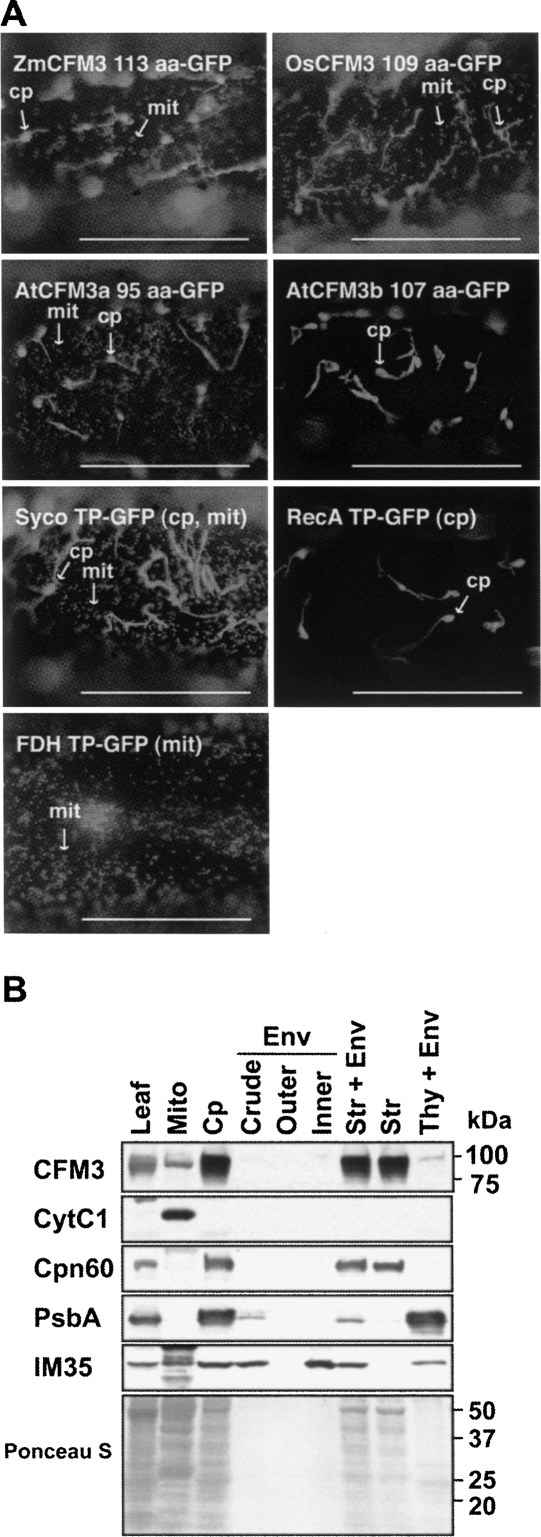
FIGURE 2.
Subcellular localization of CFM3 orthologs. (A) Localization of GFP fused to the N terminus of maize (Zm), rice (Os), or Arabidopsis CFM3 orthologs when expressed transiently in onion epidermal cells. The number of CFM3 amino acids used in each construct is indicated. GFP fused to the transit peptide of chloroplast RecA, mitochondrial FDH, or Syco (cysteinyl-tRNA synthetase) (Peeters et al. 2000) were used as controls to visualize chloroplast, mitochondrial, or dual-targeted GFP. Bars=5 μm. (B) Subcellular fractionation demonstrating the localization of ZmCFM3 to the chloroplast stroma and to mitochondria. Chloroplast (Cp) and chloroplast subfractions (Env, envelope membranes; Str, stroma; Thy, thylakoid membranes) were from a preparation described previously (Williams and Barkan 2003); each of these lanes contains material derived from the same amount of chloroplasts. Cytochrome C1 (Cyt C1), chaperonin 60 (Cpn60), the PsbA subunit of photosystem II (PsbA), and IM35 are markers for mitochondria, chloroplast stroma, thylakoid membranes, and chloroplast inner envelope membranes, respectively. The Ponceau S-stained filter is shown below.
An antibody was raised to a segment of ZmCFM3 that does not show significant similarity to other plant proteins (amino acids 76–221). This antibody detected a protein of the size expected for mature CFM3 (∼88 kDa) in chloroplast stroma and in mitochondria purified from green or etiolated maize leaf tissue, respectively (Fig. 2B). The GFP-fusion data together with the cell fractionation data demonstrate that ZmCFM3 is dual-targeted to chloroplasts and mitochondria. Likewise, genetic data described below support the notion that AtCFM3a is dual targeted.
CFM3 and CFM2 associate in vivo with distinct subsets of CAF/CRS2-dependent group II introns
As an initial screen to identify the RNA ligands of ZmCFM3, we used a “RIP-Chip” assay in which RNAs that coimmunoprecipitate with ZmCFM3 from chloroplast extract were identified by hybridization to a tiling microarray of the maize chloroplast genome (Schmitz-Linneweber et al. 2005). Immunoprecipitations were performed with antibodies from two different rabbits immunized with the CFM3 antigen. The results are summarized in Figure 3A as the log-transformed enrichment ratio for sequences corresponding to each array fragment, plotted according to chromosomal position. The previously reported data for CFM2 (Asakura and Barkan 2007) are plotted on the same graph for comparison (Fig. 3A, gray line). RNAs from six chloroplast loci showed strong enrichment in the CFM3 immunoprecipitations, in comparison with CFM2 and negative control immunoprecipitations (Fig. 3A; data not shown). The coimmunoprecipitation of these RNAs with CFM3 was validated by slot-blot hybridization of RNAs recovered from CFM3 immunoprecipitation pellets and supernatants (Fig. 3B).
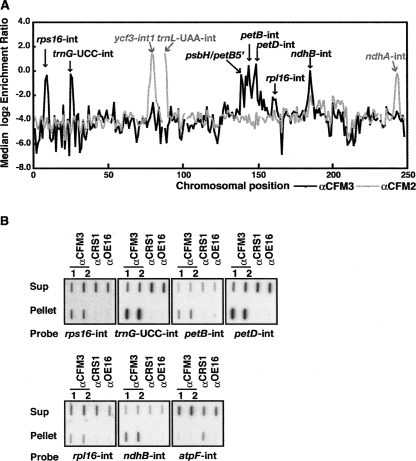
FIGURE 3.
Identification of chloroplast RNA ligands of ZmCFM3 in coimmunoprecipitation assays. (A) Summary of RIP-Chip data demonstrating the different populations of chloroplast RNAs that associate with CFM3 and CFM2. The median log2 transformed enrichment ratios (F635/F532, representing signal in the immunoprecipitation pellet/supernatant), are plotted according to chromosomal position for three replicate CFM3 RIP-Chip assays (black line). The RIP-Chip results reported previously for CFM2 (Asakura and Barkan 2007) are shown for comparison (gray line). Fragments for which fewer than two replicate spots per array yielded an F532 signal above background were excluded and appear as gaps in the curve. (B) Validation of RIP-Chip data by slot-blot hybridization. RNA purified from the pellets and supernatants of immunoprecipitations with antisera to the indicated proteins was applied to slot blots and hybridized to intron-specific probes. Antisera from different rabbits immunized with the CFM3 antigen were used in replicate assays (αCFM3 1 and 2). An immunoprecipitation with antibody to OE16, a protein in the thylakoid lumen that does not bind RNA, was used as a negative control. CRS1 is an atpF-specific chloroplast splicing factor and was analyzed as an additional control. One-third of the RNA recovered from each immunoprecipitation pellet and one-sixth of the RNA recovered from each supernatant was applied to each slot.
These experiments showed that CFM3 is associated in vivo with RNAs from the petB, petD, ndhB, rpl16, rps16, and trnG–UCC loci. Each of these genes encodes a subgroup IIB intron that requires either CRS2/CAF1 or CRS2/CAF2 for its splicing. CFM2, by contrast, associates with RNAs from the ycf3 and ndhA loci, which contain CRS2/CAF-dependent subgroup IIB introns, and the trnL–UAA locus, which contains a group I intron (Asakura and Barkan 2007). The single trans-spliced intron in land plant chloroplasts, rps12–int1, requires CRS2/CAF2 for its splicing (Ostheimer et al. 2003) but is not represented among the ligands of CFM2 and CFM3. Thus, all of the cis-spliced introns in maize that require CRS2/CAF complexes for their splicing (rps16, trnG–UCC, ycf3–int1, petB, petD, rpl16, ndhB, ndhA) associate with either CFM2 or CFM3, but not both.
