Abstract
To investigate the global expression profile of miRNAs in primary breast cancer (BC) and normal adjacent tumor tissues (NATs) and its potential relevance to clinicopathological characteristics and patient survival, the genome-wide expression profiling of miRNAs in BC was investigated using a microarray containing 435 mature human miRNA oligonucleotide probes. Nine miRNAs of hsa-miR-21, hsa-miR-365, hsa-miR-181b, hsa-let-7f, hsa-miR-155, hsa-miR-29b, hsa-miR-181d, hsa-miR-98, and hsa-miR-29c were observed to be up-regulated greater than twofold in BC compared with NAT, whereas seven miRNAs of hsa-miR-497, hsa-miR-31, hsa-miR-355, hsa-miR-320, rno-mir-140, hsa-miR-127 and hsa-miR-30a-3p were observed to be down-regulated greater than twofold. The most significantly up-regulated miRNAs, hsa-mir-21 (miR-21), was quantitatively analyzed by TaqMan real-time PCR in 113 BC tumors. Interestingly, among the 113 BC cases, high level expression of miR-21 was significantly correlated with advanced clinical stage (P = 0.006, Fisher's exact text), lymph node metastasis (P = 0.007, Fisher's exact text), and shortened survival of the patients (hazard ratio [HR]=5.476, P < 0.001). Multivariate Cox regression analysis revealed this prognostic impact (HR=4.133, P = 0.001) to be independent of disease stage (HR=2.226, P = 0.013) and histological grade (HR=3.681, P = 0.033). This study could identify the differentiated miRNAs expression profile in BC and reveal that miR-21 overexpression was correlated with specific breast cancer biopathologic features, such as advanced tumor stage, lymph node metastasis, and poor survival of the patients, indicating that miR-21 may serve as a molecular prognostic marker for BC and disease progression.
Keywords: microRNA, microarray, real time PCR, breast cancer, prognosis, miR-21
INTRODUCTION
MicroRNAs (miRNAs) are small (20–24 nucleotides [nt]) noncoding RNA gene products that post-transcriptionally modulate gene expression by negatively regulating the stability or translational efficiency of their target mRNAs (Ambros 2003). Currently, more than several hundred unique mature human miRNAs are known (http://microrna.sanger.ac.uk). Aberrant expression of miRNAs has been linked to some cancers (Caldas and Brenton 2005; Croce and Calin 2005), leading to the use of miRNA profiles in the diagnosis/prognosis of specific cancers and to distinguish certain cancer types (Calin et al. 2004, 2005; Takamizawa et al. 2004; Eis et al. 2005; Iorio et al. 2005; Lu et al. 2005).
miRNAs have been reported to be involved in tumorigenesis, acting, as one might expect, variously as either oncogenes (He et al. 2005b; Voorhoeve et al. 2006) or tumor suppressors (Johnson et al. 2005). Several other reports have described altered expression of miRNAs in cancer tissues compared to normal tissues, suggesting that these miRNAs could potentially represent novel clinical and prognostic markers (Mattie et al. 2006; Yanaihara et al. 2006). In particular, the microRNA, hsa-miR-21 (miR-21), has been found to be up-regulated in glioblastoma (Chan et al. 2005), breast cancer (Iorio et al. 2005), lung cancer (Yanaihara et al. 2006), and chronic lymphocytic leukemia (Fulci et al. 2007).
Over the past 20 years, the worldwide incidence of breast cancer (BC) has nearly doubled. As part of this situation, BC has become a serious threat to the health of women in China, with noticeably increased incidence and mortality. Compared to the year 2000, there were 470,000 more new cases and 130,000 more deaths from BC in China in 2005 (Yang et al. 2006). Recognized disease prognosticators for individuals diagnosed with BC include tumor size, histological grade, and the presence of lymph node or distant metastases (Bundred 2001). These parameters alone, or in combination, enable the identification of individuals who are at increased risk of dying of BC and also who may benefit from aggressive treatment (Wiseman et al. 2005). Moreover, identification of molecular characteristics that allow an accurate prediction of a patient survival remains an important facet in the current management of this malignancy.
In this study, we investigated the global miRNA expression profile in primary BC to evaluate their involvement in malignant progression of this tumor. To this end, we examined the up-regulation of miR-21 in 113 BC cases by means of TaqMan real-time reverse-transcription PCR (RT-PCR). We report here that miR-21 is frequently overexpressed in BC. More importantly, those patients with elevated miR-21 expression were also found to have significantly worse prognosis; and this prognostic impact appears to be independent of disease stage in multivariate Cox regression analysis.
RESULTS
miRNA expression profile alteration of BC
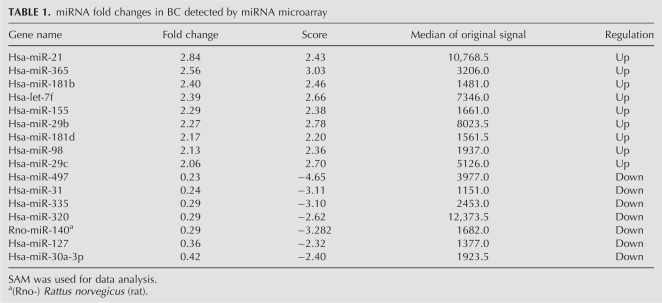
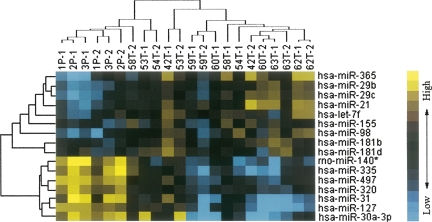
Using miRNA microarray analysis, we evaluated miRNA expression profiles of eight BC and the corresponding NAT. To identify differentially expressed miRNAs among the human, rat, and mouse miRNAs on the chip, SAM analysis generated a list of differentially expressed miRNAs at FDR=0 (0% false discovery rate) (Table 1). We identified nine miRNAs that were up-regulated greater than twofold (hsa-miR-21, hsa-miR-365, hsa-miR-181b, hsa-let-7f, hsa-miR-155, hsa-miR-29b, hsa-miR-181d, hsa-miR-98, and hsa-miR-29c) in BC compared with NAT. Seven miRNAs were down-regulated greater than twofold: hsa-miR-497, hsa-miR-31, hsa-miR-355, hsa-miR-320, rno-mir-140, hsa-miR-127, and hsa-miR-30a-3p. Cluster analysis, based on the above differentially expressed miRNA, was used to generate a tree with clear distinction between normal and cancer tissues (Fig. 1).
