Abstract
The exoribonuclease polynucleotide phosphorylase (PNPase, encoded by pnp) is a major player in bacterial RNA decay. In Escherichia coli, PNPase expression is post-transcriptionally regulated at the level of mRNA stability. The primary transcript is very efficiently processed by the endonuclease RNase III at a specific site and the processed pnp mRNA is rapidly degraded in a PNPase-dependent manner. While investigating the PNPase autoregulation mechanism we found, by UV-cross-linking experiments, that the ribosomal protein S1 in crude extracts binds to the pnp-mRNA leader region. We assayed the potential role of S1 protein in pnp gene regulation by modulating S1 expression from depletion to overexpression. We found that S1 depletion led to a sharp decrease of the amount of pnp and other tested mRNAs, as detected by Northern blotting, whereas S1 overexpression caused a strong stabilization of pnp and the other transcripts. Surprisingly, mRNA stabilization depended on PNPase, as it was not observed in a pnp deletion strain. PNPase-dependent stabilization, however, was not detected by chemical decay assay of bulk mRNA. Overall, our data suggest that PNPase exonucleolytic activity may be modulated by the translation potential of the target mRNAs and that, upon ribosomal protein S1 overexpression, PNPase protects from degradation a set of full-length mRNAs. It thus appears that a single mRNA species may be differentially targeted to either decay or PNPase-dependent stabilization, thus preventing its depletion in conditions of fast turnover.
Keywords: RNA degradation, translation regulation, ribosomal protein S1, PNPase
INTRODUCTION
Mechanisms that modulate gene expression at the level of transcription and translation efficiency have long been studied. To the contrary, the issues of mRNA maturation and degradation have been addressed only rather recently in more detail as important steps in post-transcriptional control of gene expression. It is commonly accepted that RNA translation and degradation are intimately interlinked processes and that untranslated mRNAs are generally more susceptible to degradation machinery than efficiently translated transcripts, although uncoupling between translation and degradation efficiency has been also documented (for reviews, see Stanssens et al. 1986; Yarchuk et al. 1991; Joyce and Dreyfus 1998; Nogueira et al. 2001; Komarova et al. 2005; Kaberdin and Blasi 2006). However, our present knowledge of cis-elements, trans-acting factors, and mechanisms involved in the interplay between these processes is still poor. The current model of mRNA degradation in Escherichia coli maintains that an RNase E cleavage downstream of the translation initiation region (TIR) not only functionally inactivates an mRNA, but may also trigger its degradation by subsequent downstream endonucleolytic cleavages in the ribosome-free transcript. In addition to the TIR, other sequences and secondary structures in the 5′-untranslated region (5′-UTR) can modulate the efficiency of translation initiation and the rate of the first endonucleolytic cleavage that triggers the pathways for the complete degradation of mRNA. At the 3′-end, the hairpin structures of Rho-independent transcription terminators, exonucleases such as RNase II and polynucleotide phosphorylase (PNPase, encoded by pnp gene), polyadenyl polymerase [PAP, which adds destabilizing poly(A) tails], and the RNA chaperon Hfq are important elements for the control of mRNA stability (for reviews, see Coburn et al. 1999; Régnier and Arraiano 2000; Deutscher 2006; Condon 2007).
An interesting model system for the study of the interplay between mRNA translation and decay may be the regulation of pnp gene expression. PNPase, one of the major players in E. coli mRNA degradation, is widely conserved among Bacteria, and homologous genes have been found in plants and human cells (Bermudez-Cruz et al. 2005; for reviews, see Leszczyniecka et al. 2004; Chen et al. 2007). PNPase has been shown to catalyze, both in vitro and in vivo, the processive 3′ to 5′ phosphorolytic degradation of RNA and the reverse polymerization reaction (Thang et al. 1967; Godefroy 1970; Godefroy et al. 1970; Chou and Singer 1971; Donovan and Kushner 1986; Mohanty and Kushner 2000; Mohanty and Kushner 2006) and may be found as a component of a multiprotein machine, the RNA degradosome, together with the endonuclease RNase E, the RNA helicase RhlB, and enolase (Carpousis et al. 1994; Miczak et al. 1996; Py et al. 1996). Genetic and molecular studies have involved PNPase in several cell processes, which may account for the pleiotropic effects of pnp mutations (Jarrige et al. 2002; Briani et al. 2007).
Expression of pnp gene is post-transcriptionally regulated through the control of its messenger stability and translatability. Transcription of pnp can originate from two promoters, rpsOp (formerly P1) and pnp-p (formerly P2; Portier and Régnier 1984). Both primary transcripts are rapidly processed by RNase III endonuclease 81 nucleotides (nt) upstream of the pnp start codon within a long hairpin structure (Régnier and Grunberg-Manago 1990; Robert-Le Meur and Portier 1992; Jarrige et al. 2001). This originates a mono-cistronic mRNA 2.25 kb long, which extends from the RNase III site to the pnp-t transcription termination site, and longer transcripts terminating at downstream sites (Fig. 1; Portier et al. 1987; Zangrossi et al. 2000). The processed pnp mRNAs, in the presence of PNPase, are very unstable. It has been proposed that PNPase could bind determinants at the 5′-end of pnp mRNA, thus inhibiting its translation and promoting its degradation (Robert-Le Meur and Portier 1992, 1994; Portier et al. 1987). A more recent model postulates that the short duplex remaining at the 5′-end of pnp mRNA upon RNase III processing may protect the messenger RNA from degradation. PNPase would trigger RNase E-dependent degradation of pnp mRNA by degrading the short RNA that shields the mRNA 5′-end (Jarrige et al. 2001, 2002; Regonesi et al. 2004).
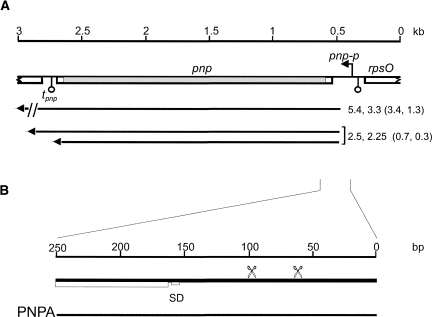
FIGURE 1.
(A) Map of the E. coli pnp region and pnp transcripts. The map of pnp gene and flanking regions is drawn to scale (kilobases on the upper line) based on the annotated E. coli genome sequence (NCBI Accession number U00096.2). (Open boxes) genes; (bent arrow) pnp-p promoter; (hanging balls) transcription terminators. The part of pnp filled in gray is deleted in frame in the Δpnp-751 allele (coordinates of the deletion are 3307070–3309180). pnp+ transcript are reported under the map and their length is indicated in kilobases (Zangrossi et al. 2000); in brackets are the length (in kilobases) of Δpnp-751 transcripts corresponding to monocistronic (0.3 and 0.7) or polycistronic (1.3 and 3.4) pnp+ RNAs. The length of Δpnp-751 mRNAs was determined by comparison to molecular weight markers and internal standards (rRNAs and transcripts with precisely mapped 5′- and 3′-ends). Their 5′-end was determined by primer extension and, as in pnp+, corresponds to RNase III processing site (coordinates 3309266; data not shown), whereas their 3′-ends were approximately deduced from transcript sizes and, for the 0.3- and 0.7-kb RNAs, confirmed by Northern hybridization with appropriate oligonucleotides. (B) The 5′ portion of pnp primary transcript from pnp-p promoter is scaled up (thick line) and the PNPA RNA substrate (thin line) is shown. (Scissors) RNase III processing sites; (SD) Shine–Dalgarno sequence; (open box) pnp ORF.
In experiments aimed at the identification of factors involved in pnp autoregulation, we found that both PNPase and the ribosomal protein S1 bind in vitro the pnp-mRNA leader region (Briani et al. 2007; this work). S1 is an “atypical” ribosomal protein whose role in translation, transcription, and control of RNA stability has been recognized but not fully understood. It is the largest (61 kDa) ribosomal protein present in the 30S subunit and consists of six repeated nonidentical motifs (S1 domains; Bycroft et al. 1997) that are typical of many RNA binding proteins including PNPase (Goldstein et al. 1990; Murzin 1993; Arcus 2002; Theobald et al. 2003). S1 has been implicated in a wide range of phenomena related to RNA transactions (Subramanian 1983). In vivo this protein seems to be required for translation (both initiation and, at least in some cases, elongation) of most mRNAs (Potapov and Subramanian 1992; Tzareva et al. 1994; Sørensen et al. 1998; Moll et al. 2002). S1 has been also purified as a poly(A) tail binding factor from E. coli cell extracts and has been shown to interact with RNase E and PNPase in Far-Western assays, suggesting that S1 could be involved in mRNA decay control (Kalapos et al. 1997; Feng et al. 2001). Recently, S1 has been implicated, albeit with ancillary roles, in trans-translation (Saguy et al. 2007) and in modulation of transcription efficiency (Sukhodolets et al. 2006). In conclusion, although S1 has been discovered over three decades ago and has been classified as a “ribosomal” protein, its multifaceted role in RNA transactions still needs clarifications.
