Abstract
RNA guanine-N7 methyltransferase catalyzes the third step of eukaryal mRNA capping, the transfer of a methyl group from AdoMet to GpppRNA to form m7GpppRNA. Mutational and crystallographic analyses of cellular and poxvirus cap methyltransferases have yielded a coherent picture of a conserved active site and determinants of substrate specificity. Models of the Michaelis complex suggest a direct in-line mechanism of methyl transfer. Because no protein contacts to the guanine-N7 nucleophile, the AdoMet methyl carbon (Cε) or the AdoHcy sulfur (Sδ) leaving group were observed in ligand-bound structures of cellular cap methyltransferase, it was initially thought that the enzyme facilitates catalysis by optimizing proximity and geometry of the donor and acceptor. However, the structure of AdoHcy-bound vaccinia virus cap methyltransferase revealed the presence of an N-terminal “lid peptide” that closes over the active site and makes multiple contacts with the substrates, including the AdoMet sulfonium. This segment is disordered in the vaccinia apoenzyme and is not visible in the available structures of cellular cap methyltransferase. Here, we conducted a mutational analysis of the vaccinia virus lid peptide (545DKFRLNPEVSYFTNKRTRG563) entailing in vivo and in vitro readouts of the effects of alanine and conservative substitutions. We thereby identified essential functional groups that interact with the AdoMet sulfonium (Tyr555, Phe556), the AdoMet adenine (Asn550), and the cap triphosphate bridge (Arg560, Arg562). The results suggest that van der Waals contacts of Tyr555 and Phe556 to the AdoMet Sδ and Cε atoms, and the electron-rich environment around the sulfonium, serve to stabilize the transition state of the transmethylation reaction.
Keywords: S-adenosylmethionine, mRNA capping, poxvirus, sulfonium
INTRODUCTION
The m7GpppN cap of eukaryal mRNA is formed by three enzymatic reactions: (1) the 5′ triphosphate end of the pre-mRNA is hydrolyzed to a diphosphate by RNA 5′ triphosphatase; (2) the diphosphate RNA end is capped with GMP by RNA guanylyltransferase; and (3) RNA (guanine-N7) methyltransferase, (cap methyltransferase) catalyzes transfer of a methyl group from S-adenosylmethionine (AdoMet) to GpppRNA to form m7GpppRNA and S-adenosylhomocysteine (AdoHcy). This pathway of mRNA capping is conserved in all eukaryal organisms and many eukaryal viruses.
Vaccinia virus cap methyltransferase is a heterodimer consisting of the carboxyl-terminal portion of the virus-encoded D1 polypeptide (aa 540–844; referred to herein as D1-C) and the 287-aa polypeptide encoded by the vaccinia D12 gene (Cong and Shuman 1992; Higman et al. 1992). The active site is located within D1-C, which has a weak intrinsic methyltransferase activity that is stimulated allosterically by D12 (Higman et al. 1994; Mao and Shuman 1994; Schwer et al. 2006). Cellular cap-methylating enzymes are monomeric proteins with primary structure similarity to vaccinia D1-C (Mao et al. 1995).
Mechanistic insights to cap methylation have emerged from crystal structures of the Encephalitozoon cuniculi cap methyltransferase Ecm1 (Fabrega et al. 2004) and the vaccinia D1-C/D12 heterodimer (De la Peña et al. 2007). These enzymes adopt similar tertiary structures and they contain several ligand-binding pockets: for the methyl donor AdoMet, for the cap guanosine methyl acceptor, and for the triphosphate bridge of the cap. Divergent primary structures of the AdoMet-binding motifs of poxvirus (VLAIDFG) versus cellular (VLDLGCG) cap methyltransferases had prompted suggestions that their AdoMet interactions might differ in functionally interesting ways (Wang and Shuman 1997; Bujnicki et al. 2001). Comparison of the crystal structures of AdoHcy-bound D1-C and Ecm1 revealed that this was indeed the case (De la Peña et al. 2007). Specifically, the adenosine nucleoside of the methyl donor in the poxvirus structure adopts a unique syn conformation (Fig. 1) unlike the anti conformation observed in Ecm1 and other AdoMet-dependent methyltransferases (De la Peña et al. 2007). The poxvirus methyltransferase also contains secondary structure elements, not found in Ecm1, which mediate its interaction with the D12 subunit. These unique features of the poxvirus enzyme have implications for antiviral drug design predicated on selectively occluding the methyl donor site of the poxvirus enzymes or interdicting D1–D12 heterodimerization.
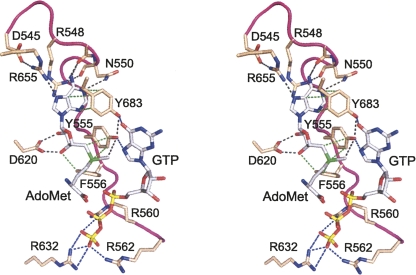
FIGURE 1.