TABLE 1.
miRNA fold changes in BC detected by miRNA microarray
FIGURE 1.
Unsupervised hierarchical cluster analysis of 16 differentially expressed miRNAs in BC versus NAT, as was determined by SAM analysis. The heat-map shows that BC segregates from NAT based on a 16-gene miRNA profile. Blue indicates low expression and yellow indicates high expression relative to the median. (Rows) Differentially expressed miRNAs; (columns) tissue samples. 42T, 53T, 54T, 58T, 59T, 60T, 62T, and 63T are 8 BC samples. 1P, 2P, and 3P are normal adjacent tissues. 1P is the NAT of 60T, 2P is a pool of NATs for 42T, 53T, 62T, and 63T, while 3P is pooled NATs from 54T, 58T, and 59T. 42T-1 and 42T-2 represent two replicates of the microarray results for sample 42T, and the rest can be deduced accordingly.
Target genes prediction of the differentially expressed miRNAs in BC
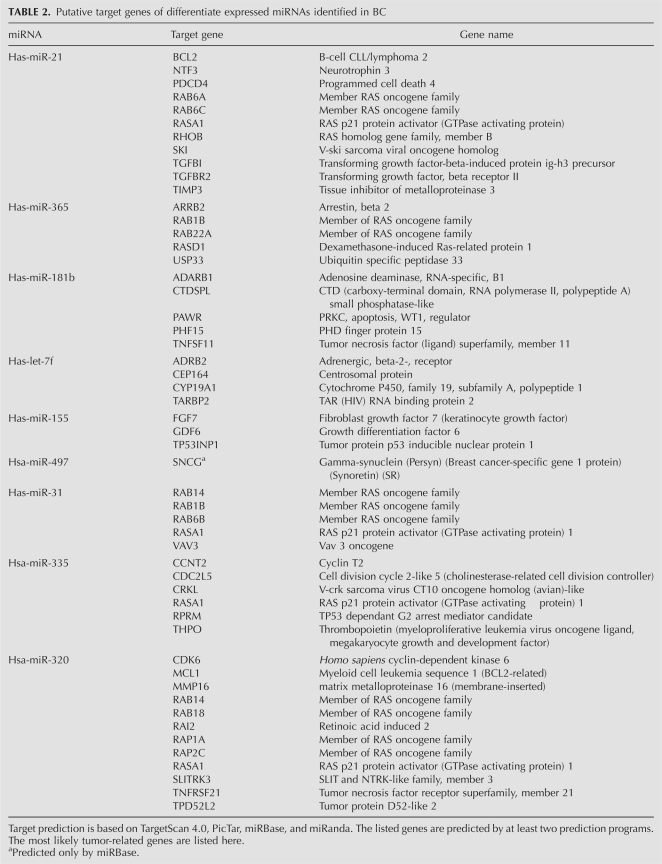
Given that biological significance of miRNA deregulation relies on the effect upon their cognate protein-coding gene targets, we analyzed the predicted targets of the most significantly up- and down-regulated miRNAs: miR-21, miR-365, miR-181b, let-7f, miR-155, miR-497, miR-31, miR-335, and 320. The analysis was done using four algorithms, TargetScan 4.0 (Iizuka et al. 2003), PicTar (Roberts and Gores 2005), miRBase (Griffiths-Jones et al. 2006), and miRanda (Lagos-Quintana et al. 2001), which are commonly used to predict human miRNA gene targets. Prediction results (Table 2) demonstrated that the putative target genes of miR-21 include the oncogenes Homo sapiens v-ski sarcoma viral oncogene homolog (SKI), RAB6A (member RAS oncogene family), RAB6C (member RAS oncogene family), and RAS homolog gene family member B (RHOB); transforming growth factor-beta-induced protein (TGFBI); transforming growth factor beta receptor II (TGFBR2); RAS p21 protein activator (RASA1); B-cell CLL/lymphoma 2 (BCL2); and the apoptosis-related gene, programmed cell death 4 (PDCD4). For miR-365, the potential target genes include members of the RAS oncogene family, such as RAB1B and RAB22A, and ubiquitin specific peptidase 33 (USP33). Tumor protein p53 inducible nuclear protein 1 (TP53INP1), fibroblast growth factor 7 (FGF7), and growth differentiation factor 6 (GDF6) are predicted as the targets of miR-155. miR-31 may target members of RAS oncogene family, such as RAB14, RAB1B, and RAB6B, and RAS p21 protein activator (RASA1). miR-21, miR-31, miR-335, and miR-320 may target the same gene, RAS p21 protein activator (RASA1).
TABLE 2.
Putative target genes of differentiate expressed miRNAs identified in BC
Validation of the microarray data by real-time RT-PCR analysis
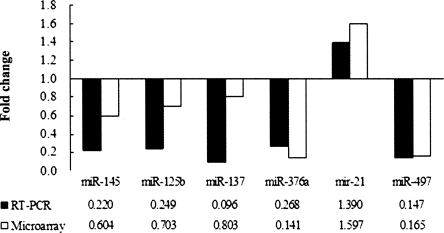
Six representative miRNAs (differentially expressed or nondifferentially expressed) (miR-21, miR-125b, miR-137, miR-145, miR-376a, and miR-497) were further verified by real-time RT-PCR. The ratio of miRNA microarray signal intensities (BC versus NAT) was compared with the RPCR results to test the consistency of the two methods. Figure 2 displays a comparison of real-time RT-PCR and microarray results for the six differentially expressed miRNAs. The RT-PCR analysis confirmed all results obtained by microarray analysis consistently, and in many cases, the differences were even stronger than that anticipated from the microarray studies.
FIGURE 2.