In this work we have analyzed the effect of S1 intracellular concentration on pnp and other model mRNAs decay. Overall, our results suggest that upon a translational stress, PNPase and S1 may cooperate in protecting full-length mRNAs from degradation pathways and imply a previously unrecognized role for these two versatile proteins in coordinating mRNA translation and decay.
RESULTS
Identification of a protein binding the pnp mRNA leader region
While studying the binding of PNPase to the 5′-untranslated region of pnp mRNA by UV-cross-linking assay, we found that, in addition to PNPase, another protein (Pnp leader binding protein [PLBP]) with an apparent MW of about 70 kDa was able to bind the RNA substrate (see Fig. 2C; Briani et al. 2007). The binding between PLBP and the pnp 5′-UTR RNA substrate (Fig. 1, PNPA) did not depend on PNPase, as it was not affected by different point mutation in pnp or by the deletion of the whole gene (Briani et al. 2007).
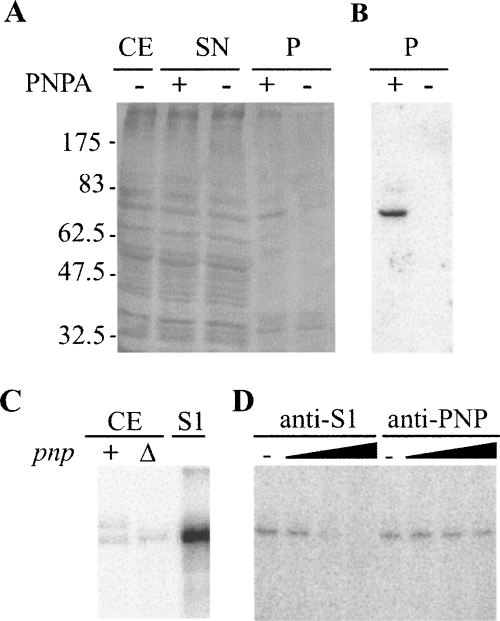
FIGURE 2.
Identification of PLBP as the ribosomal protein S1. (A,B) Affinity purification of PLBP. (A) Aliquots of fractions obtained from C-5691 during the purification procedure detailed in Materials and Methods section were separated by 10% SDS-PAGE and silver stained. (CE) crude extract (2 μg); (SN) supernatant fraction recovered after centrifugation of the mix between crude extract and streptavidin-coated beads (2 μg); (P) fraction eluted from the beads (one third of the elution volume); (−), fraction eluted from mock beads. Molecular weight of protein markers is indicated on the left. (B) Northwestern analysis. Ten microliters of P samples were run on 10% SDS-polyacrylamide gel, blotted onto a nitrocellulose sheet, and incubated with radiolabeled PNPA substrate. (C,D) S1 binding to PNPA in UV-crosslinking assays. (C) Crosslinking with crude cell extracts. We incubated 5 × 104 cpm of [32P]-labeled PNPA with 0.3 μg of C-5612/pAZ8 (+) or C-5691 (Δ) crude extract or with 1 pmol of purified S1 and UV-irradiated as detailed in Materials and Methods. The reaction products were then separated by 10% SDS-PAGE, blotted onto a nitrocellulose filter, and visualized by phosphorimaging. (D) Cross-linking with S1-immunodepleted cell extracts. We cross-linked 0.3 μg of C-1a crude extract preincubated 30 min at 37°C without (−) or with different amounts of S1 (1, 10, 100 μg) or PNPase (0.165, 1.65, 16.5 μg) antiserum to radiolabeled PNPA and analyzed it as described in C.
In order to identify PLBP, we exploited its affinity for the PNPA substrate. A biotinylated PNPA was immobilized on streptavidin-coated agarose beads and incubated with E. coli Δpnp-751 mutant crude extract. After incubation, the beads and the associated RNA–protein complexes were recovered by centrifugation and the proteins in the pellets analyzed by SDS-PAGE and silver staining of the gel. As shown in Figure 2A, we could detect, in addition to several nonspecific bands present both in the pellet of RNA-coated agarose beads and in the bare beads control sample, a specific signal that was absent in the control and corresponded to a protein with the same electrophoretic mobility as PLBP. Such a protein exhibited RNA binding activity, as assessed by Northwestern assay using [32P]-labeled PNPA as a substrate (Fig. 2B).
The 70-kDa band was extracted from the gel and analyzed by means of liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) (Maiorino et al. 2005). Ribosomal protein S1 (GI 1,787,140, calculated MW of 61 kDa; apparent MW in SDS-PAGE of ∼68 kDa; Kimura et al. 1982) turned out to be the most represented protein in the band, with 19 different peptides identified by amino acid sequencing and a SEQUEST score, a parameter correlated with protein abundance (Regonesi et al. 2006), of 290. Two additional proteins, FtsH and DnaK, were also detected with lower score values (40 and 20, respectively).
The correspondence of PLPB with S1 was confirmed by UV-cross-linking the PNPA substrate to purified S1, which gave a strong signal comigrating with the PLBP band seen in crude extract (Fig. 2C) and S1-immunodepleted crude extracts, in which the signal was absent (Fig. 2D).
Role of S1 in pnp gene regulation: Effect of modulation of S1 cellular concentration on pnp mRNA abundance and stability
To assess whether S1 might be involved in PNPase regulation, we assayed the effect of S1 cellular concentration on pnp expression.
S1 depletion
To lower S1 cellular concentration, we constructed an rpsA (the S1-encoding gene) conditional expression mutant by placing the chromosomal rpsA gene under the arabinose inducible promoter araBp. The araBp-rpsA allele was obtained in the pnp+ strain C-1a and then transferred by P1 transduction in C-5691, a Δpnp-751 derivative of C-1a. As expected, since rpsA is an essential gene (Kitakawa and Isono 1982), the growth of both C-5698 (araBp-rpsA) and C-5707 (araBp-rpsA Δpnp-751) isogenic strains turned out to be arabinose-dependent in liquid and solid media. In particular, the growth of cultures shifted from arabinose (permissive condition) to glucose (nonpermissive condition) was arrested after three or four generations, as detected by OD600 measurements, while cell viability slightly dropped (Fig. 3A; data not shown). Analysis of S1 expression by Western blotting before and at different time points after the shift to nonpermissive condition showed that the S1 abundance immediately decreased two- to threefold after a shift to glucose medium (Fig. 4A), indicating that S1 synthesis was inhibited, whereas the level of ribosomal protein L4, as detected by Western blotting with specific antibodies, remained unchanged (data not shown). It should be noted that, although in a nonpermissive condition the rpsA mRNA was undetectable by Northern blotting (data not shown), complete depletion of S1 was not reached even at late time points after the shift, thus suggesting that S1 protein is rather stable, at least under these experimental conditions, and that the observed decrease of S1 level should be mainly imputed to dilution of preexisting S1 during the residual growth after the shift to nonpermissive conditions.
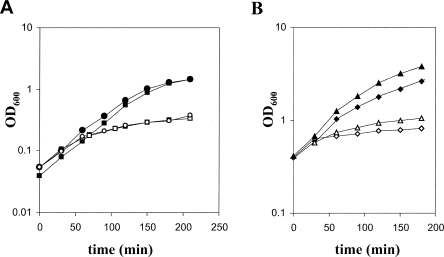
FIGURE 3.
Effect of S1 expression modulation on bacterial growth. (A) Cultures of C-5698 (pnp+; squares) and C-5707 (Δpnp-751; circles) were grown at 37°C in LD with arabinose up to OD600 = 0.2 and diluted 1:4 (time = 0 in the plot) in prewarmed LD with arabinose (closed symbols) or glucose (open symbols) as described in Materials and Methods. The incubation at 37°C was then continued and the growth was monitored spectrophotometrically by measuring the OD600 of the cultures. (B) Cultures at OD600 = 0.4 of C-1a (pnp+; diamonds) or C-5691 (Δpnp-751; triangles) carrying pREP4 and pQE31-S1 plasmids were split in two (time = 0 in the plot) and one subculture was induced with 1 mM IPTG. The incubation of noninduced (closed symbols) and IPTG-induced (open symbols) subcultures was then continued at 37°C, and the growth was monitored by measuring the OD600 of the cultures.
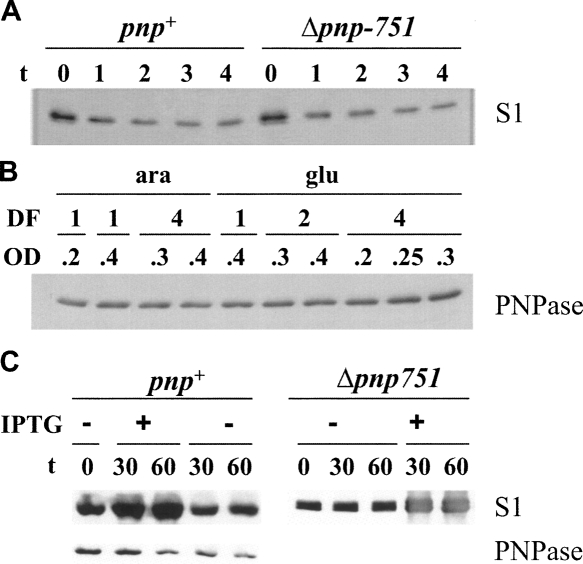
FIGURE 4.