Substrate contacts of the N-terminal lid peptide of vaccinia cap methyltransferase. The figure shows a stereo view of a model of the Michaelis complex of vaccinia D1-C active site with AdoMet and GTP bound (Zheng and Shuman 2008). The model was generated by importing the GTP ligand from the aligned structure of GTP-bound cellular cap methyltransferase (PDB ID 1ri2) and adding a methyl group to the AdoHcy ligand in the otherwise unperturbed D1-C structure (PDB ID 2vdw). The sulfur atom of the methyl donor is colored green. Actual polar atomic contacts of D1-C with the methyl donor and contacts between amino acid side chains are depicted as black dashed lines. Van der Waals contacts of Tyr555, Phe556, and Tyr683 with the methyl donor are depicted as green dashed lines. The modeled contacts between D1-C side chains and the GTP substrate are drawn as blue dashed lines.
The essential functional groups in the methyl donor and methyl acceptor sites have been defined through extensive mutational analyses of Ecm1, the homologous Saccharomyces cerevisiae cap methyltransferase Abd1, and the vaccinia virus cap methyltransferase (Mao et al. 1995, 1996; Mao and Shuman 1996; Wang and Shuman 1997; Schwer et al. 2000; Fabrega et al. 2004; Hausmann et al. 2005; Zheng et al. 2006; Zheng and Shuman 2008). Structure-function analysis of the vaccinia protein has been aided by a yeast-based genetic assay in which cell growth depends on catalysis of cap synthesis by the viral enzyme acting in lieu of yeast Abd1 (Saha et al. 2003). Complementation of the abd1Δ yeast strain requires coexpression of the catalytic D1-C and stimulatory D12 subunits. The yeast assay was used previously to identify 14 individual amino acids of D1-C as essential for cap methylation in vivo: Asn570, Lys573, Asp598, Gly600, Asp604, Lys607, Tyr608, Asp620, Arg632, Asp676, Phe679, His682, Tyr683, and Glu763 (Saha et al. 2003; Zheng and Shuman 2008). Mutational lethality in vivo correlated in most cases with severe biochemical defects in cap methylation in vitro (Mao et al. 1996; Saha et al. 2003, Zheng and Shuman 2008).
The mutational data have been interpreted in light of a model of the Michaelis complex derived from the crystal structures of AdoHcy-bound vaccinia cap methyltransferase and GTP-bound cellular cap methyltransferase (Zheng and Shuman 2008). Many of the essential amino acids comprise the binding sites for AdoMet (Lys573, Asp598, Gly600, Asp620, Tyr683), the cap guanosine (Phe679, His682, Tyr683, Glu763), or the cap triphosphate (Asn601, Arg632). Other essential D1-C residues either play structural roles or are suggested to bind to the 5′ terminal nucleotides of the RNA substrate (Zheng and Shuman 2008). The architecture of a putative mRNA docking site has not yet been elucidated for the poxvirus or cellular cap guanine-N7 methyltransferases.
Most of the essential vaccinia D1-C amino acids that contact the substrates are conserved in Ecm1 and other cellular cap methyltransferases. An important insight from the structure of the AdoHcy-bound D1-C concerns the nature and disposition of an N-terminal peptide that was disordered in the Ecm1 crystal structures (De la Peña et al. 2007). The vaccinia peptide, 545DKFRLNPEVSYFTNKRTRG563, is essential for activity insofar as its deletion is lethal in vivo (Schwer and Shuman 2006). This segment is protease sensitive in the vaccinia D1/D12 apoenzyme, but becomes protease resistant in the presence of AdoMet (Shuman 1989; De la Peña et al. 2007). The N-terminal peptide is ordered in the D1-C/D12 heterodimer, where it forms a surface loop that overlays AdoHcy and virtually buries the methyl donor and acceptor in a manner reminiscent of the structure of mammalian glycine-N-methyltransferase bound to AdoHcy (Takata et al. 2003; De la Peña et al. 2007). Several residues within the vaccinia N-terminal peptide make atomic contacts with AdoHcy and are imputed to contact the AdoMet sulfonium and the GTP methyl acceptor in the modeled Michaelis complex (Fig. 1). Initial mutational studies by Cusack and colleagues showed that Tyr555 is critical for cap methylation in vitro (De la Peña et al. 2007).
Here, we performed a comprehensive structure-guided mutational analysis of the N-terminal peptide of D1-C as well as several other residues with imputed roles at the D12 interface or as candidates to interact with RNA. Our findings shed new light on cap recognition and the catalytic mechanism of methyl transfer from AdoMet to guanine-N7.