AtCFM3a and AtCFM3b act redundantly to promote an essential step in Arabidopsis seed development
To determine whether CFM3 promotes the splicing of the RNAs with which it associates, we sought ZmCfm3 mutants in a reverse genetic screen of our collection of transposon-induced nonphotosynthetic maize mutants (Stern et al. 2004). Previous screens of this nature recovered mutations in many nuclear genes involved in chloroplast RNA splicing (caf1, caf2, crs1, rnc1, ppr4, and others), but we were unable to identify insertions in ZmCfm3. The mutant collection is near saturation for genes whose disruption results in chlorophyll-deficient seedlings (data not shown), so the failure to obtain ZmCfm3 mutants suggests that disruption of this gene may not cause such a phenotype. A mitochondrial function for ZmCFM3, as suggested by its dual-targeting to chloroplasts and mitochondria, could result in an embryo-defective mutant phenotype, which would preclude representation of the mutant in our collection. Alternatively, a functionally redundant duplicate locus might account for the absence of ZmCfm3 mutants in our collection, as partial sequence for a second cfm3-like gene can be detected in the available maize genome sequence data (data not shown).
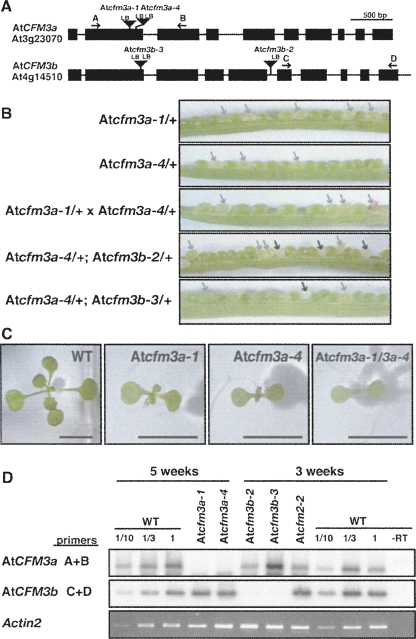
Because genetic analysis of CFM3 in maize was problematic, we obtained Arabidopsis mutants with T-DNA insertions in AtCFM3a or AtCFM3b from the SALK and SAIL collections (Sessions et al. 2002; Alonso et al. 2003). Pale green seeds segregated among the progeny of plants heterozygous for each of two exonic insertions in AtCFM3a (Atcfm3a-1 and Atcfm3a-4) (Fig. 4B). Approximately 40% of the pale seeds in each line failed to germinate; those that did germinate yielded very slow-growing seedlings (Fig. 4C) that are homozygous for the insertion (Supplemental Fig. 4A) and that lack AtCFM3a mRNA (Fig. 4D). The heteroallelic progeny of crosses between Atcfm3a-1/+ and Atcfm3a-4/+ plants exhibited these same phenotypes in similar frequencies (Fig. 4B,C; Supplemental Fig. 4B), demonstrating that these phenotypes result from disruption of AtCFM3a. Thus, loss of AtCFM3a function results in an incompletely penetrant defective seed phenotype, with the “escapes” germinating to yield severely stunted plants.
FIGURE 4.
AtCFM3a and CFM3b insertion mutants. (A) T-DNA insertions in AtCFM3a (At3g23070) and AtCFM3b (At4g14510). Exons and introns are indicated by rectangles and lines, respectively. Arrows show primers used for RT-PCR. LB, T-DNA left border. The sequences flanking each insertion are shown in Supplemental Figure 3. (B) Seed phenotypes associated with insertions in AtCFM3a and AtCFM3b. The siliques shown resulted from crosses between plants of the indicated genotypes (self-pollination where just one genotype is shown). Pale green and white seeds are indicated with gray and black arrows, respectively. Approximately two-thirds of the pale green seeds germinated to yield slow-growing seedlings (see C), whereas approximately one-third did not germinate. The white seeds did not germinate. (C) Stunted seedling phenotype caused by insertions in AtCFM3a. Plants of the indicated genotype (homozygous unless otherwise indicated) were grown for 17 d on MS medium containing 2% sucrose. Plants homozygous for the insertions in AtCFM3b have no visible phenotype (data not shown). Bar=5 mm. (D) RT-PCR demonstrating a reduction in mRNA from the disrupted loci. Total leaf RNA (200 ng) was analyzed by RT-PCR using the primer pairs indicated to the left and diagrammed in A. The age of the plants used for RNA extraction is shown at the top; the very slow growth of Atcfm3a mutants required an extended period of growth to obtain sufficient material. The Atcfm2-2 mutant was used as a control to show the mRNA levels in a different chloroplast splicing mutant. Actin2 mRNA was used an internal control. The dilution series of the wild-type sample demonstrates that the assay is roughly linear over the relevant concentration range. CFM3 RT-PCR products were detected by DNA gel blot hybridization. The actin2 RT-PCR product was detected with ethidium bromide.
Two T-DNA insertions in AtCFM3b likewise caused the loss of mRNA from the disrupted locus (Fig. 4D), but these did not cause a visible phenotype on their own. However, both insertions caused a synthetic phenotype when combined with a mutation in AtCFM3a: the progeny of self-pollinated plants that were heterozyogous for either AtCFM3b mutant allele and for Atcfm3a-4 segregated white seeds that failed to germinate (Fig. 4B). This phenotype was never observed among the progeny of plants harboring any of these mutant alleles on their own. Forty-seven germinating progeny of the double-heterozygotes were genotyped by PCR, and none of these were homozygous for both insertions (Supplemental Fig. 4C). The synthetic lethality resulting from the simultaneous disruption of both AtCFM3 co-orthologs indicates that AtCFM3a and AtCFM3b act redundantly in a process that is necessary for seed development.
AtCFM3a is required for the splicing of the chloroplast ndhB intron
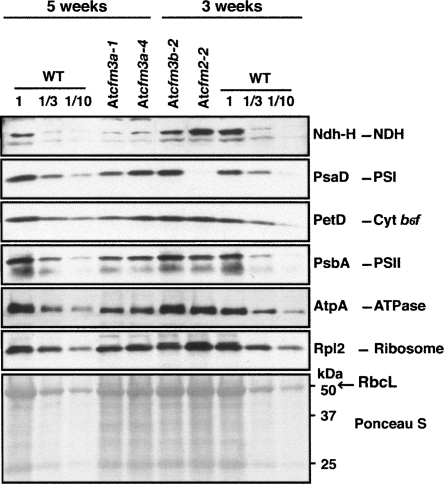
AtCFM3a homozygous mutants, although very slow growing, provided sufficient tissue for some analyses of organellar gene expression. Immunoblots were used to quantify components of the chloroplast multisubunit complexes that include plastid-encoded subunits (Fig. 5). The ribosomal protein Rpl2, the photosystem I subunit PsaD, the photosystem II subunit PsbA, the cytochrome b6f subunit PetD, the ATP synthase subunit AtpA, and the Rubisco subunit RbcL accumulate to near normal levels in AtCFM3a mutants, indicating that chloroplast gene expression as a whole is not substantially compromised. The NdhH subunit of the chloroplast NADH dehydrogenase may be reduced slightly more than these other proteins. These subtle protein defects are consistent with the fact that AtCFM3a mutants are not chlorophyll deficient, and contrast with the chlorosis and global loss of this same protein set in AtCAF1 and AtCAF2 mutants (Asakura and Barkan 2006).
FIGURE 5.
Immunoblot analyses of subunits of chloroplast protein complexes in AtCFM3 mutants. Total leaf extract (20 μg protein or dilutions as indicated) from seedlings of the ages indicated were analyzed on immunoblots by probing with antibodies for the proteins named to the right. The Ponceau S-stained membrane at the bottom illustrates sample loading and the abundance of the large subunit of Rubisco (RbcL).