S1 and PNPase expression following S1 depletion (panels A and B) or overexpression (panel C). The panels show Western blotting with S1 (A and upper part of C) or PNPase (B and lower part of C) specific antibodies of 10% SDS-polyacrylamide gels transferred onto nitrocellulose sheets and revealed with ECL Western blotting reagent. (A) Cultures of C-5698 (pnp+) or C-5707 (Δpnp-751) were grown in LD with arabinose and diluted 1:4 in LD with glucose as described in the Materials and Methods. Samples were taken for protein extraction before (t = 0) and at different time after dilution (indicated in hours on top of the lanes). One microgram of crude extract was loaded on each lane. (B) Cultures of C-5698 were grown in LD with arabinose up to OD600 = 0.2; the cells were then washed with LD, collected in a small volume of broth, resuspended in 1 vol of LD supplemented with arabinose or glucose or diluted twofold or fourfold (dilution factor [DF] of 1, 2, and 4, respectively) in the same media. Incubation at 37°C was continued and samples were taken at the OD600 reported in the figure. Crude extracts obtained from 0.1 OD600 units of cell cultures were loaded on each lane. (C) Cultures at OD600 = 0.4 of C-1a (pnp+) or C-5691 (Δpnp-751) carrying pREP4 and pQE31-S1 plasmids were split in two and one subculture was induced with 1 mM IPTG. Samples for protein extraction were taken immediately before and at the time indicated in minutes on top of the lanes after addition of IPTG. One (pnp+) or 0.5 (Δpnp-751) μg of crude extracts were loaded on each lane.
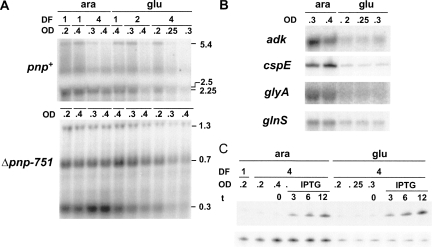
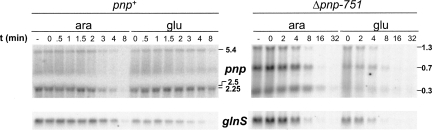
We thus monitored the PNPase and pnp mRNA expression levels upon S1 depletion. Proteins and RNA were extracted from samples taken at different OD600 from depleted and not depleted C-5698 cultures and were analyzed, respectively, by Western blotting with PNPase-specific polyclonal antibodies and by Northern blotting with the radiolabeled PNP5 riboprobe specific for the pnp 5′-end. Northern blotting was also performed on the pnp deletion mutant C-5707. Because of the in-frame deletion, C-5707 is expected to produce pnp transcripts ∼2 kb shorter than the wild-type strain. As shown in Figure 4B, we found that the amount of PNPase remained substantially unchanged (or slightly increased) up to 4 h after the shift to glucose medium. On the other hand, it appears that the level of all pnp transcripts progressively decreased concomitantly with S1 depletion both in the wild type and in the pnp mutant (Fig. 5A). It should be noted that in the Δpnp-751 mutant the pnp transcripts are more abundant than in the wild type since autogenous regulation is lacking. We tested whether this effect was specific for pnp mRNAs by performing Northern blot analysis of a set of model RNAs (adk, glnS, and glyA) with specific riboprobes (Fig. 5B; data not shown). All such transcripts are monocistronic mRNAs with a short 5′-UTR, and their abundance was not significantly affected by lack of PNPase, as shown by transcriptomic analysis (Polissi et al. 2003; T. Carzaniga, F. Falciani, and G. Dehò, unpubl.). We also included cspE, whose monocistronic mRNA was slightly decreased in a pnp defective strain (Polissi et al. 2003). As shown in Figure 5B, depletion of S1 caused a decrease in the transcript level of all such genes. The same effect was observed also in the pnp deletion mutant; in this strain the analysis was limited to the glnS (Fig. 6) and cspE (data not shown).
FIGURE 5.
Northern analysis of the pnp and other model transcripts upon S1 depletion. (A,B) Twelve micrograms of total RNA extracted from cultures of C-5698 (pnp+; upper part of panel A and panel B) and C-5707 (Δpnp-751; lower part of panel A), grown as detailed in Materials and Methods, were fractioned by 1.5% denaturating agarose gel electrophoresis, transferred onto a nylon membrane, and hybridized with the radiolabeled riboprobe specific for pnp (A) or the other mRNAs as listed on the left in B. Dilution factor (DF) of each subculture and OD600 at the sampling time are reported on the top of each lane. Length (in kilobases) of pnp transcripts is reported on the right of panel A. Only the gel portions displaying signals are shown. (C) Cultures of C-5699/pGM331 were grown at OD600 = 0.2, diluted 1:4 (DF, dilution factor) in permissive (ara) and nonpermissive (glu) condition, and incubated until OD600 = 0.2 was reached, as described in Materials and Methods. Incubation was carried on for additional 30 min (ara, OD600 = 0.4) or 60 min (glu, OD600 = 0.3) before the addition of 1 mM IPTG.. Samples were taken before (t = 0) and at different time (indicated in minutes) after the induction. Five micrograms of total RNA were fractioned on 6% denaturing polyacrylamide gel, transferred onto a nylon membrane, and hybridized with the tRNAGly-specific radiolabeled oligonucleotide (upper part) or with the cspE-specific riboprobe (lower part).
FIGURE 6.
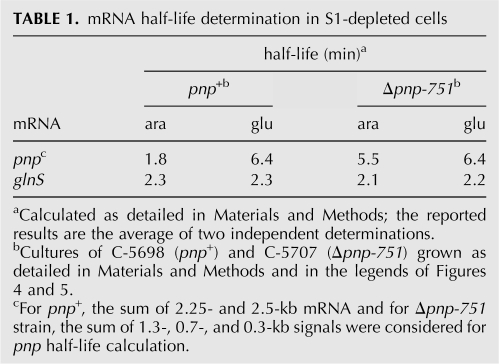
Stability of pnp and glnS transcripts upon S1 depletion. Bacterial cultures of C-5698 and C-5707 were grown up to OD600 = 0.2, diluted 1:4 in permissive (ara) or nonpermissive (glu) conditions, and further incubated until OD600 = 0.2 was reached. After an additional 60 min incubation, rifampicin (final concentration of 0.4 mg/mL) and nalidixic acid (0.03 mg/mL) were added and samples for RNA extraction were then taken at different time points (indicated above the lanes) after antibiotic addition. Twelve micrograms of RNA were run on 1.5% denaturing agarose gel, blotted onto a nylon sheet, and analyzed by Northern blotting to reveal pnp (upper part) or glnS (lower part) mRNA. Length of pnp transcripts (in kilobases) is reported on the right of the panels.
A decrease of mRNA abundance may be imputed to either mRNA faster decay or lower transcription efficiency. To probe the former hypothesis, we assayed the stability of pnp and glnS transcripts in S1-depleted cells upon rifampicin treatment, as described in Materials and Methods. Surprisingly, although the amount of pnp mRNAs was lower in S1-depleted C-5698 (pnp +) than in the nondepleted strain, this mRNA was threefold more stable (half-life of 6.4 and 1.8 min in depleted and nondepleted cells, respectively), as estimated by PhosphorImager quantification of the Northern blotting signals shown in Figure 6. On the other hand, pnp mRNA stability in C-5707 (Δpnp-751), which in the nondepleted mutant was threefold higher than in the nondepleted wild type, did not significantly change when S1 was depleted (half-life of 5.5 min in nondepleted cells and 6.4 min in depleted cells). As for glnS mRNA, although its abundance decreased upon S1 depletion, its half-life was affected by neither the absence of PNPase nor S1 depletion (Fig. 6; Table 1). Thus the decreased mRNA abundance observed upon S1 depletion cannot be imputed to a faster mRNA turnover, as it is, at best, unchanged or, paradoxically, decreases for the pnp mRNA in the presence of PNPase.
TABLE 1.
mRNA half-life determination in S1-depleted cells
It has been reported that S1 copurifies with E. coli RNA polymerase and stimulates transcription in an in vitro system (Sukhodolets and Garges 2003; Sukhodolets et al. 2006). We thus checked whether the decreased mRNA abundance in S1 depleted cells could be imputed to lower transcription initiation efficiency. To this purpose, we measured the expression of the suppressor tRNAGly reporter, which encodes a stable, untranslated RNA whose expression is therefore unaffected by factors that modulate RNA stability and translation efficiency (Sloan and Weisberg 1993; Briani et al. 1996, 2000). Plasmid pGM331, which carries the tRNAGly gene under the ptac promoter (Briani et al. 1996), was transferred in C-5699, a chloramphenicol-sensitive C-5698 derivative. Transcription from ptac was induced with IPTG in S1-depleted and nondepleted cells and expression of the reporter tRNA was monitored by Northern blotting of RNA extracted at different time points after induction. The same filter was also hybridized with a cspE specific riboprobe (Fig. 5C). As shown in Figure 5, while S1 depletion caused a decrease of cspE mRNA, the induction level of tRNAGly remained unaffected, thus suggesting that S1 depletion would not impair promoter activity in vivo.