RESULTS AND DISCUSSION
Alanine scanning of D1-C and mutational effects on cap methylation activity in vivo
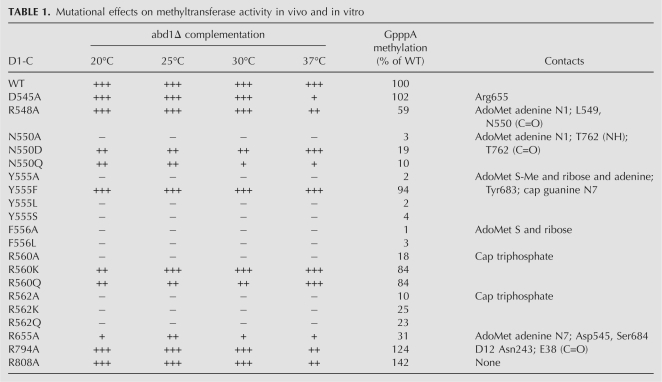
The present study focuses on the role of the N-terminal peptide of D1-C (Fig. 1). The amino acids subjected to alanine scanning were Asp545, Arg548, Asn550, Tyr555, Phe556, Arg560, and Arg562. The actual or imputed atomic contacts of these side chains with the methyl donor and acceptor in the modeled Michaelis complex are illustrated in Figure 1 and listed in Table 1. We also mutated Arg655, which interacts with Asp545 and the AdoMet adenine (Fig. 1). Two other residues were included in the alanine scan: Arg794, a component of the D1–D12 interface, and Arg808, a surface residue located distal to the cap triphosphate that seemed a plausible candidate to interact with RNA. The D1-C-Ala alleles on single-copy CEN plasmids were tested for function in vivo in the yeast abd1Δ strain by cotransformation with wild-type CEN D12. The recipient yeast bears a CEN URA3 ABD1 plasmid to sustain viability. If the vaccinia subunits associate in vivo to form a catalytically active heterodimer, the yeast cells can lose the URA3 ABD1 plasmid and grow on medium containing 5-fluoroorotic acid (FOA), a drug that selects against URA3. The results of the plasmid shuffle assay are compiled in Table 1.
TABLE 1.
Mutational effects on methyltransferase activity in vivo and in vitro
Alanine changes at five targeted positions—Asp545, Arg548, Arg655, Arg794, and Arg808—allowed for growth at 30°C on medium containing FOA, signifying that none of these five side chains was essential for cap methyltransferase activity in vivo. The five viable D1-C-Ala/D12 strains were then tested for growth on rich medium (YPD agar) at 20, 25, 30, and 37°C. Growth was scored as follows: (+++) colony size indistinguishable from strains bearing wild-type D1-C/D12; (++) reduced colony size; (+) only pinpoint colonies were formed (Table 1). Four of the D1-C-Ala mutations elicited mild temperature-sensitive growth phenotypes. The D545A, R548A, R794A, and R808A strains grew as well as wild-type D1-C cells at 20–30°C, but formed smaller colonies (R548A, R794A, R808A) or pinpoint colonies (D545A) at 37°C. In contrast, the R655A mutation resulted in a more severe phenotype entailing ++ growth at 25°C and + growth at 20, 30, and 37°C (Table 1).
Asp545 forms a salt bridge to Arg655, which, in turn, donates a hydrogen bond to the AdoMet adenine-N7 atom (Fig. 1). The Asp545/Arg655 pair is co-conserved in the chordopoxvirus capping enzymes, but is codiverged in the entomopoxvirus enzymes, where they are either Lys/Leu or Ser/Leu. The finding that Asp545 is nonessential in vivo implies that the Arg655 side chain can interact with the AdoMet adenine without being held in place by a salt bridge. Arg548 donates a trifurcated hydrogen bond from its terminal guanidinium nitrogen to the adenine N1 atom of AdoMet and the main chain carbonyls of vicinal residues 549 and 550 (Fig. 1). Arg548 is conserved in chordopoxvirus capping enzymes, but is substituted by aliphatic residues (Ile or Leu) in the entomopoxvirus homologs. The nonessential Arg794 side chain makes intersubunit hydrogen bonds to the Asn243 side chain and the Glu38 main chain carbonyl of the D12 subunit. Arg794 is invariant in chordopoxvirus D1 proteins, but is replaced by Asn or Tyr in the entomopoxvirus orthologs. Arg808 is conserved in all poxvirus capping enzymes. It is conceivable that some conserved D1 residues that are not important for cap methyltransferase activity play a role in other aspects of capping enzyme function, e.g., during transcription initiation and termination (Vos et al. 1991; Luo et al. 1995).
Lethal mutations were those that failed to support the appearance of FOA-resistant colonies at 20, 25, 30, or 37°C (scored as – at all temperatures). Alanine substitutions at five positions were lethal, thereby defining Asn550, Tyr555, Phe556, Arg560, and Arg562 as essential for methyltransferase activity in vivo (Table 1). Four of these essential residues (Asn550, Tyr555, Phe556, Arg562) are conserved in the D1 orthologs encoded by numerous diverse genera of vertebrate and invertebrate poxviruses. Arg560 is conserved in chordopoxvirus capping enzymes, but is substituted by Gln or Thr in entomopoxvirus D1 proteins.
Effects of alanine mutations on cap methyltransferase activity in vitro
Wild-type D1-C and all the D1-C-Ala mutants were produced as His6-tagged proteins in Escherichia coli cells coexpressing untagged D12. The vaccinia proteins were isolated from soluble bacterial extracts by nickel-affinity chromatography. Copurification of the untagged D12 protein with the tagged catalytic subunit reveals whether a particular mutation disrupted subunit interaction. SDS-PAGE analysis showed that the wild-type and mutant protein preparations consisted of two predominant polypeptides that corresponded to D1-C and D12, respectively (Fig. 2A). We surmise that none of the mutated amino acids is crucial for heterodimerization, not even Arg794, which participates in the homodimer interface.