Comparison of miRNA fold changes by miRNA microarray and RT-PCR. The fold change in expression was determined for 60T (BC tissue) in comparison with 1P (corresponding NAT). Microarray fold change was calculated by the ratio of miRNA signal intensities in BRAC versus NAT. RT-PCR fold change is the RPCR value (see Materials and Methods). All of the six comparisons are consistent.
Reliability of real-time PCR detection of miRNA in FFPET
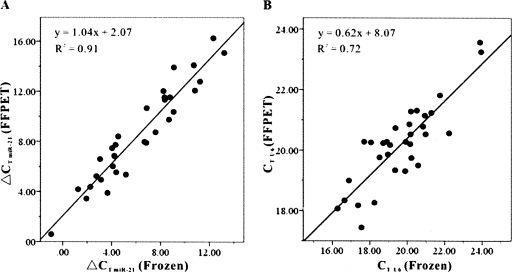
There was a significant positive correlation of miR-21 expression pattern between FFPETs and FTs, with R 2 = 0.91 (P < 0.001, linear regression) (Fig. 3A). The mean of ΔCT miR-21 of FFPETs was 8.55. The mean of ΔCT U6 of FTs was 6.22. U6 snRNA also exhibited a significant positve correlation of expression pattern between FFPETs and FTs, with R 2 = 0.72 (P < 0.001, linear regression) (Fig. 3B). These results indicate that it is reliable to detect mi-21 expression level in FFPETs by real-time RT-PCR.
FIGURE 3.
Comparison of miR-21 expression pattern using total RNA extracted from matched samples of FFPETs and frozen tissues. (A) Comparison of ΔCT values of miR-21 assays from paired FFPE and frozen tissue. ΔCT miR-21 = CT miR-21 − CT U6. Identical amount of total RNA was employed in each assay. R 2 is 0.91 between the two kinds of tissues. (B) Comparison of CT values of U6 snRNA assays from paired FFPET and frozen tissue. R 2 is 0.72 between the two kinds of tissues.
MiR-21 expression discriminates between normal and cancer breast tissues
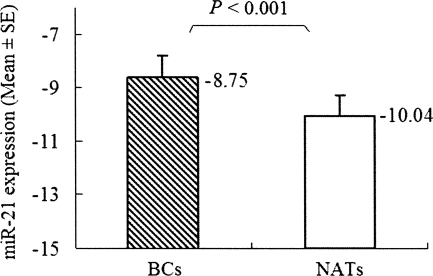
We chose miR-21, the most significantly up-regulated miRNA with high original microchip signal, for further investigation. Real-time RT-PCR was used to evaluate miR-21 expression level in 40 noncancerous NATs and 40 paired BC tissues. The result verified that miR-21 expression level in BC tissues was significantly different from the paired NATs. Consistent with the microarray data, this analysis showed that the miR-21 expression level in BC (−8.75 ± 0.80, −mean±SE) was significantly higher than in NATs (−10.04 ± 0.76, −mean±SE) (P < 0.001, t = −4.83, paired-samples t-test) (Fig. 4).
FIGURE 4.
miR-21 is differentially expressed between BC and NATs. Data obtained by real-time RT-PCR amplification of miR-21 are plotted. miR-21 is normalized by U6 snRNA. ΔCT = CT miR-21 − CT U6. It is known that the logarithm of the original concentration of the template is the inverse ratio of CT value. In order to demonstrate clearly, we used “−mean ± SE” to describe the expression level of miR-21. The ΔCT of miR-21 in BC (−8.75 ± 0.80, −mean ± SE) was significantly higher than in NATs (−10.04 ± 0.76, −mean ± SE) (P < 0.001, t = −4.84, paired-samples t-test). Boxes represent mean. Error bars represent SE.
Correlation between miR-21 expression and clinicopathological characteristics of BC
Using real-time RT-PCR detection, miR-21 was detected in all 113 (100%) BC tissues. Test of normality was used to determine whether the distribution of expression levels of miR-21 at log2 could be fitted to the normal distribution. According to this analysis, the hypothesis that the distribution of miR-21 expression levels at log2 followed a normal distribution could not be rejected (P = 0.20). The mean level of miR-21 (BC versus NAT) was 1.74 ± 5.12 (median, 2.18; minimum, −12.68; maximum, 12.51).
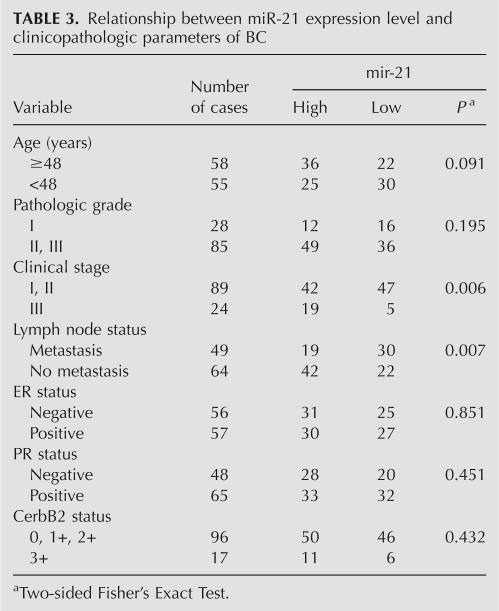
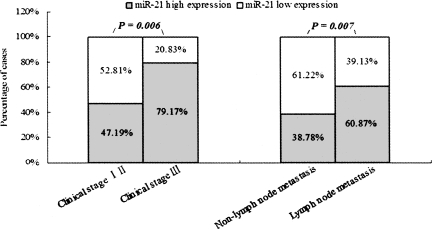
The BC patients were divided into two groups according to the mean level of miR-21 (i.e., 1.74 of the log2 value, median), miR-21 low expressers (n = 52) and miR-21 high expressers (n = 61). Correlation between miR-21 expression level and clinicopathologic characteristics of BC is summarized in Table 3. A statistically significant association between miR-21 expression level and clinical stage and metastasis of BC was observed in this study. In 24 cases presenting advanced stage III, 19 (79.17%) of the cases had high level miR-21 expression in BC tissue; whereas in 89 early stage (stages I and II), only 42 (47.19%) presented high levels of miR-21 expression (P = 0.006, Fisher's Exact Test). In the 64 cases of BC with lymph node metastasis, 42 (60.87%) presented high miR-21 expression, while only 19 (38.78%) of 49 cases of BC without lymph node metastasis presented high level miR-21 expression (P = 0.007, Fisher's Exact Test) (Fig. 5). No correlation was observed between miR-21 level and gender, histological grade, ER, PR, or CerbB2 status of BC.