As neither mRNA destabilization nor lower transcription efficiency seems to be responsible for the decreased mRNA abundance upon S1 depletion, it appears that in this condition different aliquots of an mRNA species may be directed to different fates. A fraction of mRNA may be rapidly degraded whereas the stability of the other fractions remains unchanged, as for glnS and the pnp mRNA in the absence of PNPase. It is plausible that mRNAs bound by S1 and engaged in translation represents the latter fraction whereas free mRNA rapidly decays. As for the pnp transcripts in a pnp + strain, it should be noted that their increased half-life is comparable to that observed in the pnp − strain, thus suggesting that S1 depletion alleviates autogenous regulation.
S1 overexpression
S1 overexpressing strains were obtained by transforming C-1a (pnp +) and C-5691 (Δpnp-751) with plasmids pREP4 and pQE31-S1 (Sukhodolets and Garges 2003), which carry the lacI repressor gene and rpsA under the IPTG-inducible promoter-operator pT5-lacO, respectively. S1 induction was detrimental for both strains as their growth in liquid cultures significantly slowed down within 60 min from IPTG addition (Fig. 3B) and the plating efficiency dropped about 1000-fold when IPTG was present in solid medium (data not shown). In both strains the level of S1 increased three- to fourfold by 30 min after induction and remained high at later time points (Fig. 4C) whereas the amount of ribosomal protein L4 did not change (data not shown) as shown by Western blotting with specific antibodies. Thus both S1 induction and depletion caused cell growth impairment and an unbalance of S1 to ribosomes ratio, a stressful condition henceforth indicated as translational stress.
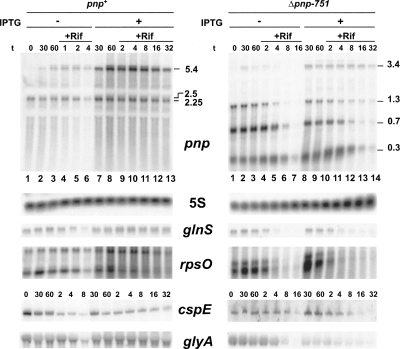
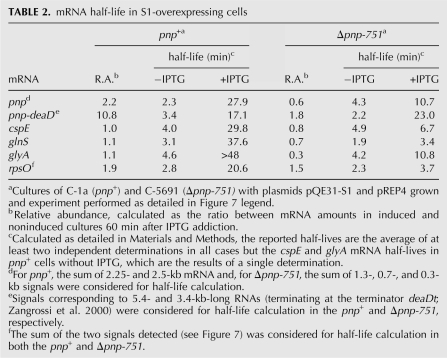
PNPase abundance in the pnp+ strain was unaffected by S1 overexpression, as assessed by Western blotting (Fig. 4C). Conversely, S1 overexpression greatly affected pnp mRNA abundance and stability. In the pnp+ strain, S1 overexpression led to an about twofold and 10-fold increase of the monocistronic (2.25 kb long) and polycistronic (5.4 kb long) pnp mRNA levels, respectively (Fig. 7, left panel, lanes 3,8) while the half-life of these transcripts was considerably extended (>10-fold for the monocistronic and about fivefold for the polycistronic mRNA; Table 2). However, in the PNPase-deficient strain, S1 overexpression did not further increase the abundance of the 0.3-, 0.7-, and 1.3-kb-long Δpnp-751 mRNAs (Fig. 7, right panel, lanes 3,9) and their half-lives increased only about twofold. To the contrary, the polycistronic 3.4 kb RNA exhibited a 10-fold half-life increase (Table 2).
FIGURE 7.
Stability of pnp and other model transcripts upon S1 overexpression. Bacterial cultures of C-1a (pnp+) and C-5691 (Δpnp-751) carrying pREP and pQE31-S1 plasmids were grown at OD600 = 0.4, split in two, and one subculture was induced with 1 mM IPTG. Sixty minutes later, rifampicin (final concentration of 0.4 mg/mL) and nalidixic acid (0.03 mg/mL) were added. Samples for RNA extraction were taken at different time points (as indicated above the lanes above pnp and cspE panels), before (no label) and after (+Rif) antibiotic addition. Twelve micrograms of RNA were run on a 1.5% denaturing agarose gel, blotted onto a nylon sheet, and analyzed by Northern blotting with a pnp or other riboprobes specific for the genes listed in the figure. For cspE, 5 μg of RNA were run on a 6% denaturing polyacrylamide gel. The size (in kilobases) of pnp mRNAs is indicated. As a loading control, the gels have been hybridized also with a 5S rRNA-specific radiolabeled oligonucleotide.
TABLE 2.
mRNA half-life in S1-overexpressing cells
We also tested whether the effect of S1 overexpression was specific for pnp or acted upon the abundance and half-life of other transcripts. The abundance of non-pnp transcripts was essentially unaffected by either absence of PNPase or S1 overexpression, as shown by Northern and/or dot blotting (data not shown). On the other hand, all assayed transcripts were strikingly (∼10-fold) stabilized in the pnp+ strain, whereas stabilization did not occur or was remarkably reduced (2.6-fold at best) in the Δpnp-751 strain (Fig. 7; Table 2). Thus, as observed for the monocistronic pnp transcript, stabilization by S1 overexpression of these mRNAs depended on PNPase.
Since the S1-induced increase of the mRNAs half-life was not accompanied by an increase in mRNA abundance, we asked whether this could be caused by inefficient transcription initiation in S1 overexpressing cells. To this end, we monitored by Northern blotting the expression level of the tRNAGly reporter from pGM384, a plasmid that harbors the tRNAGly gene under the araBp promoter, at different time points after arabinose induction. However, we did not observe any difference in the efficiency of tRNAGly expression in S1 overexpressing and nonoverexpressing cells (data not shown).
Determination of bulk RNA half-life in S1 overexpressing cells
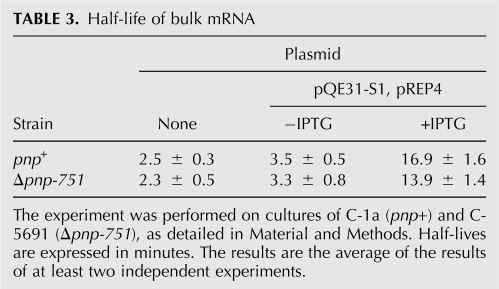
To assess whether the entire population of unstable RNA was stabilized in a PNPase-dependent manner by S1 overexpression, we measured the half-life of total pulse-labeled RNA in S1 overexpressing cultures. It should be stressed that, whereas in the Northern blot analysis described above the half-life of discrete mRNAs is evaluated, in this kind of experiment we measure the decay of the TCA-insoluble, nonstable RNA fraction, composed of both full-length RNAs and degradation products longer than 50 nt. We found (Table 3) that bulk mRNA half-life correlated with S1 expression level, being the lowest in strains carrying only the chromosomal copy of rpsA gene and the highest in S1 overexpressing cells. Conversely, lack of PNPase did not greatly affect bulk mRNA decay irrespective of S1 level of expression, indicating that the general stabilizing effect of S1 overexpression on bulk unstable RNA does not depend on PNPase.
TABLE 3.
Half-life of bulk mRNA
DISCUSSION
Studies on RNA processing and decay have revealed multiple and overlapping pathways acting on different RNA species under different physiological conditions, and many players implicated in RNA stability control have been identified (Deutscher 2006). Here we provide evidence of a novel feature of mRNA decay regulation, namely, PNPase-dependent mRNA stabilization (PDS) that occurs upon overexpression of the ribosomal protein S1. Under such conditions in which translation seems to be seriously impaired, as suggested by cell growth inhibition (Fig. 3) and disappearance of polysomes (F. Briani and G. Dehò, unpubl.), a set of mRNAs becomes very stable, with an ∼10-fold half-life increase. However, although transcription is not inhibited by S1 overexpression, this stabilization does not lead to a substantial increment of such mRNA abundance. This suggests that only a fraction of each mRNA species is stabilized, whereas extra mRNA appears to rapidly decay so that the steady-state level remains low. Surprisingly, mRNA stabilization is dependent on PNPase. Thus upon translational stress induced by S1 overexpression an enzyme thought to be principally involved in mRNA degradation (for review, see Lin-Chao et al. 2007) seems to prevent mRNA decay.