FIGURE 2.
Recombinant wild-type and mutant D1-C/D12 heterodimers. Aliquots of the nickel-agarose preparations (containing 1.8 μg of the D1-C polypeptide) were analyzed by SDS-PAGE. The polypeptides were visualized by staining with Coomassie Blue dye. The positions and sizes (kDa) of marker polypeptides are indicated on the left. Alanine mutants are shown in A; conservative mutants are shown in B.
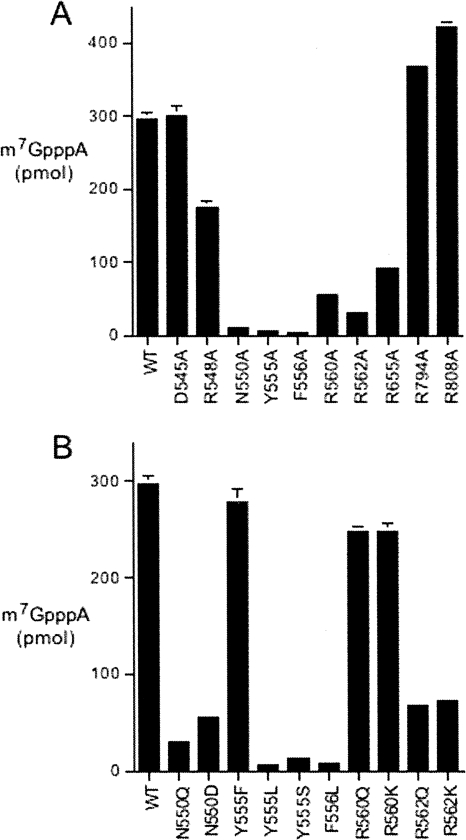
Each protein was assayed for methyl transfer from [3H-CH3]AdoMet to GpppA. The reaction products were separated by PEI-cellulose TLC and the transfer of the tritiated methyl group to generate labeled m7GpppA was quantified (Fig. 3A). The activities of the mutants were normalized to that of wild-type D1-C/D12 (see results compiled in Table 1) and interpreted in light of the phenotypes elicited in yeast. As might be expected, the mutants that displayed the highest level of function in yeast (D545A, R548A, R794A, and R808A) displayed wild-type or near wild-type methyltransferase activity in vitro. The R655A mutant that displayed a slow growth phenotype in yeast had weaker methyltransferase activity in vitro, at a level of 31% of wild type. The five mutations that resulted in unconditional lethality in yeast had even lower cap methyltransferase activity in vitro, as follows: F556A (1% of wild-type), Y555A (2%), N550A (3%), R562A (10%), and R560A (18%). Thus, for the alanine mutants, there was a fairly good correlation between the in vivo growth phenotype and catalytic function in vitro.
FIGURE 3.
Mutational effects on vaccinia virus cap methyltransferase activity. The extents of 3H-m7GpppA formation by wild-type and mutant D1-C/D12 heterodimers are plotted with alanine mutants surveyed in A and conservative mutants in B. Each datum is the average of three experiments; standard error bars are shown.
Structure-function relationships gleaned from effects of conservative mutations
Conservative amino acid substitutions were introduced in lieu of the five residues defined as essential in vivo in the present alanine scan. The 10 conservative mutants were tested by plasmid shuffle for methyltransferase activity in vivo (Table 1). The mutant His6-D1-C and wild-type D12 subunit were produced together in bacteria and affinity purified. SDS-PAGE analysis showed that the tagged D1-C and untagged D12 proteins copurified in each case (Fig. 2B). Aliquots of each mutant were assayed for methyl transfer from [3H-CH3]AdoMet to GpppA (Fig. 3B), and the activities of the mutants were normalized to wild-type D1-C/D12 activity (Table 1). Useful structure-activity relationships were thereby delineated for the essential residues and there was a good correlation in most cases between in vitro and in vivo mutational effects. The findings are summarized as follows and discussed in light of the amino acid side-chain interactions with AdoMet (Tyr555, Phe556, and Asn550) or the cap triphosphate (Arg560 and Arg562) of the modeled Michaelis complex.
We found that methyltransferase activity was restored fully in vivo when the essential Tyr555 side chain was substituted with phenylalanine, which also elicited a significant gain of catalytic function in vitro, to 94% of wild type. Thus, the Tyr555 hydroxyl group (with its imputed hydrogen bond to the cap guanine-N7 atom in the Michaelis complex, van der Waals contact to the AdoMet methyl group, and hydrogen bond to the Tyr683 side-chain hydroxyl group) is not essential for activity. In contrast, the leucine and serine changes at Tyr555 were lethal in vivo and afforded no gain of activity in vitro compared with the alanine mutant. The leucine versus phenylalanine results underscore the importance of the van der Waals contacts to AdoMet made by the terminal Tyr555 atoms: Cz (3.8 Å to the reactive Cε methyl center; 4 Å to the sulfur; 3.9 Å to the ribose C5′ atom), Cε2 (4 Å to the reactive methyl center; 4 Å to the sulfur), and Cε1 (3.6 Å to the adenine C2 atom; 3.4 Å to adenine-N3). This network of contacts, and the π-cation interaction of the aromatic tyrosine overlying the AdoMet sulfonium, likely serve to stabilize the transition state of the transmethylation reaction.