TABLE 3.
Relationship between miR-21 expression level and clinicopathologic parameters of BC
FIGURE 5.
Correlations between miR-21 expression level, clinical stage, and lymph node status of BC patients. In 24 cases with stage III, 19 (79.17%) exhibited high miR-21 expression in BC, whereas in 89 of stages I and II, only 42 (47.19%) presented high level miR-21 expression (P = 0.006, Fisher's Exact Test). In 64 cases of BC with lymph node metastases, 42 (60.87%) presented high miR-21 expression; in contrast, of the 49 cases of BC without lymph node metastasis, only 19 (38.78%) presented high miR-21 expression level (P = 0.007, Fisher's Exact Test).
Correlation between miR-21 expression level and prognosis of BC patients
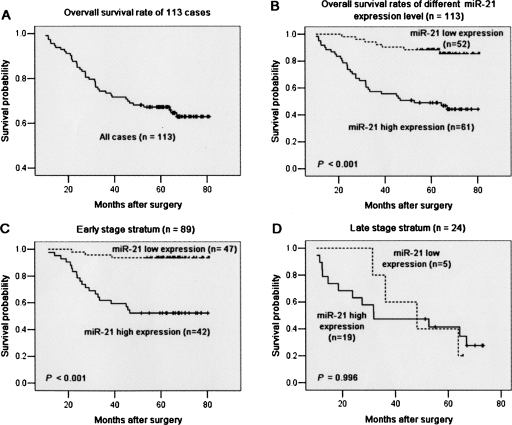
Figure 6A presents the overall survival curve of the 113 BC patients, with the 5-yr overall survival rate of 64.60% in the 113 patients. Kaplan–Meier survival analyses revealed that the BC patients with high miR-21 expression had a significantly poorer prognosis compared to those with low miR-21 expression. The 5-yr survival rate in patients with low miR-21 expression was 86.54%, which was significantly higher than those with high miR-21 expression (45.90%; P < 0.001, log-rank test; Fig. 6B). When patients were stratified by clinical stage, Kaplan–Meier survival estimates revealed that early stage patients with high miR-21 expression (n = 42) had inferior survival to those with low miR-21 expression (n = 47; P < 0.001, log-rank test; Fig. 6C). However, within the late stage cases, miR-21 expression was not related with the patients' survival (P = 0.996, log-rank test; Fig. 6D).
FIGURE 6.
Kaplan–Meier survival curve and log-rank test for BC patients classified as showing either high or low miR-21 expression. (A) Kaplan–Meier curve for 5-yr overall survival rates (64.60%) of 113 patients with BC. (B) High expression of miR-21 (n = 61), group with expression ratio ≥ mean ratio (1.741). mir-21 low expression (n = 52), group with expression ratio < mean ratio. miR-21 expression had a significant (log-rank, P < 0.001) relationship with patient survival. (C) Cases stratified by clinical stage. Within the early stage stratum, miR-21 expression exhibited a significant (P < 0.001; log-rank test) relationship with patient survival. (D) Within the late stage stratum, miR-21 expression did not show a statistical (P = 0.996; log-rank test) relationship with patient survival.
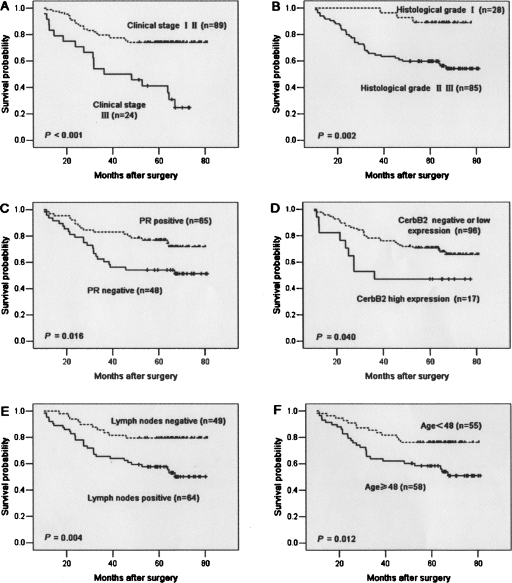
Aside from miR-21 expression, survival analysis of other clinicopathological factors also revealed that clinical stage, histological grade, PR and CerbB2 status, and lymph node metastases were associated with prognosis of the patients with BC (Fig. 7).
FIGURE 7.
Kaplan–Meier survival curves for BC patients. (A) BC patients were classified as either early or late clinical stage. Clinical stage exhibited a significant (P < 0.001, log-rank) relationship with patient survival. (B) BC patients were classified as histological grade I and II or grade III. Histological grade showed a significant (P = 0.002, log-rank) relationship with patient survival. (C) Kaplan–Meier survival curve for BC patients classified as PR positive or negative. PR expression status was significantly associated with patient survival (log-rank, P = 0.016). (D) Kaplan–Meier survival curve for BC patients classified as showing either high or low CerbB2 expression. CerbB2 status was found to be strongly associated (log-rank, P = 0.04) with patient survival. (E) Kaplan–Meier survival curve for BC patients classified as either lymph node metastasis positive or negative. Lymph node metastasis status was found to be strongly associated (log-rank, P = 0.004) with patient survival. (F) Kaplan–Meier survival curve for BC patients classified as older (≥ median age, 48 yr) or younger (<48 yr). A significant difference in patient survival was observed between the different age groups (log-rank, P = 0.012). (x-axis) Survival time after surgery; (y-axis) percentage of survivors.
Univariate Cox proportional hazard regression analysis revealed that high miR-21 expression was the most significant predictive factor for poor prognosis of patients with BC = (P < 0.001, Hazard ratio [HR] = 5.476). Other clinicopathologic parameters, including clinical stage (P < 0.001, HR = 3.598), histological grade (P = 0.006, HR = 5.238), PR (P = 0.019, HR = 0.471), CerbB2 (P = 0.045, HR = 2.142), lymph nodes status (P = 0.006, HR = 2.752), and age (P = 0.014, HR = 0.437) were also found to be predictive factors for poor prognosis of BC patients (Table 4).