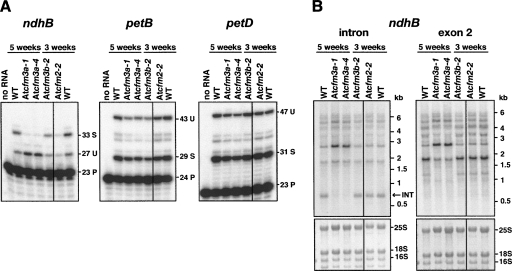
The splicing of each chloroplast RNA whose ortholog coimmunoprecipitated with ZmCFM3 was analyzed in Atcfm3 mutants. Poisoned-primer extension assays showed that the ratio of spliced to unspliced RNAs from the ndhB locus was dramatically reduced (Fig. 6A). RNA gel blot data showed that unspliced ndhB transcripts accumulate to increased levels, whereas the excised intron fails to accumulate in AtCFM3a mutants, confirming a defect in the splicing of the ndhB intron (Fig. 6B). The splicing of the five other introns orthologous to ZmCFM3′s ligands, and of the three chloroplast introns that are found in Arabidopsis but not in maize (clpP–int1, clpP–int2, rpoC1), were unaffected in AtCFM3a mutants (Fig. 6A; Supplemental Fig. 5). A chloroplast defect specifically in the splicing of the ndhB intron in Atcfm3a mutants is consistent with their lack of chlorosis and their lack of gross plastid protein deficiencies, as an Arabidopsis mutant that fails to express ndhB due to a different RNA processing defect lacks a visible phenotype (Hashimoto et al. 2003). In contrast, the failure to splice any of the other chloroplast RNAs that associate with CFM3 would be expected to reduce the accumulation of one or more photosynthetic enzyme complex, compromise photosynthesis, and lead to chlorosis.
FIGURE 6.
Chloroplast ndhB splicing defect in AtCFM3a mutants. (A) Poisoned-primer extension assays monitoring splicing of several introns. Ten micrograms of total leaf RNA from plants homozygous for the indicated alleles was analyzed. The length in nucleotides of each product is shown to the right. U, unspliced; S, spliced; P, primer. (B) RNA gel blots confirming an ndhB splicing defect in AtCFM3a mutants. Total leaf RNA (4 μg) was probed with an intron-specific or exon 2-specific probe. The same blot stained with methylene blue is shown below, with rRNA bands marked. INT, excised intron.
These results demonstrated that AtCFM3a is necessary for the splicing of the group II intron in the chloroplast ndhB pre-mRNA. Because AtCFM3b and AtCFM3a are, in part, functionally redundant, these results do not preclude a role for At CFM3a/b in the splicing of additional introns. There is strong evidence that chloroplast translation is essential for embryogenesis in Arabidopsis (see, e.g., Stern et al. 2004; Duchene et al. 2005; Schmitz-Linneweber et al. 2006). Thus, a redundant role for AtCFM3a and AtCFM3b in the splicing of any of its ligands that function in translation (e.g., trnG–UCC or rpl16) could contribute to the defective seed phenotype of Atcfm3a/Atcfm3b double mutants.
OsCFM3 is required for the splicing of several CRS2/CAF-dependent introns
Mutants lacking plastid ribosomes in graminaceous plants (e.g., maize, barley, rice) have been reported frequently; these germinate to yield albino seedlings that die after several weeks, when seed reserves are exhausted (e.g., Walbot and Coe 1979; Hess et al. 1993; Jenkins et al. 1997; Williams and Barkan 2003; Gothandam et al. 2005; Schmitz-Linneweber et al. 2006). Therefore, we initiated a genetic analysis of the single CFM3 ortholog in rice to bypass the difficulties introduced by the redundant embryo-essential functions of CFM3a and CFM3b in Arabidopsis.
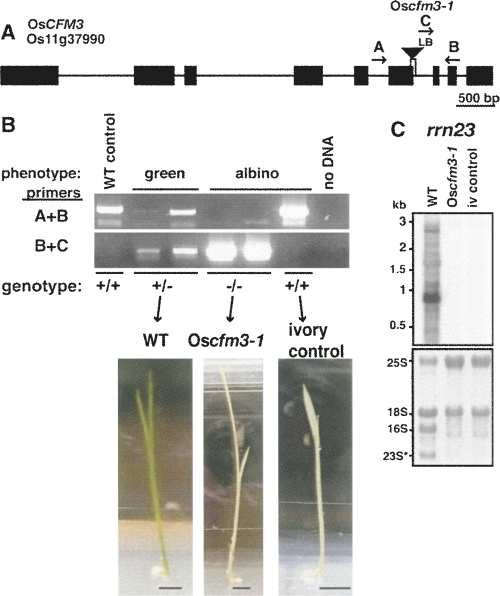
We obtained a small number of seeds from the Postech insertion collection (Jeong et al. 2006) from a rice line with a T-DNA insertion in OsCFM3. Five of the seeds germinated, two of which were homozygous for the insertion in OsCFM3 (Fig. 7B). The homozygous mutants had albino leaves (Fig. 7B) and contained little, if any, plastid rRNA (Fig. 7C). Albino leaves that lack plastid ribosomes would be expected to result from the failure to splice the CFM3 ligands trnG–UCC, rps16, and/or rpl16, each of which is essential for translation, and thus for synthesis of ribosomes themselves. A third rice seed gave rise to an albino plant that did not harbor the insertion in OsCFM3 (Fig. 7B). This plant did, however, lack plastid rRNAs (Fig. 7C) and served as a control in subsequent experiments to reveal pleiotropic defects resulting from a plastid ribosome deficiency. This plant is referred to as the “ivory” control in experiments described below.
FIGURE 7.
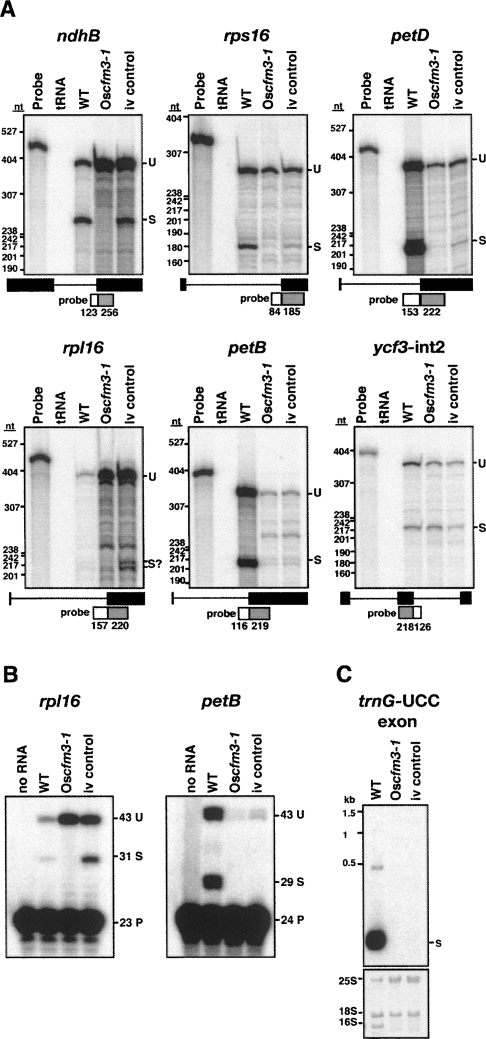
OsCFM3 T-DNA insertion mutant. (A) T-DNA insertion in OsCFM3. Exons and introns are indicated by rectangles and lines, respectively. A deletion of a 3′ splice site accompanied the T-DNA insertion, as shown by the vertical lines below the triangle. Arrows show the primers used for PCR genotyping. LB, T-DNA left border. (B) Phenotype and genotype of the five germinating plants from the Postech T-DNA line 3A-16097. Rice seedlings were grown for 3 wk on MS medium containing 2% sucrose. Bar=5 mm. The primer pairs used are indicated to the left and diagrammed in A. The inferred OsCFM3 genotype is shown below. Two of the albino seedlings were homozygous for the insertion in OsCFM3. The third albino seedling lacked this insertion, and was used as a control for the RNA analyses (ivory control). (C) RNA gel blot showing loss of plastid 23S rRNA in Oscfm3-1 mutant. The probe is from the 5′ portion of the 23S rRNA gene and detects just one of the two 23S rRNA fragments that accumulates in vivo. Shown below is the same blot stained with methylene blue. 16S is the plastid 16S rRNA. 23S* marks the smaller of the two 23S rRNA fragments; the larger fragment comigrates with cytosolic 18S rRNA.