No restoration of function in vivo or in vitro was achieved by replacing Phe556 with leucine, again emphasizing the key contributions of the phenyl group via the van der Waals contacts to AdoMet made by the terminal Phe556 atoms: Cz (3.8 Å to the sulfur; 4 Å to the ribose C3′ atom) and Cε1 (3.7 Å to the ribose O3′ atom).
The lethality of the N550A mutation was ameliorated by aspartate, which conferred ++ growth at 25° and 30°C and +++ growth at 37°C (Table 1) and also increased catalytic activity to 19% of wild type, compared with 3% for N550A. However, the N550Q mutant was less active in vivo (+ growth) and in vitro (10% of wild type) than N550D. These results are interpretable in light of the atomic contacts of Asn550, including: (1) a bidentate hydrogen bond from Asn550 Oδ and Nδ to the main-chain amide and carbonyl of residue Thr726, and (2) a hydrogen bond from Asn550 Nδ to the adenine N1 atom of AdoMet. The contacts to Thr726 help tether the N-terminal peptide to a β strand of the cap guanine binding pocket. The hierarchy of conservative mutational effects at Asn550 reveals the importance of the contact of the Oδ of Asn550 (or its isosteric Asp replacement) to the Thr726 amide and an apparent steric constraint on this interaction that limits the efficacy of the longer glutamine side chain. Although the Asn550 Nδ is needed for optimal methyltransferase activity in vitro, as inferred from the fivefold difference between the wild-type and N550D proteins, we suspect that the Asn550 Nδ contact to AdoMet is partly redundant to that of the nonessential Arg548 side chain, which also donates a hydrogen bond to AdoMet adenine-N1 (Fig. 1).
The cap-binding cleft in the D1–D12 crystal structure includes a sulfate anion coordinated by the guanidinium nitrogens of Arg560, Arg562, and Arg632 (De la Peña et al. 2007). Our model of the Michaelis complex superimposes the γ-phosphate of the GTP methyl acceptor on this sulfate. We reported previously that Arg632 was essential for vaccinia cap methyltransferase activity in vitro and in vivo. Because lysine could functionally substitute for Arg632, whereas glutamine could not, we surmised that the electrostatic interactions with the cap triphosphate bridge are most relevant. Here, we found that the Arg560 and Arg562 ligands for the cap triphosphate are also essential, albeit with different structure-activity relationships inferred from the effects of conservative substitutions. Replacing Arg560 with either lysine or glutamine restored function in yeast and revived methyltransferase activity in vitro (to 84% of wild type) compared with the lethal alanine mutant (18%). Thus, we infer that hydrogen bonding to the cap triphosphate is the key contribution of this side chain. Note again that Arg560 is replaced by potential hydrogen-bond donors glutamine or threonine in entomopoxvirus D1 orthologs. In contrast, replacing Arg562 with either lysine or glutamine was lethal in vivo in yeast. The R562K and R562Q proteins had 23%–25% of wild-type cap methyltransferase activity in vitro, a result that, at first glance, might appear not to jive with lethality in vivo. (The same issue arises for the lethal R560A mutant, which had 18% of wild-type methyltransferase activity in vitro.)
As noted previously (Zheng and Shuman 2008), the in vitro cap methyltransferase screening assay tends to obscure mutational effects on substrate binding because the GpppA concentration used is >10-fold higher than the K m for GpppA of the wild-type enzyme. Thus, the R560A, R562K, and R562Q mutants might have diminished affinities for the cap that explain their poor performance in vivo, as documented previously for the R632Q mutant (Zheng and Shuman 2008). To address this issue, we determined kinetic parameters for the wild-type D1-C/D12 heterodimer and the R562K, R652Q, and R560A mutants by titrating GpppA at a fixed concentration of AdoMet (40 μM). K m and k cat values were calculated from double-reciprocal plots of the data (data not shown). The results are summarized as follows. The wild-type enzyme had a K m of 35 μM GpppA and a k cat of 1.2 min−1. The R560A, R562K, and R562Q mutants had K m values of 360, 150, and 1300 μM GpppA, respectively. The k cat values of the R560A, R562K, and R562Q mutants were 0.29, 0.24, and 0.29 min−1, respectively. If one calculates catalytic efficiency as k cat/K m, then the R560A, R562K, and R562Q mutations elicited 43-fold, 20-fold, and 150-fold reductions compared with the wild-type methyltransferase. These effects on in vitro catalysis can account for the lethality in vivo. With respect to structure-activity relationships at Arg562, the results underscore that optimal affinity for the methyl acceptor is dependent on electrostatic interactions of the arginine side chain with the cap triphosphate bridge.