TABLE 4.
Univariate and multivariate Cox regression analyses of the relationship between expression levels of miR-21 and clinicopathologic characteristics of BC
Multivariate Cox proportional hazard regression analysis revealed that high-level miR-21 expression (P = 0.001, HR = 4.133, 95% confidence interval [CI]: 1.799–9.499) was also the most significantly unfavorable prognostic factor independent of other clinicopathological factors, including clinical stage (P = 0.013, HR = 2.266, 95% CI: 1.187–4.323), histological grade (P = 0.033, HR = 3.681, 95% CI: 1.112–12.182), PR (P = 0.029, HR = 0.493, 95% CI: 0.261–0.930), and age (P = 0.049, HR = 0.511, 95% CI: 0.263–0.996; Table 4).
DISCUSSION
In the present study, BC miRNA microarray expression analysis identified that nine miRNAs were up-regulated greater than twofold, including hsa-miR-21, hsa-miR-365, hsa-miR-181b, hsa-let-7f, hsa-miR-155, hsa-miR-29b, hsa-miR-181d, hsa-miR-98, and hsa-miR-29c in BC. Whereas seven miRNAs were down-regulated greater than twofold, including hsa-miR-497, hsa-miR-31, hsa-miR-355, hsa-miR-320, rno-mir-140, hsa-miR-127, and hsa-miR-30a-3p. These results differ from a similar, recently reported BC study (Iorio et al. 2005). Using miRNA microarray analysis, Iorio et al. (2005) reported that miR-10b, miR-125b, and miR-145 were down-regulated, whereas only two miRNAs of miR-21 and miR-155 were up-regulated in BC. The difference between the two BC miRNA profiles might be due to the difference in sample origin, different analytical approaches, or the different technical platform of the studies. Our results enrich the global miRNA expression profiling of BC, providing more clues into the functional and mechanistic study of miRNAs in the carcinogenesis of BC.
At present, the lack of knowledge regarding miRNA gene targets hampers a full understanding of the biological functions that may be disregulated by aberrant miRNA expression. We made use of presently available computational approaches to predict gene targets (Lagos-Quintana et al. 2001; Iizuka et al. 2003; Roberts and Gores 2005; Griffiths-Jones et al. 2006) in an attempt to partially overcome this limitation. Various cancer-associated genes, which are potentially regulated by miRNAs in BC, were predicted by at least two of the predicting software applications, and are summarized in Table 2. For the up-regulated miR-21, its potential target genes include the tumor suppressors TIMP3 and PDCD4. It is noteworthy that TPM1 has recently been shown to be a target of miR-21 (Zhu et al. 2007), which is not shown in any of the four target predictors. Interestingly, miR-21, miR-31, miR-335, and miR-320 may target the same gene, RAS p21 protein activator (RASA1). In BC, the anticipated targets of down-regulated miRNAs are expected to be oncogenes or genes encoding proteins with potential oncogenic functions. For example, miR-31 and miR-320 may, respectively, target RAB1B, RAB6B, RAB14, and RAB18 (members of the RAS oncogene family). However, with few exceptions, the physiologic targets of miRNAs remain to be identified. Our results indicate that differentially expressed miRNAs may play an important role in pathogenesis and development of BC by inducing deregulation of specific target genes. Further investigative focus on these miRNAs and their functions in the pathogenesis of BC is warranted.
Interestingly, miR-21 was found to be significantly up-regulated in BC in both our and previous miRNA expression profiles (Iorio et al. 2005). In addition, miR-21 and its precursor have been reported to be up-regulated in other kinds of tumors, including malignant cholangiocytes (Schmittgen et al. 2004), glioblastomas (Chan et al. 2005), and malignancies of the colon, lung, pancreas, prostate, and stomach (Ciafre et al. 2005; Iorio et al. 2005; Roldo et al. 2006; Volinia et al. 2006; Bloomston et al. 2007). However, data concerning the expression level of miR-21 and its relevance to clinicopathologic behavior of BCRA is unclear.
Our present study next focused on the potential relationship between the expression level of the most significantly up-regulated miR-21 and various BC clinicopathological characteristics, as well as survival of the patients. It is worth noting that high levels of miR-21 appear to be significantly correlated with advanced clinical stage, lymph node metastases, and poor prognosis of the patients with BC. miR-21 was up-regulated in BCs presenting lymph node metastases, suggesting that its up-regulation was acquired in the course of tumor progression and, in particular, during the acquisition of metastatic potential. Multivariate Cox proportional hazard regression analysis revealed that miR-21 overexpression had a significantly worse prognostic impact (P = 0.001) on survival of BC patients independent of clinical stage (P = 0.013), PR status (P = 0.029), histological grade (P = 0.033), and age (P = 0.049). These results indicate that, as an independent risk factor, miR-21 could serve as a prognostic marker for survival of BC patients.
The precise molecular mechanisms behind the altered expression of miR-21 in BC are unclear. To our knowledge, this is the first report to describe the significance of miR-21 to clinical stage, lymph node metastasis, and prognosis of BC patients. It was recently reported that miR-21 mediates MCF-7 cell growth; suppression of this miRNA, which is also overexpressed in MCF-7 cells, was associated with increased apoptosis and decreased cell proliferation (Si et al. 2007). A function for miR-21 in cancer is further supported by knockdown experiments in glioblastoma cells, demonstrating that this miRNA has an anti-apoptotic function (Chan et al. 2005). Furthermore, a recent study reported two additional direct miR-21 targets, programmed cell death 4 (PDCD4) and maspin, both of which reduced invasiveness of metastatic breast cancer MDA-MB-231 cells like TPM1. The results suggest that, as an oncogenic miRNA, miR-21 has a role not only in tumor growth but also in invasion and tumor metastasis by targeting multiple tumor/metastasis suppressor genes (Zhu et al. 2008). Taken together, these studies may explain why miR-21 overexpression associated with poor prognosis of BC patients make a strong case for the role of the miRNA in cancer. Given that a single miRNA has multiple targets and different miRNAs might target the same gene, it has been speculated that increased miR-21 expression might block a panel of genes that regulate cell proliferation and apoptosis. Additional studies to investigate the molecular mechanisms of both the cause and effects of altered expression of miR-21 in the development and/or progression of BC are essential.