The number of experiments that could be performed with the Oscfm3 mutant was limited by the very small amount of tissue available. Nonetheless, by using sensitive ribonuclease-protection assays, we were able to monitor the splicing of each RNA whose maize ortholog coimmunoprecipitated with ZmCFM3 (Fig. 8A). The results demonstrated that CFM3 is required for ndhB splicing in rice, as in Arabidopsis. The results also provided strong evidence that OsCFM3 is necessary for the splicing of the petD and rps16 RNAs: these spliced RNAs were virtually absent in the Oscfm3 mutant, and were reduced less so in the ivory control mutant. The ribonuclease protection data further suggested that rpl16 splicing relies on OsCFM3, but noise in the data precluded an unambiguous interpretation. However, a poisoned-primer extension assay (Fig. 8B) confirmed the specific loss of spliced rpl16 transcripts in the Oscfm3 mutant. Ribonuclease protection and poisoned-primer extension assays demonstrated the absence of spliced petB RNA (Fig. 8A,B), and RNA gel blot hybridization demonstrated the absence of spliced trnG–UCC RNA (Fig. 8C) in the Oscfm3 mutant, consistent with a requirement for CFM3 in their splicing. However, the control albino mutant also lacked these spliced RNAs so firm conclusions could not be made regarding a direct role for CFM3. The ycf3–int2 intron is spliced normally in the Oscfm3 mutant (Fig. 8A), correlating with its lack of coimmunoprecipitation with ZmCFM3 (Fig. 3).
FIGURE 8.
Chloroplast splicing defects in the Oscfm3-1 mutant. RNA was analyzed from a normal seedling (WT), a Oscfm3-1 homozygote, and from an OsCFM3+ albino mutant segregating in the same background (iv control). (A) Ribonuclease protection assays. Total leaf RNA (2 μg) was analyzed using probes spanning either the 3′ or 5′ splice junction, as diagrammed. The length in nucleotides of probe segments corresponding to intron and exon sequences are shown in the diagrams. The positions of DNA size standards are shown to the left. U, unspliced; S, spliced. (B) Poisoned-primer extension assays. Total leaf RNA (4 μg) was used in reverse-transcription reactions using primers mapping several nucleotides downstream of the indicated intron. A dideoxynucleotide present in each reaction terminates reverse-transcription after different distances on spliced and unspliced RNA templates. The length in nucleotides of each product is indicated. U, unspliced; S, spliced; P, primer. (C) RNA gel blot showing loss of trnG-UCC in Oscfm3-1 mutant. Total leaf RNA (2 μg) was probed with a maize trnG-UCC exon probe. The same blot stained with methylene blue is shown in below.
The genetic data from Arabidopsis and rice in conjunction with the RNA coimmunoprecipitation data from maize together provide strong evidence that CFM3 associates with and promotes the splicing of the chloroplast ndhB, rpl16, rps16, and petD introns. CFM3 also coimmunoprecipitates with the trnG–UCC and petB introns, and the genetic data are consistent with a requirement for CFM3 in their splicing.
CFM3 is associated in vivo with the group II intron splicing factors CAF1, CAF2, and RNC1
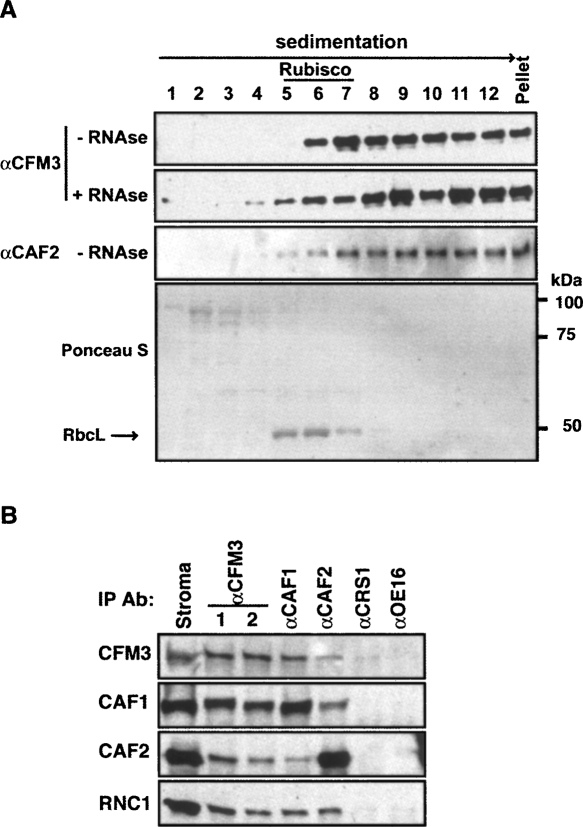
Among CFM3′s RNA ligands are introns that also associate with CAF1, CAF2, and/or the RNAse III-domain splicing factor RNC1 (Ostheimer et al. 2003; Schmitz-Linneweber et al. 2005; Watkins et al. 2007). Therefore, it was of interest to determine whether CFM3 is found in the same ribonucleoprotein particles as these previously identified splicing factors. After sedimentation of stromal extract through a sucrose gradient, ZmCFM3 was found in a broad high molecular weight peak that sediments more rapidly than Rubisco (∼550 kDa) (Fig. 9A) and that coincides with the peaks reported previously for RNC1, CAF1, and CAF2 (Watkins et al. 2007). Furthermore, antibodies to CFM3 coimmunoprecipitate each of these proteins from chloroplast extract and vice versa (Fig. 9B). Together, these cosedimentation and coimmunoprecipitation data provide strong evidence that CFM3, a CAF/CRS2 complex, and RNC1 associate simultaneously with their shared intron ligands.
FIGURE 9.
CFM3 is associated with CAF1 and CAF2 in large particles in the chloroplast stroma. (A) Untreated (−RNase) and ribonuclease-A-treated (+ RNase) maize stromal extracts were sedimented through sucrose gradients under conditions in which particles greater than ∼70S pellet. An equal portion of each fraction was analyzed on immunoblots by probing with CFM3 or CAF2 antiserum. The CFM3 blot stained with Ponceau S is shown below to illustrate the position of Rubisco in the gradient (RbcL, large subunit of Rubisco). The modest effect of RNAse-treatment on the sedimentation of CFM3 particles is characteristic of group II intron ribonucleoprotein partices, which are resistant to RNase treatment (Till et al. 2001; Asakura and Barkan 2007; Watkins et al. 2007). CAF1 and RNC1 were shown previously to be recovered in analogous gradient fractions (Watkins et al. 2007). (B) CFM3 coimmunoprecipitates with CAF1 and CAF2. Chloroplast stroma was used for immunoprecipitations with the antibody indicated at the top. Antisera from two rabbits immunized with the CFM3 antigen were analyzed (lanes 1,2). Immunoblots of the immunoprecipitated proteins were probed with the antibodies indicated to the left.
Functions for CFM3 in mitochondria
GFP-fusion and cell fractionation data indicated that ZmCFM3 is dual targeted to chloroplasts and mitochondria. GFP-fusion data likewise suggested that AtCFM3a is dual-targeted. A mitochondrial function for AtCFM3a is supported by the slow-growth phenotype of AtCFM3a mutants because mutations in Arabidopsis that solely disrupt photosynthesis cause only a small reduction in growth rate when seeds are sown on medium containing sugars. For example, AtCAF1, AtCRS1, and AtCAF2 mutants are nearly albino, are severely deficient for numerous chloroplast-encoded proteins, yet grow much more rapidly than AtCFM3a mutants on MS medium (Asakura and Barkan 2006). The only molecular defect detected in the chloroplasts of AtCFM3a mutants was in ndhB splicing; this defect cannot account for their stunted growth, as Arabidopsis mutants lacking solely the chloroplast NDH complex have no visible phenotype (Hashimoto et al. 2003; Kotera et al. 2005).