Apparently, results of total pulse-labeled mRNA decay analysis conflict with decay rates of specific mRNAs obtained by Northern blot analysis. In fact, the half-life of bulk nonstable RNA increased about fivefold upon S1 overexpression both in the presence and in the absence of PNPase. The two sets of data can be reconciled, however, by considering that by Northern blotting we evaluate the decay of discrete full-length mRNAs, whereas bulk unstable RNA is composed of any RNA species longer than 50 nt, including prematurely terminated and partially degraded molecules. In addition, PNPase-dependent stabilization could be a property of a specific subset of full-length transcripts and thus may not contribute significantly to the stability of bulk RNA. In agreement with this idea, stabilization of the long polycistronic pnp mRNA does not seem to be PNPase dependent. Finally, the mRNA stabilized in a PNPase-dependent manner represents a fraction of the mRNA produced for any specific gene, whereas most mRNA appears to be degraded by a PNPase-independent pathway(s), as suggested by the fact that the strong mRNA stabilization is not followed by an increase of mRNA abundance. Interestingly, Marujo et al. (2000) have shown that RNase II overproduction may increase rpsO mRNA half-life about twofold, as compared with an RNase II-defective strain. Although RNase II-dependent stabilization is lower than the up to 10-fold increase of mRNA half-life observed under PDS, this is another instance of an RNA-protecting role exerted by an RNA-degrading enzyme.
The biological significance of PDS remains to be assessed. It seems unlikely that under physiological conditions the absolute amount of S1 ribosomal protein could significantly change, given its tight autoregulation (Skouv et al. 1990; Boni et al. 2000, 2001). However, it is not implausible that the intracellular distribution of the protein (free versus ribosome associated) could vary as a consequence of changes in the efficiency of protein synthesis, as it may occur, for example, upon amino acid starvation or cold shock, thus creating an S1 to ribosomes unbalance. In facts, PNPase-dependent functional stabilization of mRNA has also been observed upon cold shock (Yamanaka and Inouye 2001). While PNPase is required to degrade mRNAs encoding cold shock proteins (CSPs) at the end of the cold acclimation phase, it enhances the functional half-life of a number (24 out of the 27 measured) of mRNAs coding for non-CSPs. Our data and the above considerations suggest that PDS might represent a response to transient inactivation of protein synthesis that, under specific conditions, preserves a fraction of full-length mRNAs.
As a general working model for PDS, we suggest that upon S1 overexpression, PNPase remains bound to the 3′-end of full-length mRNA at the Rho-independent terminator hairpin, thus protecting mRNA from other exonucleolytic degradation pathways (e.g., PAP I-dependent mRNA decay), while excess S1 and/or ribosomes stalled on the mRNA could help prevent endonucleolytic decay mechanisms. In particular, S1 could protect mRNA from endonucleolytic RNase E digestion, as S1 binding sites may overlap RNase E cleavage sites (Kaberdin and Blasi 2006). As for mRNAs not engaged with S1 and/or translating ribosomes, binding of Rho factor to the unprotected nascent mRNA may lead to premature Rho-dependent transcription termination and generate RNA fragments lacking the 3′-end hairpin, thus allowing degradation via alternative decay pathways.
It has been shown that S1 binds also to poly(A) tracts (Kalapos et al. 1997; Feng et al. 2001) and physically interacts with both PNPase and RNase E. However, the consequences of such interactions on RNase E and PNPase activities have not been detected in vitro nor have they been tested in vivo (Feng et al. 2001). To the contrary, CspE, a “cold shock domain” protein, also binds to poly(A) tracts and impedes PNPase and RNase E-mediated degradation of polyadenylated RNA in vitro (Feng et al. 2001). We speculate that S1 overexpression indirectly inhibits PNPase phosphorolytic activity at the 3′-poly(A) ends of specific mRNAs and that PNPase stalled at such 3′-ends protects the mRNA from PNPase-independent decay pathways. The ability of S1 to bind both to mRNA 5′-ends and poly(A) tails could link together the two mRNA extremities and provide a structural basis for PDS upon S1 overexpression.
A phenomenon seemingly related to PDS appears to occur upon S1 depletion, where the abundance of both pnp and glnS mRNAs (and other mRNAs) conflicts with their stability. Also in these conditions, as in PDS, two populations of mRNA may exist for a single mRNA species, one that is probably not translated and doomed to rapid decay pathways and the other preserved to normal turnover by the interaction with the residual translation proficient ribosomes.
This study has been primed by the finding that both PNPase and S1 bind to the pnp mRNA 5′-UTR. Our data suggest that both S1 overexpression and depletion may interfere with PNPase autoregulation. In particular, in S1 overexpressing strains, inhibition of pnp autogenous regulation appears to be distinct from and overlapping to PDS. Unlike the other tested mRNAs, the abundance of pnp mRNAs upon overexpression of S1 in the pnp + strain sharply increases, reaching levels comparable to pnp transcripts in a pnp− strain (not shown). This is possibly a consequence of autogenous control inhibition, since in the absence of PNPase the abundance of pnp mRNAs does not further increase following S1 induction. The half-life increase of the pnp monocistronic mRNAs, however, is PNPase dependent. Thus pnp post-transcriptional regulation is subject to both PDS and autogenous control. We suggest that S1 interference with PNPase autogenous regulation might depend on competition for binding with the pnp 5′-UTR.
Many aspects of PDS remain to be established, such as defining how a specific mRNA is targeted (or avoids being targeted) to PDS, which are the rate limiting factors that set the fraction of a given RNA species that is committed to decay versus stabilization, whether PDS after S1 overexpression or cold shock share common mechanisms and involve the same factors. Addressing these questions will contribute to shedding light on post-transcriptional control of gene expression and on the interplay between mRNA degradation and translation.
MATERIALS AND METHODS
Bacterial strains and plasmids
E. coli sequence coordinates throughout are from NCBI Accession Number U00096.2. E. coli strains C-1a (prototrophic, pnp +) (Sasaki and Bertani 1965), C-5612 (C-1a pnp-7∼Tn10) (Sasaki and Bertani 1965; Piazza et al. 1996), and C-5691 (C-1a Δpnp-751) (Regonesi et al. 2006) were described previously. Δpnp-751 is an in frame deletion encompassing ∼95% of pnp coding sequence.
C-5698 [Φ (cat rrnBt araBp-rpsA)] is a C-1a derivative with the rpsA gene under control of the inducible promoter araBp. Upstream of the araBp promoter, the rrnBt transcription termination region and an FRT-flanked cat cassette are located. This conditional expression mutant was obtained essentially as described (Datsenko and Wanner 2000; Morita et al. 2004), by transforming C1a/pKD46, which expresses λ Red function, with a PCR fragment amplified from pTM26 (Morita et al. 2004) with oligonucleotides FG1573 (CCCGCTGTCATTCCATTGCAACGGGGGTACTGCAAATTCATGGAGAAACAGTAGAGAG, coordinates 961152–961114 in italics, followed by a pTM26 specific primer overlapping araBp) and FG1574 (CAATACGCGCGCCAGAAATTGGCTCTCGCATAAGCGACCGTGTAGGCTGGAGCTGCT, 961075–961104, followed by pTM26-specific primer overlapping the FRT site upstream of the cat gene). Transformants were selected at 37°C on minimal M9 medium agar plates supplemented with chloramphenicol (15 μg/mL), 0.2% L-arabinose, and 0.2% glycerol and tested for both the loss of the pKD46 helper plasmid and for the presence of the cat rrnBt araBp-rpsA construct by PCR and sequencing. C-5699 [Φ (rrnBt araBp-rpsA)] is a C-5698 derivative lacking the cat gene. It was obtained by transforming C-5698 with the temperature sensitive plasmid pCP20 (Datsenko and Wanner 2000), which encodes the FLP recombinase, thus promoting recombination between the FRT sites. A few transformants were purified at 42°C and tested for the loss of both the CamR cassette and plasmid pCP20. C-5707 (Δpnp-751 Φ [cat rrnBt araBp-rpsA]) was obtained by transferring the cat rrnBt araBp-rpsA construct from C-5698 into C-5691 by P1 transduction.
Plasmid pAZ8 (pnp +; 3309406–3306791) was described previously (Piazza et al. 1996). pGM331 is a pGZ119EH (Lessl et al. 1992) derivative carrying the suppressor tRNAGly (McClain et al. 1991) downstream of ptac (Briani et al. 1996). pGM384 is a pGM743 (Zangrossi et al. 2000) derivative carrying the suppressor tRNAGly downstream of the araBp promoter. It was obtained by cloning in pGM743 digested with SmaI the NarI (Klenow filled)-SmaI fragment of pBAD24 (Guzman et al. 1995). Plasmids pQE31-S1 and pREP4 (Sukhodolets and Garges 2003) were kindly provided by M.V. Sukhodolets (Lamar University, Beaumont, Texas). pREP4 (Qiagen) is a pACYC (Chang and Cohen 1978) derivative carrying the lacI gene. pQE31-S1 is a pQE31 (Qiagen) derivative obtained by cloning the coding sequence of the rpsA gene (NCBI Gene ID 945536: 961221–962898) downstream of the pT5-lacO region. The recombinant S1 protein expressed from the plasmid allele carries an N-terminal His6 tag and exhibits properties comparable to the native S1 in different in vitro assays (Sukhodolets and Garges 2003; data not shown; M.V. Sukhodolets, pers. comm.).
LD broth (Ghisotti et al. 1992) was supplemented with chloramphenicol (30 μg/mL), ampicillin (100 μg/mL), kanamycin (50 μg/mL), 0.4% glucose, or 1% L-arabinose when needed.