An article, published in Nature Methods first reported that miRNAs could be isolated and profiled from FFPE tissue (Nelson et al. 2004). Subsequently, another method reported that small RNA molecules appear to be less affected by tissue processing, and miRNAs demonstrate stable expression levels in FFPE compared with snap frozen paired samples (Li et al. 2007). Our results confirm that it is reliable to detect miR-21 by real-time PCR with total RNA isolated from FFPETs. This foundational work opened up new opportunities for the analysis of miRNAs from archival tissues and supports the reliability of our discovery regarding the relationships between miRNA expression level in FFPE tissues and BC patient survival.
In summary, our results have added to the as yet wanting body of knowledge concerning alterations of the miRNA expression profile in human BC. Our principal finding is that overexpression of miR-21, one of the most significantly altered miRNAs in BC, is associated with progression and poor prognosis of the patients. Although the precise molecular mechanism surrounding miR-21 up-regulation requires further clarification, our data indicate that miR-21 may be a good candidate as a molecular prognostic marker. Future study on the role of miR-21 in BC progression will no doubt enrich of knowledge of this nascent field.
MATERIALS AND METHODS
Samples and cases
For the miRNA microarray analysis, fresh samples from eight cases of human BC and paired normal adjacent tissues (NATs, >2 cm from cancer tissue) were obtained from Sun Yat-sen University Cancer Center (SYSUCC) (Guangzhou, China) between January 2006 and March 2006. The fresh specimens were stored at 4°C for 24 h in RNA Later (Ambion Inc.), then at −80°C liquid nitrogen until further use. The samples used were not subjected to preoperative radiotherapy and/or chemotherapy and were diagnosed as infiltrating carcinoma by pathology. Before RNA extraction, sections were stained with H&E for histological diagnosis and tumor cell evaluation. Only those cases with >70% tumor cell population in the section were used in this study.
For comparison of the reliability of real-time PCR for detection of miRNA in formalin-fixed paraffin-embedded tissues (FFPETs), 32 pairs of samples (including 24 BC and 8 NATs) of FFPETs and FTs were collected from March 2007 to December 2007 at SYSUCC.
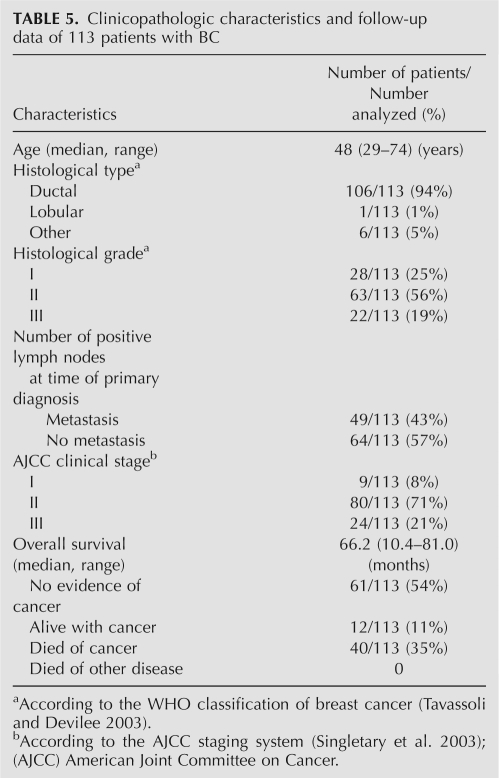
For further miR-21 quantitative analysis, FFPETs of 113 BC and 40 paired noncancerous NATs were prepared. The BC cases were selected randomly from January 2000 to December 2002 at SYSUCC with informed consent and agreement and clinicopathologic information available. The diagnosis and histological grade of each case were independently confirmed by two pathologists based on WHO classification (Tavassoli and Devilee 2003). The clinical stage was classified according to the American Joint Committee on Cancer (AJCC) tumor-lymph node-metastasis (TNM) classification system (Singletary et al. 2003; Table 5). The clinicopathologic characteristics of the patients and follow-up data are summarized in Table 5.
TABLE 5.
Clinicopathologic characteristics and follow-up data of 113 patients with BC
The median age of the BC patients at diagnosis was 48 yr (range, 29–74 yr). Among the 113 invasive BC, 106 (94%) were diagnosed as ductal carcinoma, 1 (1%) lobular carcinoma, and 6 (5%) other types. Histological grade consisted of grade I (n = 28, 25%), grade II (n = 63, 56%), and grade III (n = 22, 19%). Lymph node involvement was found in 64 (57%) patients. At the time of surgery, 9 (8%) patients were classified as TNM stage I, 80 (71%) stage II, and 24 (21%) stage III. The median follow-up time was 66.2 mo (range, 10.4–81.0 mo) after the primary surgical treatment. During the follow-up period, 12 (11%) patients presented disease recurrence (1 loco regional, 11 distant metastases) and 40 (35%) patients died of BC.
RNA extraction
For miRNA microarray analysis, total RNA from fresh BC tissues was isolated using Trizol reagent (Invitrogen) according to the manufacturer's instructions. Low molecular weight (LMW) RNA (<200 bases) was separated from the total RNA using an miRNA Isolation Kit (Ambion, Inc.) according to the manufacturer's protocol.