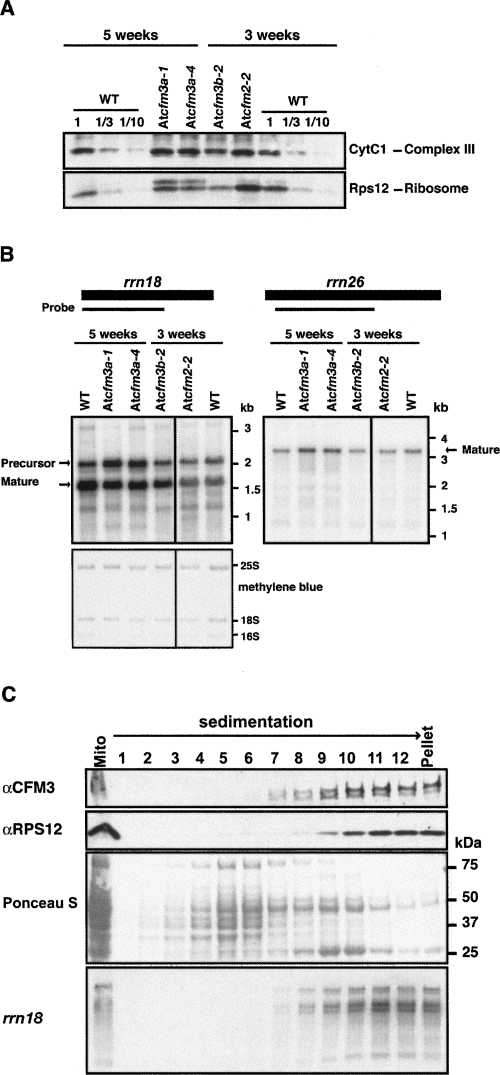
To gain insight into the role of AtCFM3a in mitochondria, the abundance of several mitochondrial proteins was assayed in those Atcfm3a mutants that succeeded to germinate (Fig. 10A). Cytochrome c1, a nucleus-encoded subunit of the cytochrome bc1 complex, accumulated to normal levels in these mutants. However, an antibody to the mitochondrially encoded ribosomal protein RPS12 detected an aberrant, slowly migrating isoform specifically in Atcfm3a mutants (Fig. 10A). The metabolism of mitochondrial 18S rRNA is also altered in these mutants: a large rRNA isoform (∼2.1 kb), presumably a precursor, is overrepresented, whereas the presumed mature form (∼1.7 kb) is underrepresented slightly (Fig. 10B). These results suggest that AtCFM3a influences the biogenesis of the mitochondrial small ribosomal subunit. Furthermore, ZmCFM3 cosediments with mitochondrial small ribosomal subunits, as indicated by its cosedimentation with 18S rRNA and RPS12 when mitochondrial extract was fractionated by sucrose gradient sedimentation (Fig. 10C). However, antibodies to ZmCFM3 did not unambiguously coimmunoprecipitate 18S rRNA from mitochondrial extract (data not shown). These results suggest that ZmCFM3 either associates with a low abundance intermediate in ribosome assembly, or that it influences ribosome metabolism indirectly and that its cosedimentation with ribosomes is coincidental. Clarification of the role of CFM3 in mitochondria will require further study.
FIGURE 10.
Analyses of CFM3 function in mitochondria. (A) Immunoblots of total leaf extract were probed with antibodies to cytochrome c1 or mitochondrial Rps12. The corresponding Ponceau S stained blot is shown in the bottom panel of Figure 5. (B) RNA gel blot analysis of total leaf RNA (0.5 μg), using probes from the mitochondrial rrn18 and rrn26 genes. The forms marked as “mature” are based on the annotated termini of these rRNAs. The methylene blue-stained blot is shown below. (C) Sucrose-gradient fractionation of maize mitochondrial extract. Maize mitochondria were solubilized with 1% NP-40 and sedimented through a sucrose gradient under conditions in which particles greater than ∼70S pellet. An equal portion of each fraction was analyzed on immunoblots, by probing with CFM3 or RPS12 antiserum. RNA purified from the same fractions was analyzed by RNA gel blot hybridization, using a probe for rrn18 (bottom panel).
DISCUSSION
The results presented here elucidate the nature of the chloroplast RNA splicing machinery and its relationship to the expansion and diversification of the CRM domain protein family in the plant lineage. The CRM domain family is derived from a single-domain prokaryotic protein whose extant representative in Escherichia coli, YhbY, functions in ribosome maturation (Barkan et al. 2007). The duplication of this ancestral gene to yield a family of single- and multidomain proteins appears to be confined to plants. All characterized members of the family in plants function in the splicing of chloroplast introns: 13 of the group II introns and the sole group I intron in angiosperm chloroplasts have been shown to require one or more CRM domain protein for their splicing (Fig. 11). Both the acquisition of group II introns in land plant chloroplasts and the expansion of the CRM domain family occurred after the divergence of land plants and chlorophytes (green algae) but prior to the divergence of vascular and nonvascular plants, suggesting that these two events may be evolutionarily linked.
The first three CRM domain splicing factors to be characterized, CRS1, CAF1, and CAF2, act on largely nonoverlapping intron subsets, consistent with the possibility that different CRM proteins interact with their intron substrates in similar locations and play similar roles in splicing. However, with the recent characterization of CFM2 and CFM3 that scenario is no longer plausible. CFM2 interacts with and promotes the splicing of two introns that also require CRS2/CAF complexes (ycf3–int1 and ndhA) (Asakura and Barkan 2007). Data presented here show that CFM3 associates with all but one of the CRS2/CAF-dependent introns that do not associate with CFM2, and that there is no overlap among the intron sets that coimmunoprecipitate with CFM2 and CFM3 (see summary in Fig. 11A). Genetic data demonstrate that CFM3 promotes the splicing of four of the RNAs with which it associates (ndhB, rpl16, rps16, and petD), and are consistent with a role for CFM3 in the splicing of its remaining two ligands, the trnG and petB RNAs. However, the redundancy of the two CFM3 co-orthologs in Arabidopsis and the absence of these spliced RNAs in a different albino rice mutant preclude firm conclusions about trnG and petB. It seems likely, however, that CFM3 does promote the splicing of these introns, given the perfect correspondence between the set of introns that coimmunoprecipitate with each previously characterized CRM splicing factor (CRS1, CAF1, CAF2, CFM2) and the set that fails to splice in the corresponding mutants (Ostheimer et al. 2003; Schmitz-Linneweber et al. 2005; Asakura and Barkan 2007).
The division of labor among the CAF1/2 and CFM2/3 CRM proteins in the splicing of chloroplast subgroup IIB introns is summarized in Figure 11A. CRS2 functions in concert with either CAF1 or CAF2 to promote the splicing of all but one of the subgroup IIB introns. CRS2/CAF complexes, in turn, function in concert with either CFM2 or CFM3 to promote the splicing of all but one of the CRS2/CAF-dependent introns. The genetic disruption of either the CAF or CFM subunit of each ribonucleoprotein particle blocks splicing, indicating that these subunits play nonredundant roles. These results suggest that the closely related paralogs CAF1 and CAF2 play similar biochemical roles in splicing but bind different introns, and that the situation is analogous for the closely related paralogs CFM2 and CFM3. CRS1, which is in the same CRM subfamily as CFM2 and CFM3, has been shown to interact with domains 1 and 4 in its atpF intron substrate (a subgroup IIA intron), and to enhance intron folding (Ostersetzer et al. 2005); it will be interesting to determine whether CFM2 and CFM3 function analogously. CAF1 and CAF2, in contrast, recruit the splicing factor CRS2 to specific introns, and this may be their primary role. CRS2 is a peptidyl-tRNA hydrolase homolog that plays an unknown but essential role in the splicing of all but one of the subgroup IIB introns in the chloroplast.
The rps12–int1 and ycf3–int2 introns are the only plant chloroplast subgroup IIB introns that associate with neither CFM2 nor CFM3 (Fig. 11A). These introns are noncanonical in other ways as well. rps12–int1 is the sole plant chloroplast intron that is trans-spliced, and this intron requires a dedicated splicing factor called PPR4 (Schmitz-Linneweber et al. 2006). ycf3–int2 is the only chloroplast subgroup IIB intron that splices independently of a CAF/CRS2 complex. This intron also relies on a dedicated splicing factor, called OTP51 (de Longevialle et al. 2008). Interestingly, both PPR4 and OTP51 consist of pentatricopeptide repeats (PPRs) followed by an accessory RNA binding domain: an RRM domain in the case of PPR4 and a LAGLIDADG domain in the case of OTP51. Although CRS1-like CRM splicing factors have not been identified for rps12–int1, ycf3–int2, or for most of the chloroplast subgroup IIA introns (Fig. 11B), two members of the CRS1 subfamily have not yet been analyzed.
Our results show that CFM3 is a dual-targeted protein that localizes not only to chloroplasts, but also to mitochondria. GFP fused to the N terminus of ZmCFM3, AtCFM3a, or OsCFM3 localized to both chloroplasts and mitochondria in transient expression assays. Furthermore, ZmCFM3 was detected in purified chloroplast and mitochondrial fractions by immunoblotting. Finally, the AtCFM3a mutant phenotype points to a mitochondrial function: Atcfm3a mutants grow very slowly even when supplied with exogenous sugars, unlike mutants whose defects are limited to photosynthesis. Furthermore, aberrant mitochondrial 18S rRNA metabolism and an aberrant mitochondrial RPS12 protein isoform were detected in these mutants. Elucidation of the precise role of CFM3 in mitochondria and whether this function is conserved among species will require further study.