Identification of PLBP
The biotinylated PNPA RNA substrate, which corresponds to the first 247 nt of the nonprocessed pnp transcript from promoter pnp-p (Jarrige et al. 2001), was synthesized by in vitro transcription by incubating the template DNA (Regonesi et al. 2004) 2 h at 37°C in the following mix: 1× transcription buffer (Promega), 10 mM DTT, 1 U/μL RNasin (Promega), 0.5 mM (each) rNTPs, 50 μM biotin-11-CTP (Perkin Elmer), and 0.2 U/μL T7 RNA polymerase (Promega). After phenol-chloroform extraction and ethanol precipitation, the RNA was resuspended at 1 pmol/μL in diethylpyrocarbonate (DEPC)-treated water. Two hundred microliters of streptavidin-coated 4% agarose beads (Sigma) were preequilibrated in 5 mM Tris-HCl (pH 7.4), 0.5 mM EDTA, 1 M NaCl and resuspended in 40 μL of the same buffer. Forty microliters of biotinylated PNPA were mixed with the beads and incubated 16 h at 4°C with gentle agitation. Nine samples were prepared as described above; a 10th sample with 40 μL of DEPC-treated water without biotinylated RNA substrate was also set up as a negative control. The 10 samples were processed in parallel as detailed below. After incubation, the beads were spun down in a microfuge, washed twice in DEPC-treated water and twice in Binding buffer without glycerol, and then resuspended in 50 μL of a mix composed of 50 mM Tris-HCl (pH 7.4), 250 mM NaCl, 2.5 mM DTT, 0.75% NP40 (Fluka), and 6 ng/μL poly(A). Two hundred microliters (3 mg) of C-5691 crude extract were added to the mix. After 30 min at 21°C with occasional shaking to keep the beads in suspension, the samples were centrifuged down in a microfuge and washed three times with 50 mM Tris-HCl at pH 7.4, 50 mM NaCl to eliminate the unbound proteins. The pellets were resuspended in 30 μL of 2× SDS buffer (100 mM Tris-HCl at pH 6.8, 200 mM DTT, 4% SDS, 0.2% bromophenol blue, 20% glycerol), heated 5 min at 95°C, and spun down in a microfuge. The supernatants were recovered and pooled (with the exception of the negative control that was kept separated) and aliquots were analyzed by 10% SDS-PAGE and silver staining of the gel and by Northwestern assay. The rest of the samples was concentrated on a Microcon Centrifugal Filter Unit (Millipore) and loaded on a preparative 10% SDS-polyacrylamide gel. The gel was stained with Coomassie and the protein of interest was eluted from the gel. PLBP was then in-gel digested using trypsin and the resulting peptide mixture analyzed by means of liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS, based on LCQXP ion trap mass spectrometer) (Maiorino et al. 2005).
UV-cross-linking assay
32P-labeled PNPA RNA substrate, which corresponds to the first 249 nt of the nonprocessed pnp transcript from the pnp-p promoter region (coordinates 3309348–3309100), was synthesized by in vitro transcription with T7 RNA polymerase and [α-32P] CTP, as described (Regonesi et al. 2004). We incubated 50,000 cpm of substrate for 20 min at 21°C in Binding Buffer (50 mM Tris-HCl at pH 7.4, 50 mM NaCl, 0.5 mM DTT, 0.025% NP40 [Fluka] and 10% glycerol) with 300 ng of crude extract in a final volume of 10 μL. The samples were UV-irradiated (254 nm, 2.8 J/cm2) in a UV Stratalinker 2400 (Stratagene) and treated with RNase A. After addition of one volume of 2× SDS buffer, the samples were run on 10% SDS-PAGE and the gel was dried and analyzed by phosphorimaging.
Northwestern analysis
Following electrophoresis on 10% SDS-PAGE, proteins were blotted onto a nitrocellulose filter and allowed to refold by incubation of the filter in TEN buffer (10 mM Tris-HCl at pH 7.4, 50 mM NaCl, 0.1 mM EDTA) for 3 d at 4°C. The filter was then incubated for 2 h at 44°C with [32P]-labeled PNPA RNA (3 × 107 cpm) in 20 mL TEN, 1× Denhardt's solution and 10 mg/mL herring sperm DNA. Unbound RNA was then removed by extensive washing in TEN buffer and the membrane analyzed by phosphorimaging.
S1 expression modulation in E. coli cultures
Bacterial cultures were grown at 37°C in LD broth supplemented with sugars and/or antibiotics when needed, as detailed in the Results and legends of Figures 3, 4, 5, and 6. In S1 overexpression experiments, the cultures were grown in LD broth in a reciprocating waterbath until OD600 = 0.4 was reached. The cultures were then split in two, 1 mM IPTG was added to one of the subcultures, and incubation was continued at 37°C. Samples of the subcultures were taken at different times for RNA and/or protein extraction. In S1 depletion experiments, conditional expression rpsA mutants were inoculated in LD broth supplemented with 1% arabinose and incubated at 37°C in a reciprocating waterbath until OD600 = 0.2 was reached. The cultures were then filtered on a 0.4 μm Millipore membrane, washed with one volume of LD, resuspended in 0.1 vol of LD, and diluted as indicated in the figure legends in LD with 1% arabinose (permissive condition) or 0.4% glucose (nonpermissive-depletion condition). Incubation at 37°C was then continued for such subcultures and growth was monitored by measuring OD every 30 min. Samples of nondepleted and depleted cultures were taken at different times for RNA or protein extraction.
Preparation of E. coli crude extracts, SDS-PAGE of proteins, and immuno-assays
E. coli crude extracts were obtained as described (Regonesi et al. 2004). Protein content was determined using Coomassie Plus Protein Assay Reagent (Pierce). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described (Laemmli 1970) on 10% resolving gels containing 0.1% SDS. Rainbow high molecular weight markers (Amersham Biosciences) were used as size references. The proteins on the gel were revealed through Coomassie (Sambrook et al. 1989) or silver staining with the SilverQuest Silver Staining kit (Invitrogen). For immunological detection of PNPase or S1, the gels were blotted onto a nitrocellulose (Hybond C) sheet and incubated with polyclonal anti-PNPase (Fontanella et al. 1999) or anti-S1 (kindly provided by U. Blasi, Vienna Biocenter, Vienna, Austria) antibodies (Sambrook et al. 1989). Immunoreactive bands were revealed using ECL Western blotting reagent (Amersham Pharmacia Biotech).
Northern blotting
Basic procedures for RNA extraction, Northern blot analysis, and synthesis of radiolabeled riboprobes by in vitro transcription with T7 RNA polymerase were previously described (Dehò et al. 1992; Briani et al. 2007). The riboprobes were specific for the following regions of adk, cspE, glnS, glyA, pnp, rpsA, and rpsO mRNAs. Coordinates of the antisense riboprobes are, from NCBI accession number U00096.2, as follows: ADK, 496468–496623; CSPE, 656576–656704; GLNS, 705425–705615; GLYA, 2683218–2683368; PNP5, 3309141–3309268; RPSA, 961264-961412; RPSO, 3309466–3309682. The oligonucleotides used to detect the tRNAGly (Sloan and Weisberg 1993; Zangrossi et al. 2000) and 5S rRNA (oligo 1842, GTTCGGCATGGGGTCAGG, complementary to nucleotides 30–47 of 5S rRNA) were 5′-end labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP (Sambrook et al. 1989). The conditions of hybridization with oligonucleotides were described previously (Briani et al. 1996). Autoradiographic images and densitometric analysis of Northern blots were obtained by phosphorimaging using ImageQuant software (Molecular Dynamics). In the experiments of mRNA decay analysis, mRNA half-lives were estimated by regression analysis of curves obtained by plotting the percentage of mRNA remaining (calculated as the densitometric signal at a given time after rifampicin addition divided by the signal at time = 0) versus time after rifampicin addition.
Chemical decay of bulk mRNA assay
The chemical decay of pulse-labeled RNA was assayed as previously described (Donovan and Kushner 1986). Bacterial cultures of different strains were inoculated in LD supplemented with antibiotics, whenever needed, and incubated at 37°C in a reciprocating waterbath until OD600 = 0.4 was reached. For C-1a and C-5691 carrying plasmids pREP4 and pQE31-S1, the cultures at OD600 = 0.2 were split in two aliquots and 1 mM IPTG was added to one of the two. Incubation at 37°C was continued for 60 min for the IPTG-treated culture (the final OD600 was ∼0.35–0.4) whereas the other one was grown up to OD600 = 0.4 (about 30 min). Cells were then pulse-labeled with [5,6-3H]-uridine (final concentration 3 μCi/mL, corresponding to ∼80 nM) for 1.5 min. Labeling was thus quenched by adding a Stop mix composed of rifampicin, nalidixic acid, and uridine (final concentrations of 800 μg/mL, 60 μg/mL, and 0.82 mM, respectively). Samples of 0.5 mL were drawn immediately before (time 0) and at different time points after Stop mix addition and mixed with 3 mL of ice-cold 20% TCA. The acid-precipitated samples were collected on Whatman GF/C filters and washed three times with 5 mL of 10% TCA and once with 5 mL of 95% ethanol/0.1 M HCl. The fraction of [5,6-3H]-uridine incorporated into mRNA was calculated as the radioactivity at a given time minus the plateau level radioactivity remaining at late time points after the addition of the Stop mix. The latter may be imputed to [5,6-3H]-uridine incorporated in stable RNA. mRNA half-life was estimated by regression analysis of curves obtained by plotting the percentage of [3H]-mRNA remaining versus time after Stop mix addition For half-life calculation, time 0 was set at the time point of maximum incorporated radioactivity (from 0 to 120 sec after Stop mix addition in different experiments). Only curves with a least-square fit of ≥0.90 were considered.