For real-time PCR analysis of miRNA, total RNA from FFPETs was isolated using Trizol reagent (Invitrogen) according to the manufacturer's instructions. Briefly, plugs were punched out (1.5 mm × 1.5 mm) of a paraffin block. Samples were deparaffinized three times in 1 mL ACS grade xylene with incubation at 60°C for 10 min, followed by a wash with 100% ACS grade ethanol and air drying at room temperature. Samples were then incubated with proteinase K (Merck) at 55°C overnight, shaking every 2 h. RNA samples were resuspended in RNase-free water after the final precipitation step. RNA quality and quantity were assessed using a biophotometer (Eppendorf). The paraffin plugs were enriched for tumor tissue under microscope control using H&E-stained sections of the same sample for guidance.
miRNA microarray experiments
Total RNA samples were analyzed by CapitalBio (CapitalBio Corp.) for miRNA microarray experiments. Procedures were performed as described in detail on the website of CapitalBio (http://www.capitalbio.com). Briefly, miRNA was separated from 30–50 μg total RNA using the Ambion miRNA Isolation Kit. Fluorescein-labeled miRNA (Thomson et al. 2004) were used for hybridization on each miRNA microarray chip containing 509 probes in triplicate, corresponding to 435 human (including 122 predicted miRNAs), 261 mouse, and 196 rat miRNAs found in the miRNA Registry (http://microrna.sanger.ac.uk/sequences/; accessed October 2005). Image intensities were measured as a function of the median of foreground minus the background, as previously described (Castoldi et al. 2006). Raw data were normalized and analyzed in GenePix Pro 4.0 software (Axon Instruments). Expression data were median-centered using the global median normalization function of the Bioconductor package (http://www.bioconductor.org). Clustering analysis was performed by Cluster 3.0 (Chiang et al. 2001). Statistical comparisons were performed with the SAM software (SAM version 2.1, http://www-stat.stanford.edu/∼tibs/SAM/index.html) (He et al. 2005a).
Target genes prediction of the differentially expressed miRNAs
The analysis of miRNA predicted targets was determined using the algorithms of TargetScan 4.0 (Iizuka et al. 2003), PicTar (Roberts and Gores 2005), miRBase (Griffiths-Jones et al. 2006), and miRanda (Lagos-Quintana et al. 2001).
Validation of miRNA microarray results
Real-time PCR analysis was performed on 60T (fresh breast cancer sample No. 60) and 1P (NAT of 60T) to validate the miRNA microarray results in this study. The specific stem–looped RT-PCR primers to representatives of differentially expressed miRNAs (miR-21, miR-125b, miR-137, miR-145, miR-376a, and miR-497) were designed according to a previous study (Thomson et al. 2004), and are summarized in Table 6. The RT-PCR reaction was performed according to a previous report (Li et al. 2007). PCR primers for amplification of the human mature miRNAs are summarized in Table 6.
TABLE 6.
PCR primers for amplification of the human mature miRNAs
Real-time PCR analysis was performed on an ABI 7900HT instrument (ABI Inc.) with 20 μL volume reaction containing 1 μL reverse transcription product, 2 μL HSS (Roche), 0.3 μm paired specific primers (0.3 μM), 3 mM MgCl2, and H2O. The reactions were incubated in 96-well plates at 95°C for 10 min, following by 40 cycles (95°C for 15 s, 60°C for 1 min), then ramped from 70°C to 97°C to obtain the melting curve (Li et al. 2007). U6 snRNA was measured by the same method and used for normalization. The relative quantity of each miRNA in BC tissues, normalized to U6 RNA and relative to the expression in NAT, was calculated using the equation RQ = 2−ΔΔCT, where ΔΔCT = (CT miRNA − CT U6 RNA)BC − (CT miRNA − CT U6 RNA)NAT and CT is the threshold cycle to detect fluorescence (Gramantieri et al. 2007).
Reliability analysis of miRNA detection by real-time PCR in FFPETs
Total RNA extracted from the 32 matched FFPETs and FTs samples was used to investigate whether miR-21 could be detected reliably in archival specimens by real-time PCR. The relevance of expression level of U6 snRNA and miR-21 in mathced FFPETs and FTs samples was analyzed. The miR-21 relative to U6 snRNA was determined using the 2−ΔCT method, where ΔCT = CT(FFPET) − CT (FT) (Livak and Schmittgen 2001).
Real-time PCR for quantitative analysis of miR-21
Quantitative analysis of miR-21 expression in 113 BCs were assayed using Hairpin-it miRNA real-time PCR Quantitation Kit (GenePharma). Each sample was analyzed in triplicate. U6 snRNA was used for normalization; no template and no reverse transcription were included as negative controls. The quantity of miR-21 in each BC relative to the average expression in 40 NATs was calculated using the equation RQ = 2−ΔΔCT, where ΔΔCT = (CT miRNA − CTU6 RNA)BC − (CT miRNA − CT U6 RNA)MeanNAT (Gramantieri et al. 2007).
Immunohistochemical staining of BC samples
Hormonal receptors were evaluated with the 1D5 antibody for estrogen receptor α (ER) and antibody PGR-1A6 for the progesterone receptor (PR; Dako Inc.). CerbB2 was detected with CB11 (Dako). Staining procedures were performed per the manufacturer's instructions. Only tumor tissues with distinct nuclear staining for ER and PR in >10% of the tumor cells were recorded as positive (Iorio et al. 2005). CerbB2 expression was scored semi-quantitatively according to the degree and the proportion of membrane staining using the following scale: 0, no staining or membrane staining in <10% of tumor cells; 1+, faint or partly stained membranes in >10% of tumor cells; 2+, weak to moderate complete membrane staining in >10% of tumor cells; 3+, strong, complete membrane staining in >10% of tumor cells (Rhodes et al. 2002). The represent features of ER, PR, and CerbB2 immunohistochemical staining of breast cancer is shown in Supplemental Figure 1.
Statistical analysis
For microarray data, a two-tailed t-test was performed by using Significance Analysis of Microarray (SAM), with the default settings of random seed number and 100 permutations for false discovery rate (FDR) analysis. The genes showing significant expression changes (≥ twofold change) and FDR = 0 were considered to be significantly differentially expressed in BC compared with NAT. For RT-PCR data, statistical analysis was performed in SPSS 13.0 for Windows (SPSS Inc.). The ratio of miR-21 expression level in BC to that in the corresponding NAT was log2 transformed. A Fisher's exact text was used to analyze the relationship between miR-21 expression level and various clinicopathologic characteristics. The survival curves were estimated by the Kaplan–Meier method, and the resulting curves were compared using the log-rank test. The joint effect of covariables was examined using the Cox proportional hazard regression model. All tests were two-tailed, and the significance level was set at P < 0.05.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported in part by the Chinese State Key Basic Research Project (No. 2004CB518708), the Program for New Century Excellent Talents in University (NCET), and a key grant from 985-II Project.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1034808.