To explore the functions of CFM3 we took advantage of three different organisms (maize, rice, and Arabidopsis). The information obtained from each was incomplete, but together the data provide a compelling picture for the role of CFM3 in chloroplast splicing. The consistency of the findings among the three species strongly suggests that CFM3′s functions in the chloroplast are largely conserved among monocots and dicots, like those of CRS1, CAF1, and CAF2 (Asakura and Barkan 2006). Despite the powerful genetic resources available for Arabidopsis, its utility in this study was limited by the lineage-specific CFM3 gene duplication and by the embryo-lethality resulting from the disruption of both CFM3 co-orthologs. The embryo essentiality of chloroplast translation in Arabidopsis complicates the genetic analysis of nuclear genes that are necessary for the expression of any essential chloroplast ribosomal protein, tRNA or rRNA (Williams and Barkan 2003; Asakura and Barkan 2006; Schmitz-Linneweber et al. 2006; Asakura and Barkan 2007). Several such RNAs are included among CFM3′s RNA ligands (rpl16, rps16, trnG–UCC); an essential role for AtCFM3 in the metabolism of any of these RNAs could account for the embryo-lethality associated with simultaneous disruption of both AtCFM3a and AtCFM3b. These introns fail to splice in the Oscfm3 mutant, and in crs2 and caf1 mutants in maize (Jenkins et al. 1997; Ostheimer et al. 2003); these and other mutants lacking plastid ribosomes in the grasses germinate to yield albino, seedling lethal mutants whose molecular defects can be analyzed. Thus, the use of a variety of model organisms will be necessary to achieve a comprehensive understanding of the mechanisms underlying chloroplast gene expression and its control in higher plants.
MATERIALS AND METHODS
Sequence analysis of maize Cfm3 cDNAs
The nucleotide sequence of OsCfm3 (Os11g37990) was used to identify closely related maize cDNAs at the POGs (http://plantrbp.uoregon.edu/) and Plant GDB (http://www.plantgdb.org/) databases. Corresponding maize cDNAs from the Arizona full-length cDNA collection (Accession numbers; DR823593.1 and EB158689.1) were obtained and sequenced. EB158689.1 was truncated at the 5′ end, and DR823593.1 included an intron with an in-frame stop codon. To confirm the sequence of exon 1, a DNA fragment was amplified from our maize seedling leaf cDNA library (inbred line B73) by nested PCR using primers as follows: first PCR, YA654Cfm35′ (5′-AGGGTCAAGGTCTCTGGCAC-3′) and YA1191Cfm3′ (5′-AGCAAAGTGAGGAGGCAGT-3′); nested PCR, YA680Cfm35′ (5′-GCTCAATATGCGCCTCTTCC-3′) and YA1153Cfm33′ (5′-TTGGTAGTGTCCCTTCGGCT-3′). The PCR product was cloned into pGEM-T (Promega) and its sequence was determined. The ZmCfm3 cDNA sequence deposited in GenBank (Accession No. EU084957) is the composite sequence derived from these three cDNAs.
Plant growth and extract preparation
Maize chloroplasts were purified from the leaves of seedlings (inbred line B73; Pioneer HiBred) grown in soil in a growth chamber at 28°C under a 16-h light (400 μE m−2 S−1)/8-h dark cycle for several days until the coleoptiles emerged, transferred to the dark for ∼6 d until two leaves have emerged, and then shifted to the light for ∼12 h prior to harvest. Maize chloroplasts and stromal extract were prepared as described previously (Ostheimer et al. 2003). Maize chloroplast subfractions were from the same preparation described in Williams and Barkan (2003).
Maize mitochondria were purified from the leaves of seedlings grown in soil in a growth chamber under a 16-h light (28°C; 400 μE m−2 S−1)/8-h dark cycle for 4 d, then incubated in the dark at 28°C for 6 to 7 d. Mitochondria were purified on sucrose step gradients as described in Newton (1994). Mitochondrial extract for sucrose gradient analysis was obtained by suspending the mitochondria in a minimal volume of 1% NP-40, 50 mM Tris-HCl at pH 8.0, 7 mM MgCl2, 150 mM KOAc, 2 mM DTT, 2 μg/mL aprotinin, 2 μg/mL leupeptin, 1 μg/mL pepstein, 1 mM phenylmethylsulfonyl fluoride, 20 μL/mL RNasin [Promega]), and subjecting them to three freeze–thaw cycles. After addition of glycerol to 10% (v/v), the lysate was divided into small aliquots and stored at −80°C.
Arabidopsis T-DNA insertion lines were obtained from the Salk Institute Genomic Analysis Laboratory: SALK_125653 (Atcfm3a-1), SAIL_595_F03 (Atcfm3a-4), SALK_039803 (Atcfm3b-2), and SAIL_23_C02 (Atcfm3b-3). Atcfm2-2 (SAIL_67_E05), which conditions pale green seedling-lethal mutants with several chloroplast splicing defects (Asakura and Barkan 2007), was used as a mutant control. Arabidopsis plants used for RNA and protein analysis were 3-wk-old (Atcfm3b-2 and Atcfm2-2) or 5-wk-old (Atcfm3a-1 and Atcfm3a-4) seedlings grown on MS plates containing 2% sucrose, as described previously (Asakura and Barkan 2006). Total Arabidopsis leaf protein and RNA were extracted as described previously for maize (Barkan 1998).
The rice T-DNA insertion line 3A-16097 was obtained by Maureen Hanson (Cornell University) from POSTECH (Jeong et al. 2006). An albino mutant plant that segregated among the progeny but that lacked the T-DNA insertion in OsCFM3 was used as a control for RNA analyses. The cultivar “Dongjing,” which is the background from which this line was derived, was used as the source of wild-type RNA. The growth of the rice seedlings was performed at Cornell University by John Robbins. Rice seeds were surface sterilized and planted on Murashige and Skoog medium (0.5% [w/v] Caisson agar [Caisson Laboratories, North Logan, Utah], 1× Murashige and Skoog salts [Sigma-Aldrich], 1× Gamborg's B5 vitamin [Sigma-Aldrich], and 2% [w/v] sucrose at pH 5.8) until the third leaf stage under 12-h light (400 μE m−2 S−1; 28°C)/12-h dark (21°C) cycle.
Production of anti-CFM3 antisera
A segment of ZmCFM3 (amino acids 76–221) was expressed in Escherichia coli (BL21 Star (DE3), Invitrogen) as a 6xHis-tagged fusion protein from vector pET-28b(+) (Novagen). Protein expression was induced by the addition of 0.5 mM isopropyl-thio-β-D-galactopyranoside for 3 h at 37°C. The recombinant CFM3 fragment was recovered in inclusion bodies, which were solubilized with 8 M urea, 50 mM Tris-HCl at pH 7.5, and 300 mM NaCl. The protein was affinity purified with Ni-NTA agarose beads (Qiagen) according to the manufacturer's protocol. Ten milligrams of purified protein was dialyzed into PBS containing 2 M urea. Polyclonal antisera were generated in two rabbits at Alpha Diagnostic Inc. Antisera were affinity purified against the antigen coupled to HiTrap NHS-activated column (GE Healthcare Life Science).