Mechanistic implications
Here, we determined the essential functions of the N-terminal lid peptide of vaccinia cap methyltransferase by mutational analysis guided by the crystal structure and a model of the Michaelis complex. Four of the five essential lid residues have clear nonstructural roles as components of the substrate-binding pockets for the cap-triphosphate (Arg560 and Arg562) or the AdoMet sulfonium and ribose (Tyr555 and Phe556). The essential Asn550 appears to play a structural role in tethering the lid domain over the active site, while also promoting methyltransferase activity via its contact to the AdoMet adenine.
As noted above, Arg560 and Arg562 participate with Arg632 in binding the cap triphosphate. Arg632 is conserved among poxvirus and cellular cap methyltransferases and is essential in all cases where its role has been tested (Wang and Shuman 1997; Saha et al. 1999; Hausmann et al. 2005; Zheng et al. 2006). Because the segment corresponding to the D1 lid peptide is disordered in the crystal structures of the E. cuniculi and human cap methyltransferases (Fabrega et al. 2004; Wu et al. 2007), it is not clear whether the cellular homologs share the three-arginine binding mode for the cap triphosphate bridge. We suspect they might, insofar as most cellular cap methyltransferases have an RxR, RxK, or RxQ tripeptide immediately upstream of the N-terminal helix in the crystal structures, which is a potential counterpart of the 560RxR562 tripeptide in the vaccinia D1 lid.
Our results reinforce the invoked similarities between cap guanine-N7 methyltransferase and glycine N-methyltransferase (GNMT) (Bujnicki et al. 2001; Takata et al. 2003; Fabrega et al. 2004; De la Peña et al. 2007). Comparisons of the crystal structure of the GNMT apoenzyme with a ternary complex of GNMT bound to AdoMet and acetate (an analog of the glycine methyl acceptor) revealed that substrate binding to GNMT triggers closure of an N-terminal loop over the active site (Takata et al. 2003) in a manner roughly analogous to the lid seen in the vaccinia D1 structure (De la Peña et al. 2007). Indeed, superposition of the D1 and GNMT structures shows that Tyr21 in the GNMT “lid” (GVAAEGIPDQY21ADGEAARV) occupies the same position as the essential Tyr555 in the vaccinia D1 lid. Like vaccinia Tyr555, the Tyr21 in GNMT makes contacts—via its OH, Cz, and Cε atoms—to the methyl carbon, sulfur, and ribose C5′ and C3′ atoms of AdoMet. Takata et al. (2003) proposed a mechanism for GNMT in which the charge-dipole interaction between AdoMet Sδ and Tyr21 OH facilitates an SN2 transfer reaction at the methyl carbon center. However, they also noted that mutating Tyr21 to phenylalanine elicited a relatively modest (fourfold) decrement in k cat, which would weigh against a significant role for the tyrosine OH (and its charge-dipole with Sδ) in reaction chemistry. Our mutational analysis of Tyr555 in vaccinia D1 affirms the inessentiality of the tyrosine OH, while emphasizing—via the differences between an active Y555F mutant and an inactive Y555L mutant—the critical role of the AdoMet contacts made by the terminal atoms of the phenyl ring. We propose that these contacts of Tyr555 (and its π-cation interaction with the sulfonium), together with those of Phe556, facilitate transmethylation by providing an electron-rich environment surrounding the two cationic centers (N7 and Sδ) in the predicted SN2 transition state.
The lid peptide of GNMT has no other primary structure similarity to the vaccinia D1 lid, which is not surprising given that the methyl acceptor glycine is smaller than the RNA cap, and there is no need in GNMT for analogs of D1 cap-binding residues such as Arg560 and Arg562. Indeed, the α-helix following the lid peptide is laterally displaced in GNMT so that it effectively occludes the binding groove for the triphosphate that is present in the cap guanine-N7 methyltransferases. It is notable that the position of the trigonal acetate moiety in the acceptor site of GNMT (which mimics glycine except for the amine) appears to overlie the trigonal guanine-N9 center of the cap in the modeled Michaelis complex of vaccinia D1. Adding a nitrogen to the acetate to simulate the glycine–AdoMet Michaelis complex of GNMT, as modeled by Takata et al. (2003), places the attacking glycine nitrogen in a position corresponding the cap guanine-N7 in the model of the D1 active site. Thus, the fundamentals of catalysis though substrate proximity, orientation, and transition-state stabilization are likely to be similar for GNMT and mRNA cap guanine-N7 methyltransferase.
MATERIALS AND METHODS
Mutations of the D1-C subunit of vaccinia cap methyltransferase
Missense mutations were introduced into the D1(540–844) gene by the PCR-based two-stage overlap extension method and the mutated genes were inserted into yeast expression vector pYN132 (CEN TRP1), where expression of D1-C is under the control of the yeast TPI1 promoter (Saha et al. 2003). The inserts were sequenced completely to ensure that no unwanted mutations were introduced during amplification and cloning. The mutated D1-C genes were excised from the yeast vectors by digestion with EcoRI/XhoI and then inserted into the bacterial expression plasmid pET28a, where they are fused to a 5′ leader sequence encoding a His6 tag.