REFERENCES
- Ambros V. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Bloomston M., Frankel W.L., Petrocca F., Volinia S., Alder H., Hagan J.P., Liu C.G., Bhatt D., Taccioli C., Croce C.M. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- Bundred N.J. Prognostic and predictive factors in breast cancer. Cancer Treat. Rev. 2001;27:137–142. doi: 10.1053/ctrv.2000.0207. [DOI] [PubMed] [Google Scholar]
- Caldas C., Brenton J.D. Sizing up miRNAs as cancer genes. Nat. Med. 2005;11:712–714. doi: 10.1038/nm0705-712. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Liu C.G., Sevignani C., Ferracin M., Felli N., Dumitru C.D., Shimizu M., Cimmino A., Zupo S., Dono M., et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M., et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Castoldi M., Schmidt S., Benes V., Noerholm M., Kulozik A.E., Hentze M.W., Muckenthaler M.U. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.A., Krichevsky A.M., Kosik K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Chiang D.Y., Brown P.O., Eisen M.B. Visualizing associations between genome sequences and gene expression data using genome-mean expression profiles. Bioinformatics. 2001;17(Suppl 1):S49–S55. doi: 10.1093/bioinformatics/17.suppl_1.s49. [DOI] [PubMed] [Google Scholar]
- Ciafre S.A., Galardi S., Mangiola A., Ferracin M., Liu C.G., Sabatino G., Negrini M., Maira G., Croce C.M., Farace M.G. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Croce C.M., Calin G.A. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Eis P.S., Tam W., Sun L., Chadburn A., Li Z., Gomez M.F., Lund E., Dahlberg J.E. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulci V., Chiaretti S., Goldoni M., Azzalin G., Carucci N., Tavolaro S., Castellano L., Magrelli A., Citarella F., Messina M., et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- Gramantieri L., Ferracin M., Fornari F., Veronese A., Sabbioni S., Liu C.G., Calin G.A., Giovannini C., Ferrazzi E., Grazi G.L., et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Jazdzewski K., Li W., Liyanarachchi S., Nagy R., Volinia S., Calin G.A., Liu C.G., Franssila K., Suster S., et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. 2005a;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005b;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka N., Oka M., Yamada-Okabe H., Nishida M., Maeda Y., Mori N., Takao T., Tamesa T., Tangoku A., Tabuchi H., et al. Oligonucleotide microarray for prediction of early intrahepatic recurrence of hepatocellular carcinoma after curative resection. Lancet. 2003;361:923–929. doi: 10.1016/S0140-6736(03)12775-4. [DOI] [PubMed] [Google Scholar]
- Iorio M.V., Ferracin M., Liu C.G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Li J., Smyth P., Flavin R., Cahill S., Denning K., Aherne S., Guenther S.M., O'Leary J.J., Sheils O. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Mattie M.D., Benz C.C., Bowers J., Sensinger K., Wong L., Scott G.K., Fedele V., Ginzinger D., Getts R., Haqq C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P.T., Baldwin D.A., Scearce L.M., Oberholtzer J.C., Tobias J.W., Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods. 2004;1:155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- Rhodes A., Jasani B., Anderson E., Dodson A.R., Balaton A.J. Evaluation of HER-2/neu immunohistochemical assay sensitivity and scoring on formalin-fixed and paraffin-processed cell lines and breast tumors: A comparative study involving results from laboratories in 21 countries. Am. J. Clin. Pathol. 2002;118:408–417. doi: 10.1309/97WN-W6UX-XJWT-02H2. [DOI] [PubMed] [Google Scholar]
- Roberts L.R., Gores G.J. Hepatocellular carcinoma: Molecular pathways and new therapeutic targets. Semin. Liver Dis. 2005;25:212–225. doi: 10.1055/s-2005-871200. [DOI] [PubMed] [Google Scholar]
- Roldo C., Missiaglia E., Hagan J.P., Falconi M., Capelli P., Bersani S., Calin G.A., Volinia S., Liu C.G., Scarpa A., et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J. Clin. Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Jiang J., Liu Q., Yang L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004;32:e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si M.L., Zhu S., Wu H., Lu Z., Wu F., Mo Y.Y. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Singletary S.E., Allred C., Ashley P., Bassett L.W., Berry D., Bland K.I., Borgen P.I., Clark G.M., Edge S.B., Hayes D.F., et al. Staging system for breast cancer: Revisions for the 6th edition of the AJCC Cancer Staging Manual. Surg. Clin. North Am. 2003;83:803–819. doi: 10.1016/S0039-6109(03)00034-3. [DOI] [PubMed] [Google Scholar]
- Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Tavassoli F., Devilee P., editors. Pathology and genetics of tumours of the breast and female genital organs. IARC Press; Lyon: 2003. [Google Scholar]
- Thomson J.M., Parker J., Perou C.M., Hammond S.M. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve P.M., le Sage C., Schrier M., Gillis A.J., Stoop H., Nagel R., Liu Y.P., van Duijse J., Drost J., Griekspoor A., et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- Wiseman S.M., Makretsov N., Nielsen T.O., Gilks B., Yorida E., Cheang M., Turbin D., Gelmon K., Huntsman D.G. Coexpression of the type 1 growth factor receptor family members HER-1, HER-2, and HER-3 has a synergistic negative prognostic effect on breast carcinoma survival. Cancer. 2005;103:1770–1777. doi: 10.1002/cncr.20970. [DOI] [PubMed] [Google Scholar]
- Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R.M., Okamoto A., Yokota J., Tanaka T., et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Yang L., Li L.D., Chen Y.D., Parkin D.M. Time trends, estimates and projects for breast cancer incidence and mortality in China. Zhonghua Zhong Liu Za Zhi [Chinese journal of oncology] 2006;28:438–440. [PubMed] [Google Scholar]
- Zhu S., Si M.L., Wu H., Mo Y.Y. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J. Biol. Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- Zhu S., Wu H., Wu F., Nie D., Sheng S., Mo Y.Y. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]