ACKNOWLEDGMENTS
We are indebted to U. Bläsi for the generous gift of purified S1 protein and S1-specific antibodies. We also thank M.V. Soukhodolets for providing plasmids pREP4 and pQE31-S1, C.O. Gualerzi for L4-specific antibodies, and S. Zangrossi for skillful technical assistance. This work has been supported by joint grants from Ministero dell'Università e della Ricerca and Università degli Studi di Milano (PRIN 2006).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1123908.
REFERENCES
- Arcus V. OB-fold domains: A snapshot of the evolution of sequence, structure and function. Curr. Opin. Struct. Biol. 2002;12:794–801. doi: 10.1016/s0959-440x(02)00392-5. [DOI] [PubMed] [Google Scholar]
- Bermudez-Cruz R.M., Ramirez F., Kameyama-Kawabe L., Montanez C. Conserved domains in polynucleotide phosphorylase among eubacteria. Biochimie. 2005;87:737–745. doi: 10.1016/j.biochi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Boni I.V., Isaeva D.M., Musychenko M.L., Tzareva N.V. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991;19:155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni I.V., Artamonova V.S., Dreyfus M. The last RNA-binding repeat of the Escherichia coli ribosomal protein S1 is specifically involved in autogenous control. J. Bacteriol. 2000;182:5872–5879. doi: 10.1128/jb.182.20.5872-5879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni I.V., Artamonova V.S., Tzareva N.V., Dreyfus M. Non-canonical mechanism for translational control in bacteria: Synthesis of ribosomal protein S1. EMBO J. 2001;20:4222–4232. doi: 10.1093/emboj/20.15.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briani F., Zangrossi S., Ghisotti D., Dehò G. A Rho-dependent transcription termination site regulated by bacteriophage P4 RNA immunity factor. Virology. 1996;223:57–67. doi: 10.1006/viro.1996.0455. [DOI] [PubMed] [Google Scholar]
- Briani F., Ghisotti D., Dehò G. Antisense RNA-dependent transcription termination sites that modulate lysogenic development of satellite phage P4. Mol. Microbiol. 2000;36:1124–1134. doi: 10.1046/j.1365-2958.2000.01927.x. [DOI] [PubMed] [Google Scholar]
- Briani F., Del Favero M., Capizzuto R., Consonni C., Zangrossi S., Greco C., De Gioia L., Tortora P., Deho G. Genetic analysis of polynucleotide phosphorylase structure and functions. Biochimie. 2007;89:145–157. doi: 10.1016/j.biochi.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Bycroft M., Hubbard T.J., Proctor M., Freund S.M., Murzin A.G. The solution structure of the S1 RNA binding domain: A member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- Carpousis A.J., Van Houwe G., Ehretsmann C., Krisch H.M. Copurification of E. coli RNAase E and PNPase: Evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- Chang A.C., Cohen S.N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.W., Koehler C.M., Teitell M.A. Human polynucleotide phosphorylase: Location matters. Trends Cell Biol. 2007;17:600–608. doi: 10.1016/j.tcb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Chou J.Y., Singer M.F. Deoxyadenosine diphosphate as a substrate and inhibitor of polynucleotide phosphorylase of Micrococcus luteus. 3. Copolymerization of adenosine diphosphate and deoxyadenosine diphosphate. J. Biol. Chem. 1971;246:7505–7513. [PubMed] [Google Scholar]
- Coburn G.A., Miao X., Briant D.J., Mackie G.A. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes & Dev. 1999;13:2594–2603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C. Maturation and degradation of RNA in bacteria. Curr. Opin. Microbiol. 2007;10:271–278. doi: 10.1016/j.mib.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehò G., Zangrossi S., Sabbattini P., Sironi G., Ghisotti D. Bacteriophage P4 immunity controlled by small RNAs via transcription termination. Mol. Microbiol. 1992;6:3415–3425. doi: 10.1111/j.1365-2958.1992.tb02209.x. [DOI] [PubMed] [Google Scholar]
- Deutscher M.P. Degradation of RNA in bacteria: Comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan W.P., Kushner S.R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. 1986;83:120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Huang H., Liao J., Cohen S.N. Escherichia coli poly(A)-binding proteins that interact with components of degradosomes or impede RNA decay mediated by polynucleotide phosphorylase and RNase E. J. Biol. Chem. 2001;276:31651–31656. doi: 10.1074/jbc.M102855200. [DOI] [PubMed] [Google Scholar]
- Fontanella L., Pozzuolo S., Costanzo A., Favaro R., Dehò G., Tortora P. Photometric assay for polynucleotide phosphorylase. Anal. Biochem. 1999;269:353–358. doi: 10.1006/abio.1999.4042. [DOI] [PubMed] [Google Scholar]
- Ghisotti D., Chiaramonte R., Forti F., Zangrossi S., Sironi G., Dehò G. Genetic analysis of the immunity region of phage-plasmid P4. Mol. Microbiol. 1992;6:3405–3413. doi: 10.1111/j.1365-2958.1992.tb02208.x. [DOI] [PubMed] [Google Scholar]
- Godefroy T. Kinetics of polymerization and phosphorolysis reactions of Escherichia coli polynucleotide phosphorylase. Evidence for multiple binding of polynucleotide in phosphorolysis. Eur. J. Biochem. 1970;14:222–231. doi: 10.1111/j.1432-1033.1970.tb00281.x. [DOI] [PubMed] [Google Scholar]
- Godefroy T., Cohn M., Grunberg-Manago M. Kinetics of polymerization and phosphorolysis reactions of E. coli polynucleotide phosphorylase. Role of oligonucleotides in polymerization. Eur. J. Biochem. 1970;12:236–249. doi: 10.1111/j.1432-1033.1970.tb00843.x. [DOI] [PubMed] [Google Scholar]
- Goldstein J., Pollitt N.S., Inouye M. Major cold shock protein of Escherichia coli . Proc. Natl. Acad. Sci. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L.M., Belin D., Carson M.J., Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrige A.C., Mathy N., Portier C. PNPase autocontrols its expression by degrading a double-stranded structure in the pnp mRNA leader. EMBO J. 2001;20:6845–6855. doi: 10.1093/emboj/20.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrige A., Brechemier-Baey D., Mathy N., Duche O., Portier C. Mutational analysis of polynucleotide phosphorylase from Escherichia coli . J. Mol. Biol. 2002;321:397–409. doi: 10.1016/s0022-2836(02)00645-9. [DOI] [PubMed] [Google Scholar]
- Joyce S.A., Dreyfus M. In the absence of translation, RNase E can bypass 5′ mRNA stabilizers in Escherichia coli . J. Mol. Biol. 1998;282:241–254. doi: 10.1006/jmbi.1998.2027. [DOI] [PubMed] [Google Scholar]
- Kaberdin V.R., Blasi U. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol. Rev. 2006;30:967–979. doi: 10.1111/j.1574-6976.2006.00043.x. [DOI] [PubMed] [Google Scholar]
- Kalapos M.P., Paulus H., Sarkar N. Identification of ribosomal protein S1 as a poly(A) binding protein in Escherichia coli . Biochimie. 1997;79:493–502. doi: 10.1016/s0300-9084(97)82741-1. [DOI] [PubMed] [Google Scholar]
- Kimura M., Foulaki K., Subramanian A.R., Wittmann-Liebold B. Primary structure of Escherichia coli ribosomal protein S1 and features of its functional domains. Eur. J. Biochem. 1982;123:37–53. doi: 10.1111/j.1432-1033.1982.tb06495.x. [DOI] [PubMed] [Google Scholar]
- Kitakawa M., Isono K. An amber mutation in the gene rpsA for ribosomal protein S1 in Escherichia coli . Mol. Gen. Genet. 1982;185:445–447. doi: 10.1007/BF00334137. [DOI] [PubMed] [Google Scholar]
- Komarova A.V., Tchufistova L.S., Dreyfus M., Boni I.V. AU-rich sequences within 5′-untranslated leaders enhance translation and stabilize mRNA in Escherichia coli . J. Bacteriol. 2005;187:1344–1349. doi: 10.1128/JB.187.4.1344-1349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lessl M., Balzer D., Lurz R., Waters V.L., Guiney D.G., Lanka E. Dissection of IncP conjugative plasmid transfer: Definition of the transfer region Tra2 by mobilization of the Tra1 region in trans . J. Bacteriol. 1992;174:2493–2500. doi: 10.1128/jb.174.8.2493-2500.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczyniecka M., DeSalle R., Kang D.C., Fisher P.B. The origin of polynucleotide phosphorylase domains. Mol. Phylogenet. Evol. 2004;31:123–130. doi: 10.1016/j.ympev.2003.07.012. [DOI] [PubMed] [Google Scholar]
- Lin-Chao S., Chiou N.T., Schuster G. The PNPase, exosome and RNA helicases as the building components of evolutionarily conserved RNA degradation machines. J. Biomed. Sci. 2007;14:523–532. doi: 10.1007/s11373-007-9178-y. [DOI] [PubMed] [Google Scholar]