Yeast-based assay of vaccinia cap methyltransferase function in vivo
Saccharomyces cerevisiae strain YBS40 is deleted at the chromosomal ABD1 locus encoding the yeast cap methyltransferase. Growth of YBS40 depends on the maintenance of plasmid p360-ABD1 (CEN URA3 ADE2 ABD1). abd1Δ cells were cotransformed with CEN TRP1 D1-C and CEN HIS3 D12 plasmids encoding wild-type or mutated versions of the D1-C subunit and the wild-type D12 subunit, respectively. Individual Trp+ His+ transformants were streaked on agar medium containing 0.75 mg/mL 5-fluoroorotic acid (FOA). Growth was scored after 7 d of incubation at 20, 25, 30, and 37°C. Lethal mutants were those that failed to form colonies on FOA at any temperature. Individual FOA-resistant colonies with viable D1-C mutants were transferred to YPD agar medium. Two isolates of each mutant were tested for growth on YPD agar at 20, 25, 30, and 37°C. Growth was assessed as follows: (+++) colony size indistinguishable from strains bearing wild-type D1-C; (++) slightly reduced colony size; (+) only pinpoint colonies were formed; (–) no growth.
Wild-type and mutant methyltransferase heterodimers
pET-His6-D1-C plasmids encoding tagged wild-type and mutant catalytic subunits were transformed into E. coli BL21-CodonPlus(DE3) together with plasmid pET-D12 encoding the nontagged stimulatory subunit. Cultures (500 mL) derived from single transformants were grown at 37°C in LB medium containing 50 μg/mL kanamycin, 100 μg/mL ampicillin, and 50 μg/mL chloramphenicol until the A 600 reached ∼0.6. The cultures were placed on ice for 30 min and then adjusted to 0.2 mM IPTG and 2% ethanol and incubation was continued for 20 h at 17°C with constant shaking. Cells were harvested by centrifugation and stored at −80°C. The His-tagged vaccinia proteins were purified from soluble bacterial lysates by Ni-agarose chromatography as described previously (Zheng and Shuman 2008). The concentrations of D1-C protein were determined by SDS-PAGE analysis of serial dilutions of the protein preparations in parallel with serial dilutions of a BSA standard. The gels were stained with Coomassie Blue, and the staining intensities of the D1-C and BSA polypeptides were quantified using a BIO-RAD Molecular Imager ChemiDoc gel densitometry analysis system. D1-C concentrations were calculated by interpolation to the BSA standard curve.
Methyltransferase assay
Reaction mixtures (20 μL) containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 700 μM GpppA (New England Biolabs), 40 μM [3H-CH3]AdoMet (Perkin Elmer Life Sciences), and aliquots of the Ni-agarose preparations of wild-type or mutant recombinant D1-C/D12 (containing 0.3 μg of the D1-C polypeptide) were incubated for 30 min at 30°C. Aliquots (4 μL) were spotted on PEI-cellulose TLC plates, which were developed with 0.05 M ammonium sulfate. The AdoMet- and m7GpppA-containing portions of the lanes were cut out and the radioactivity in each was quantified by liquid scintillation counting.
ACKNOWLEDGMENTS
This work was supported by NIH Grant GM42498. S.S. is an American Cancer Society Research Professor.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1201308.
REFERENCES
- Bujnicki J.M., Feder M., Radlinska M., Rychlewski L. mRNA:guanine-N7 methyltransferases: Identification of novel members of the family, evolutionary analysis, homology modeling, and analysis of sequence-structure-function relationships. BMC Bioinformatics. 2001;2:2. doi: 10.1186/1471-2105-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong P., Shuman S. Methyltransferase and subunit association domains of vaccinia virus mRNA capping enzyme. J. Biol. Chem. 1992;267:16424–16429. [PubMed] [Google Scholar]
- De la Peña M., Kyrielis O., Cusack S. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyltransferase. EMBO J. 2007;26:4913–4925. doi: 10.1038/sj.emboj.7601912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrega C., Hausmann S., Shen V., Shuman S., Lima C.D. Structure and mechanism of cap (guanine-N7) methyltransferase. Mol. Cell. 2004;13:77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- Hausmann S., Zheng S., Fabrega C., Schneller S.W., Lima C.D., Shuman S. Encephalitozoon cuniculi mRNA cap (guanine-N7) methyltransferase: Methyl acceptor specificity, inhibition by AdoMet analogs, and structure-guided mutational analysis. J. Biol. Chem. 2005;280:20404–20412. doi: 10.1074/jbc.M501073200. [DOI] [PubMed] [Google Scholar]
- Higman M.A., Bourgeois N., Niles E.G. The vaccinia virus mRNA (guanine-N7-) methyltransferase requires both subunits of the mRNA capping enzyme for activity. J. Biol. Chem. 1992;267:16430–16437. [PubMed] [Google Scholar]