Transient expression of GFP fusion proteins
Sequences encoding the N-terminal 113 amino acids of ZmCFM3 were amplified by PCR from the ZmCfm3 cDNA with the primers YA1Cfm35′ (SpeI) (5′-CGTACTAGTATGGCCATGGCGTCCTC-3′) and YA339Cfm33′ (KpnI) (5′-CGTGGTACCCGACGTCCTCGACCGACCCCC-3′). Sequences encoding the N-terminal 109 amino acids of OsCFM3 were amplified by nested PCR from rice leaf DNA with the following primers: first PCR, YAOs257Cfm35′ (5′-AAAATATCCTCACCCGCTAAACC-3′) and YAOs1089Cfm33′ (5′-AGCAATTCAGCATTTTCCCGCT-3′); nested PCR, YAOsCFM3-F (SpeI) (5′-CTTACTAGTATGGCCGCCGCGGCCATGGCC-3′) and YAOsCFM3-R (KpnI) (5′-CTTGGTACCGGAGCTCCGGGGGGGAGTAGGA-3′). Sequences encoding 95 and 107 amino acids of AtCFM3a and AtCFM3b, respectively, were amplified by nested PCR with the following primers: AtCFM3a, first PCR, AtCfm3–7 5′ (5′-TTCTATGTGAATAAACCAACAAT-3′) and AtCfm3–8 3′ (5′-TTAGCTCTGATTCCGGAAGAGT-3′); nested PCR, YAAtCfm3GFPF (SpeI) (5′-CTTACTAGTATGGCGATGAAGCCCAGTCTC-3′) and YAAtCfm3GFPR (KpnI) (5′-CTTGGTACCTTTCTTTTCGATAATTCACAAC-3′); AtCFM3b, first PCR, AtCfm4-11 5′ (5′-GCGTAGCTGAAGGAATCAAAGGT-3′) and Atcfm4-6 (5′-GTTGCTGCCACCTTTGTAATTG-3′); nested PCR, YAAtCfm4GFPF (SpeI) (5′-CTTACTAGTATGGCGATTAATTCAAGTC-3′) and YAAtCfm4GFPR (KpnI) (5′-CTTGGTACCTCTCACCACTTATAACCCCTAA-3′). PCR products were digested with SpeI and KpnI and cloned into pOL-LT, which was kindly supplied by Ian Small (University of Western Australia). The particle bombardment into onion epidermal tissues was performed as described previously (Barkan et al. 2007). Samples were observed 2–3 d after the bombardment using an Axoplan microscope (Zeiss) at 40× magnification with a FITC filter.
DNA isolation and PCR-genotyping
DNA was isolated from Arabidopsis as described previously (Asakura and Barkan 2006). T-DNA insertions were confirmed by PCR, using T-DNA left border primer LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) for Atcfm3a-1 and Atcfm3b-2, or LB(SAIL) (5′-TTCATAACCAATCTCGATACAC-3′) for Atcfm3a-4 and and Atcfm3b-3. Gene specific primer pairs were as follows: Atcfm3a-1 and Atcfm3a-4, AtCfm3-5(787) (5′-GGGAGATTTTCTGGGTCGGA-3′) and cfm32 (5′-CCGGCCAGTCCATGAACCTA-3′); Atcfm3b-2, Atcfm4-3 (5′-GAAGTTGGAAAGGAAACTTGCT-3′), and Atcfm4-4 (5′-CGTTTTTGAAGCTCAAGAGAA-3′); Atcfm3b-3, Atcfm4–5 (5′-ATTGGAATAGAACACAGAAGC-3′), and Atcfm4–6 (5′-GTTGCTGCCACCTTTGTAATTG-3′). DNA was isolated from rice using Plant DNAzol reagent (Invitrogen) according to the manufacturer's instructions. The T-DNA insertion was confirmed by PCR using the T-DNA left border primer OsLB2(pGA2715_Postec) (5′-AAACGTCCGCAATGTGTTATT-3′), and the gene-specific primer pair OsYA4702Cfm35′ (5′-CAGGCAGGAGAGGAGTTTTTGAT-3′) and OsYA5411Cfm33′ (5′-CACATCTTCCTCGTCACTGGAAT-3′).
Protein analyses
Antisera to the chloroplast proteins NDH-H, IM35, Cpn60, and RPL2 were kindly provided by Tsuyoshi Endo (Kyoto University), Danny Schnell (University of Massachusetts), Harry Roy (Rensselaer Polytechnic Institute), and A. Subramanian (University of Arizona), respectively. Antisera to maize mitochondrial RPS12, yeast cytochrome c1, and maize MDH were kindly provided by Michael Mulligan (University of California–Irvine), Masato Nakai (Osaka University), and Kathy Newton (University of Missouri), respectively. Antisera against maize CAF1, CRS1, CAF2, RNC1, PsbA, PsaD, PetD, and AtpA were generated by us and described previously (Roy and Barkan 1998; Till et al. 2001; Ostheimer et al. 2003; Watkins et al. 2007). Immunoblots and coimmunoprecipitations were performed as described in Ostheimer et al. (2003) except that immunoprecipitated proteins were detected on immunoblots using the One-Step Western Kit (GenScript Corporation).
RNA analyses
RNA-coimmunoprecipitation and the analyses of coimmunoprecipitated RNAs by RIP-Chip and slot-blot hybridization were carried out as described in Barkan (2008). Three replicate immunoprecipitations were performed for CFM3 RIP-Chip assays: two with antiserum from one rabbit, and one with antiserum from a second rabbit. RNA extraction, RNA gel blot hybridization and ribonuclease protection assays were performed as described previously (Jenkins et al. 1997). The probes for RNA-gel blot hybridization of the Arabidopsis genes were amplified from Arabidopsis leaf DNA by PCR with the following primers: ndhB intron, YAAtndhBF2 (5′-CGTTTGAGAAATTCCTACCTCTCT-3′) and YAAtndhBR3 (5′-AAAAGAGGATTCCTCACTTCTTTC-3′); ndhB exon 2, YAAtndhB Ex F3 (5′-TCTCCCACTCCAGTCGTTGCTTT-3′) and YAAtndhB Ex R4 (5′-AAAAAGGGTATCCTGAGCAATCG-3′); rrn18, YAAt mit rrn18F1 (5′-ATCATAGTCAAAAGAAGAGT-3′) and YAAt mit rrn18R1 (5′-TCAATTACTAGGTCTGGCAC-3′); rrn26, YAAt mit rrn26F1 (5′-TTAGTGTAGGCGCTTTCCAA-3′), and YAAt mit rrn26R1 (5′-GTTTACCGGGGCTTCCATTC-3′). Poisoned primer extension assays were performed as described previously (Asakura and Barkan 2006). RT-PCR analysis of cytosolic mRNA levels was performed using 200 ng of total leaf RNA, as described previously (Asakura and Barkan 2007). Primers for RT-PCR were as follows: AtCFM3a, same primer pair as for PCR genotyping; AtCFM3b, AtCfm4-9 5′ (5′-ACCACACTGAGGCCTAAAAAC-3′) and AtCfm4-10 3′ (5′-AAGTTCCTCATTCTGCAGTTCTA-3′); Actin2, YAAt actin2 F (5′-GAAGCTCTCCTTTGTTGCTGTT-3′) and YAAt actin2 R (5′-TTAGAAACATTTTCTGTGAACGA-3′). Probes used to detect RT-PCR products on DNA gel blots were amplified from Arabidopsis leaf DNA using the same primers as used for RT-PCR, and were radiolabeled by random hexamer priming in the presence of [α-32P]dCTP.
Rice RNA was extracted from the leaves of plants grown for 3 wk on MS medium containing 2% sucrose, using Trizol reagent. The probes for ribonuclease-protections were amplified from rice total DNA, and cloned into pBluescript SK+ (Stratagene). The primers used for PCR were as follows: ndhB, YAOs_ndhBF (SpeI) (5′-CTTACTAGTAATCAAAAAAGAAAGAAGACAA-3′) and YAOs_ndhBR (HindIII) (5′-CTTAAGCTTTTGAGTCTCCAACAATTATTCC-3′); petB, YAOs_petBF2 (SmaI) (5′-CTTCCCGGGTTGAACCTTTTTTCTAATATCC-3′) and YAOs_petBR (HindIII) (5′-CTTAAGCTTCTCGGTCATTATGTATTGAACCGA-3′); petD, YAOs_petDF (SpeI) (5′-CTTACTAGTCTTACTTAGAGCCATTGAGCCA-3′) and YAOs_petDR (HindIII) (5′-CTTAAGCTTGGTAATATTTCCAGAGGAGTTG-3′); rps16, YAOs_rps16F (SpeI) (5′-CTTACAGTGCAAGTCATATTCTCGAGCCGT-3′) and YAOs_rps16R (HindIII) (5′-CTTAAGCTTTTCCGCCTTCCTTAAAATATCA-3′); rpl16, YAOs_rpl16F (SpeI) (5′-CTTACTAGTCTATTAATGTGTATCTATCCGT-3′) and YAOs_rpl16R (HindIII) (5′-CTTAAGCTTTTATTGTAACCGGTTTGTCGGG-3′); ycf3 intron 2, YAOs_ycf3-2F (SpeI) (5′-CTTACTAGTCGAATTCAGTTAAGACAAGGGA-3′) and YAOs_ycf3-2R (XhoI) (5′-CTTCTCGAGGCGCTTCGTAATCTTCAACCAG-3′). Poisoned primer extension assays for rice used the following primers and dideoxynucleotide: rpl16, YAOs_rpl16-R5PPE, 5′-TGTTGTTTACGAAATCTGGTTCT-3′, ddCTP; petB, YAOs_petB-R2PPE, 5′-ACGTTCCTCAAACCAATCATATAC-3′, ddCTP. The RNA gel blot probe for trnG–UCC was the maize cDNA probe described previously (Watkins et al. 2007). The rrn18 probe was amplified from rice total DNA using primers; YAOs_rrn18-F (5′-AAAATCTGAGTTTGATCCTGGC-3′) and (5′-AAAATCTGAGTTTGATCCTGGC-3′).