- Maiorino M., Roveri A., Benazzi L., Bosello V., Mauri P., Toppo S., Tosatto S.C., Ursini F. Functional interaction of phospholipid hydroperoxide glutathione peroxidase with sperm mitochondrion-associated cysteine-rich protein discloses the adjacent cysteine motif as a new substrate of the selenoperoxidase. J. Biol. Chem. 2005;280:38395–38402. doi: 10.1074/jbc.M505983200. [DOI] [PubMed] [Google Scholar]
- Marujo P.E., Hajnsdorf E., Le Derout J., Andrade R., Arraiano C.M., Régnier P. RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli . RNA. 2000;6:1185–1193. doi: 10.1017/s135583820000073x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W.H., Foss K., Jenkins R.A., Schneider J. Rapid determination of nucleotides that define tRNAGly acceptor identity. Proc. Natl. Acad. Sci. 1991;88:6147–6151. doi: 10.1073/pnas.88.14.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miczak A., Kaberdin V.R., Wei C.L., Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B.K., Kushner S.R. Polynucleotide phosphorylase, RNase II and RNase E play different roles in the in vivo modulation of polyadenylation in Escherichia coli . Mol. Microbiol. 2000;36:982–994. doi: 10.1046/j.1365-2958.2000.01921.x. [DOI] [PubMed] [Google Scholar]
- Mohanty B.K., Kushner S.R. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34:5695–5704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I., Grill S., Gualerzi C.O., Blasi U. Leaderless mRNAs in bacteria: Surprises in ribosomal recruitment and translational control. Mol. Microbiol. 2002;43:239–246. doi: 10.1046/j.1365-2958.2002.02739.x. [DOI] [PubMed] [Google Scholar]
- Morita T., Kawamoto H., Mizota T., Inada T., Aiba H. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli . Mol. Microbiol. 2004;54:1063–1075. doi: 10.1111/j.1365-2958.2004.04329.x. [DOI] [PubMed] [Google Scholar]
- Murzin A.G. OB(oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for nonhomologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira T., de Smit M., Graffe M., Springer M. The relationship between translational control and mRNA degradation for the Escherichia coli threonyl-tRNA synthetase gene. J. Mol. Biol. 2001;310:709–722. doi: 10.1006/jmbi.2001.4796. [DOI] [PubMed] [Google Scholar]
- Piazza F., Zappone M., Sana M., Briani F., Dehò G. Polynucleotide phosphorylase of Escherichia coli is required for the establishment of bacteriophage P4 immunity. J. Bacteriol. 1996;178:5513–5521. doi: 10.1128/jb.178.18.5513-5521.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polissi A., De Laurentis W., Zangrossi S., Briani F., Longhi V., Pesole G., Dehò G. Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 2003;154:573–580. doi: 10.1016/S0923-2508(03)00167-0. [DOI] [PubMed] [Google Scholar]
- Portier C., Régnier P. Expression of the rpsO and pnp genes: Structural analysis of a DNA fragment carrying their control regions. Nucleic Acids Res. 1984;12:6091–6102. doi: 10.1093/nar/12.15.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier C., Dondon L., Grunberg-Manago M., Régnier P. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5′-end. EMBO J. 1987;6:2165–2170. doi: 10.1002/j.1460-2075.1987.tb02484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapov A.P., Subramanian A.R. Effect of E. coli ribosomal protein S1 on the fidelity of the translational elongation step: Reading and misreading of poly(U) and poly(dT) Biochem. Int. 1992;27:745–753. [PubMed] [Google Scholar]
- Py B., Higgins C.F., Krisch H.M., Carpousis A.J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- Régnier P., Arraiano C.M. Degradation of mRNA in bacteria: Emergence of ubiquitous features. Bioessays. 2000;22:235–244. doi: 10.1002/(SICI)1521-1878(200003)22:3<235::AID-BIES5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Régnier P., Grunberg-Manago M. RNase III cleavages in non-coding leaders of Escherichia coli transcripts control mRNA stability and genetic expression. Biochimie. 1990;72:825–834. doi: 10.1016/0300-9084(90)90192-j. [DOI] [PubMed] [Google Scholar]
- Regonesi M.E., Briani F., Ghetta A., Zangrossi S., Ghisotti D., Tortora P., Dehò G. A mutation in polynucleotide phosphorylase from Escherichia coli impairing RNA binding and degradosome stability. Nucleic Acids Res. 2004;32:1006–1017. doi: 10.1093/nar/gkh268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regonesi M.E., Del Favero M., Basilico F., Briani F., Benazzi L., Tortora P., Mauri P., Dehò G. Analysis of the Escherichia coli RNA degradosome composition by a proteomic approach. Biochimie. 2006;88:151–161. doi: 10.1016/j.biochi.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Robert-Le Meur M., Portier C. E. coli polynucleotide phosphorylase expression is autoregulated through an RNase III-dependent mechanism. EMBO J. 1992;11:2633–2641. doi: 10.1002/j.1460-2075.1992.tb05329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Le Meur M., Portier C. Polynucleotide phosphorylase of Escherichia coli induces the degradation of its RNase III processed messenger by preventing its translation. Nucleic Acids Res. 1994;22:397–403. doi: 10.1093/nar/22.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saguy M., Gillet R., Skorski P., Hermann-Le Denmat S., Felden B. Ribosomal protein S1 influences trans-translation in vitro and in vivo . Nucleic Acids Res. 2007;35:2368–2376. doi: 10.1093/nar/gkm100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1989. Molecular cloning. [Google Scholar]
- Sasaki I., Bertani G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J. Gen. Microbiol. 1965;40:365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- Skouv J., Schnier J., Rasmussen M.D., Subramanian A.R., Pedersen S. Ribosomal protein S1 of Escherichia coli is the effector for the regulation of its own synthesis. J. Biol. Chem. 1990;265:17044–17049. [PubMed] [Google Scholar]
- Sloan S.B., Weisberg R.A. Use of a gene encoding a suppressor tRNA as a reporter of transcription: Analyzing the action of the Nun protein of bacteriophage HK022. Proc. Natl. Acad. Sci. 1993;90:9842–9846. doi: 10.1073/pnas.90.21.9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen M.A., Fricke J., Pedersen S. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo . J. Mol. Biol. 1998;280:561–569. doi: 10.1006/jmbi.1998.1909. [DOI] [PubMed] [Google Scholar]
- Stanssens P., Remaut E., Fiers W. Inefficient translation initiation causes premature transcription termination in the lacZ gene. Cell. 1986;44:711–718. doi: 10.1016/0092-8674(86)90837-8. [DOI] [PubMed] [Google Scholar]
- Subramanian A.R. Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 1983;28:101–142. doi: 10.1016/s0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- Sukhodolets M.V., Garges S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42:8022–8034. doi: 10.1021/bi020638i. [DOI] [PubMed] [Google Scholar]
- Sukhodolets M.V., Garges S., Adhya S. Ribosomal protein S1 promotes transcriptional cycling. RNA. 2006;12:1505–1513. doi: 10.1261/rna.2321606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thang M.N., Thang D.C., Leautey J. Separation and identification of polynucleotide phosphorylase by electrophoresis on polyacrylamide gel. C.R.Acad.Sci.Hebd.Seances Acad.Sci. D. 1967;265:1823–1826. [PubMed] [Google Scholar]
- Theobald D.L., Mitton-Fry R.M., Wuttke D.S. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzareva N.V., Makhno V.I., Boni I.V. Ribosome-messenger recognition in the absence of the Shine–Dalgarno interactions. FEBS Lett. 1994;337:189–194. doi: 10.1016/0014-5793(94)80271-8. [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Inouye M. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli . J. Bacteriol. 2001;183:2808–2816. doi: 10.1128/JB.183.9.2808-2816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarchuk O., Iost I., Dreyfus M. The relation between translation and mRNA degradation in the lacZ gene. Biochimie. 1991;73:1533–1541. doi: 10.1016/0300-9084(91)90188-7. [DOI] [PubMed] [Google Scholar]
- Zangrossi S., Briani F., Ghisotti D., Regonesi M.E., Tortora P., Dehò G. Transcriptional and post-transcriptional control of polynucleotide phosphorylase during cold acclimation in Escherichia coli . Mol. Microbiol. 2000;36:1470–1480. doi: 10.1046/j.1365-2958.2000.01971.x. [DOI] [PubMed] [Google Scholar]