- Higman M.A., Christen L.A., Niles E.G. The mRNA (guanine-7-) methyltransferase domain of the vaccinia virus mRNA capping enzyme: Expression in Escherichia coli and structural and kinetic comparison to the intact capping enzyme. J. Biol. Chem. 1994;269:14974–14981. [PubMed] [Google Scholar]
- Luo Y., Mao X., Deng L., Cong P., Shuman S. The D1 and D12 subunits are both essential for the transcription termination factor activity of vaccinia virus capping enzyme. J. Virol. 1995;69:3852–3856. doi: 10.1128/jvi.69.6.3852-3856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Shuman S. Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit: Identification of amino acid residues in the D1 protein required for subunit association and methyl group transfer. J. Biol. Chem. 1994;269:24472–24479. [PubMed] [Google Scholar]
- Mao X., Shuman S. Vaccinia virus mRNA (guanine-7) methyltransferase: Mutational effects on cap methylation and AdoHcy-dependent photocrosslinking of the cap to the methyl acceptor site. Biochemistry. 1996;35:6900–6910. doi: 10.1021/bi960221a. [DOI] [PubMed] [Google Scholar]
- Mao X., Schwer B., Shuman S. Yeast mRNA cap methyltransferase is a 50-kDa protein encoded by an essential gene. Mol. Cell. Biol. 1995;15:4167–4174. doi: 10.1128/mcb.15.8.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Schwer B., Shuman S. Mutational analysis of the Saccharomyces cerevisiae ABD1 gene: Cap methyltransferase activity is essential for cell growth. Mol. Cell. Biol. 1996;16:475–480. doi: 10.1128/mcb.16.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha N., Schwer B., Shuman S. Characterization of human, Schizosaccharomyces pombe and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J. Biol. Chem. 1999;274:16553–16562. doi: 10.1074/jbc.274.23.16553. [DOI] [PubMed] [Google Scholar]
- Saha N., Shuman S., Schwer B. Yeast-based genetic system for functional analysis of poxvirus mRNA cap methyltransferase. J. Virol. 2003;77:7300–7307. doi: 10.1128/JVI.77.13.7300-7307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Shuman S. Genetic analysis of poxvirus mRNA cap methyltransferase: Suppression of conditional mutations in the stimulatory D12 Subunit by second-site mutations in the catalytic D1 subunit. Virology. 2006;352:145–156. doi: 10.1016/j.virol.2006.03.050. [DOI] [PubMed] [Google Scholar]
- Schwer B., Saha N., Mao X., Chen H.W., Shuman S. Structure-function analysis of yeast mRNA cap methyltransferase and high-copy suppression of conditional mutants by AdoMet synthase and the ubiquitin conjugating enzyme Cdc34p. Genetics. 2000;155:1561–1576. doi: 10.1093/genetics/155.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Hausmann S., Schneider S., Shuman S. Poxvirus cap methyltransferase: Bypass of the requirement for the stimulatory subunit by mutations in the catalytic subunit and evidence for intersubunit allostery. J. Biol. Chem. 2006;281:18953–18960. doi: 10.1074/jbc.M602867200. [DOI] [PubMed] [Google Scholar]
- Shuman S. Functional domains of vaccinia virus mRNA capping enzyme. Analysis by limited tryptic digestion. J. Biol. Chem. 1989;264:9690–9695. [PubMed] [Google Scholar]
- Takata Y., Huang Y., Komoto J., Yamada T., Konishi K., Ogawa H., Gomi T., Fujioka M., Takusagawa F. Catalytic mechanism of glycine N-methyltransferase. Biochemistry. 2003;34:8394–8402. doi: 10.1021/bi034245a. [DOI] [PubMed] [Google Scholar]
- Vos J.C., Sasker M., Stunnenberg H.G. Vaccinia virus capping enzyme is a transcription initiation factor. EMBO J. 1991;10:2553–2558. doi: 10.1002/j.1460-2075.1991.tb07795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.P., Shuman S. Structure-function analysis of the mRNA cap methyltransferase of Saccharomyces cerevisiae . J. Biol. Chem. 1997;272:14683–14689. doi: 10.1074/jbc.272.23.14683. [DOI] [PubMed] [Google Scholar]
- Wu H., Lunin V.V., Zeng H., Antoshenko T., MacKenzie F., Weigelt J., Arrowsmith C.H., Edwards A.M., Bochkarev A., Min J., et al. The crystal structure of human RNA (guanine-7-) methyltransferase in complex with SAH. RCSB Protein Data Bank ID code 3bgv . 2007 http://www.rcsb.org/pdb/explore.do?structureId=3bgv.
- Zheng S., Shuman S. Structure-function analysis of vaccinia virus mRNA cap methyltransferase. RNA. 2008;14:696–705. doi: 10.1261/rna.928208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Hausmann S., Liu Q., Ghosh A., Schwer B., Lima C.D., Shuman S. Mutational analysis of Encephalitozoon cuniculi mRNA cap (guanine-N7) methyltransferase, structure of the enzyme bound to sinefungin, and evidence that cap methyltransferase is the target of sinefungin's antifungal activity. J. Biol. Chem. 2006;281:35904–35913. doi: 10.1074/jbc.M607292200. [DOI] [PubMed] [Google Scholar]