Sucrose gradient fractionation of chloroplast and mitochondrial extract
Three hundred microliters of maize mitochondrial extract (∼0.9 mg protein) was precleared by centrifugation at 21,000g for 30 min at 4°C. The supernatant was applied to a 10 to 30% sucrose gradient containing KEX buffer [30 mM HEPES-KOH at pH 8, 150 mM KOAc, 10 mM Mg(OAc)2, and 5 mM DTT] and centrifuged in a Beckman SW55.1 rotor at 50,000 rpm for 4 h at 4°C. The sucrose-gradient fractionated maize chloroplast stroma was the same preparation described previously (Watkins et al. 2007).
Accession number
Sequence data for the maize CFM3 cDNA can be found in the GenBank data library under Accession No. EU084957.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
NOTE ADDED IN PROOF
AtCFM3a mutants fail to splice the mitochondrial nad1-intron 2 group 2 intron, confirming that AtCFM3a plays a role in mitochondrial gene expression.
ACKNOWLEDGMENTS
We are grateful to John Robbins and Maureen Hanson for obtaining and propagating the rice OsCFM3 mutant. We thank Susan Belcher for preparing the mitochondrial and stromal extracts used in this work, Jana Prikryl for help with RIP-Chip assays, and Kenny Watkins for help purifying the CFM3 antibody and for comments on the manuscript. We also thank Mike Mulligan, Masato Nakai, Danny Schnell, Alap Subramanian, Harry Roy, Kathy Newton, and Tsuyoshi Endo for providing antisera, and Ian Small for providing plasmids used for GFP fusion protein analysis. Arabidopsis and rice insertion lines were obtained from the ABRC and Postech, respectively. This work was supported by grant DBI-0421799 from the National Science Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1223708.
REFERENCES
- Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana . Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Asakura Y., Barkan A. Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol. 2006;142:1656–1663. doi: 10.1104/pp.106.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y., Barkan A. A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant Cell. 2007;19:3864–3875. doi: 10.1105/tpc.107.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 1998;297:38–57. [Google Scholar]
- Barkan A. Genome-wide analysis of RNA-protein interactions in plants. In: Belostotsky D., editor. Plant systems biology. Series: Methods in Molecular Biology. Humana Press; New York: 2008. [DOI] [PubMed] [Google Scholar]
- Barkan A., Klipcan L., Ostersetzer O., Kawamura T., Asakura Y., Watkins K. The CRM domain: An RNA binding module derived from an ancient ribosome-associated protein. RNA. 2007;13:55–64. doi: 10.1261/rna.139607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L. cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion. 2008;8:26–34. doi: 10.1016/j.mito.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Bonen L., Vogel J. The ins and outs of group II introns. Trends Genet. 2001;17:322–331. doi: 10.1016/s0168-9525(01)02324-1. [DOI] [PubMed] [Google Scholar]
- de Longevialle A., Hendrickson L., Taylor N., Delannoy E., Lurin C., Badger M., Millar A.H., Small I. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana . Plant J. 2008 doi: 10.1111/j.1365-313X.2008.03581.x. in press. [DOI] [PubMed] [Google Scholar]
- Duchene A.M., Giritch A., Hoffmann B., Cognat V., Lancelin D., Peeters N.M., Zaepfel M., Marechal-Drouard L., Small I.D. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana . Proc. Natl. Acad. Sci. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Heijne G.v. Prediction of organellar targeting signals. Biochim. Biophys. Acta. 2001;1541:114–119. doi: 10.1016/s0167-4889(01)00145-8. [DOI] [PubMed] [Google Scholar]
- Gothandam K.M., Kim E.S., Cho H., Chung Y.Y. OsPPR1, a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis. Plant Mol. Biol. 2005;58:421–433. doi: 10.1007/s11103-005-5702-5. [DOI] [PubMed] [Google Scholar]
- Hashimoto M., Endo T., Peltier G., Tasaka M., Shikanai T. A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis . Plant J. 2003;36:541–549. doi: 10.1046/j.1365-313x.2003.01900.x. [DOI] [PubMed] [Google Scholar]
- Hess W.R., Prombona A., Fieder B., Subramanian A.R., Borner T. Chloroplast rps15 and rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: Evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J. 1993;12:563–571. doi: 10.1002/j.1460-2075.1993.tb05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins B., Kulhanek D., Barkan A. Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell. 1997;9:283–296. doi: 10.1105/tpc.9.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.H., An S., Park S., Kang H.G., Park G.G., Kim S.R., Sim J., Kim Y.O., Kim M.K., Kim J., et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- Kotera E., Tasaka M., Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005;433:326–330. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- Meierhoff K., Felder S., Nakamura T., Bechtold N., Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Umesono K., Ozeki H. Comparative and functional anatomy of group II catalytic introns—A review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- Newton K. Procedures for isolating mitochondria and mitochondrial DNA and RNA. In: Freeling M., Walbot V., editors. The maize handbook. Springer-Verlag; New York: 1994. pp. 549–556. [Google Scholar]
- Ostersetzer O., Watkins K., Cooke A., Barkan A. CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell. 2005;17:241–255. doi: 10.1105/tpc.104.027516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer G., Williams-Carrier R., Belcher S., Osborne E., Gierke J., Barkan A. Group II intron splicing factors derived by diversification of an ancient RNA binding module. EMBO J. 2003;22:3919–3929. doi: 10.1093/emboj/cdg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters N., Chapron A., Giritch A., Grandjean O., Lancelin D., Lhomme T., Vivrel A., Small I. Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J. Mol. Evol. 2000;50:413–423. doi: 10.1007/s002390010044. [DOI] [PubMed] [Google Scholar]
- Pyle A., Lambowitz A. Group II introns: Ribozymes that splice RNA and invade DNA. In: Gesteland R., Cech T., Atkins J., editors. The RNA world. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2006. pp. 469–506. [Google Scholar]
- Roy L.M., Barkan A. A secY homologue is required for the elaboration of the chloroplast thylakoid membrane and for normal chloroplast gene expression. J. Cell Biol. 1998;141:385–395. doi: 10.1083/jcb.141.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Barkan A. Cell and molecular biology of plastids. Springer; Berlin: 2007. RNA splicing and RNA editing in chloroplasts; pp. 213–248. [Google Scholar]
- Schmitz-Linneweber C., Williams-Carrier R., Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′-region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Williams-Carrier R.E., Williams-Voelker P.M., Kroeger T.S., Vichas A., Barkan A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 Pre-mRNA. Plant Cell. 2006;18:2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., Burke E., Presting G., Aux G., McElver J., Patton D., Dietrich B., Ho P., Bacwaden J., Ko C., et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I., Peeters N., Legeai F., Lurin C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- Stern D., Hanson M., Barkan A. Genetics and genomics of chloroplast biogenesis: Maize as a model system. Trends Plant Sci. 2004;9:293–301. doi: 10.1016/j.tplants.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Till B., Schmitz-Linneweber C., Williams-Carrier R., Barkan A. CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA. 2001;7:1227–1238. doi: 10.1017/s1355838201010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Boerner T., Hess W. Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res. 1999;27:3866–3874. doi: 10.1093/nar/27.19.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V., Coe E.H. Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays . Proc. Natl. Acad. Sci. 1979;76:2760–2764. doi: 10.1073/pnas.76.6.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins K., Kroeger T., Cooke A., Williams-Carrier R., Friso G., Belcher S., Wijk K.v., Barkan A. A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell. 2007;19:2606–2623. doi: 10.1105/tpc.107.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P., Barkan A. A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 2003;36:675–686. doi: 10.1046/j.1365-313x.2003.01915.x. [DOI] [PubMed] [Google Scholar]