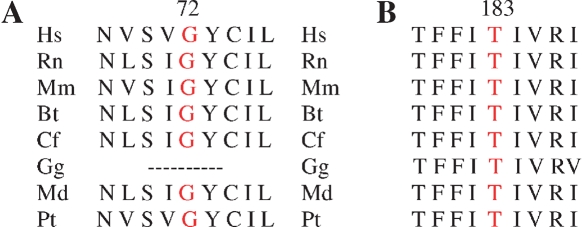To the Editor:
Bardet–Biedl syndrome (BBS) is a rare genetically heterogeneous disorder presenting with retinal dystrophy, postaxial polydactyly, obesity, renal malformations, learning disabilities, and male hypogenitalism [Beales et al., 1999]. To date, 12 genes have proven to be implicated in the disease, accounting for the mutational load in 70–90% of the patients [Blacque and Leroux, 2006; Stoetzel et al., 2007]. BBS is inherited as an autosomal recessive disease though in some instances, digenic triallelic inheritance has been suggested [Katsanis et al., 2001]. Although mutations in many different BBS genes have been described in Caucasians, BBS1 and BBS10 are the two major genes accounting for greater than 50% of the BBS patients [Mykytyn et al., 2002; Stoetzel et al., 2006a]. Mutational findings in the remaining known genes have predominately been reported in non-Caucasian patients [Ansley et al., 2003; Chiang et al., 2004; Stoetzel et al., 2006b]. BBS5 is a minor contributor to BBS as only 2% of families from various ethnic backgrounds harbor BBS5 mutations [Li et al., 2004].
We studied five BBS patients from two nonconsanguineous families residing in Denmark: A Somali family (Family 1) with five siblings of whom four were affected, and an affected boy from Sri Lanka (Family 2). The Sri Lankan patient was a single adopted child and no further information of his biological parents was available. The patients were identified from the files of the Retinitis Pigmentosa Registry at the National Eye Clinic, Hellerup, Denmark [Haim, 2002]. Diagnosis was based on an ERG-verified panretinal photoreceptor dystrophy in association with three or more systemic manifestations; that is, postaxial polydactyly, obesity, cognitive impairment, renal signs, and male hypogenitalism [Beales et al., 1999]. DNA from the patients was collected for mutation analysis. The control group consisted of 43 East Africans (Kenya) and 58 West Africans (Nigeria) for family 1 [Rotimi et al., 2001], and 54 Indian individuals for family 2. Appropriate informed consent was obtained from the patients and their families.
As part of a larger study screening of BBS1, BBS2, BBS4, MKKS, and BBS10 was done by denaturing high performance liquid chromatography (DHPLC, Varian, Inc., Palo Alto, CA) followed by DNA sequencing of aberrant products on an ABI3100 automated capillary sequencer using Big Dye Terminator v.3.1 (Applied Biosystems, Foster City, CA; authors' unpublished data, manuscript submitted). BBS5 mutational analysis was likewise performed by DHPLC. Primer sequences and PCR protocols for BBS5 are available upon request.
Genotyping of the two families was done separately. For Family 1 we used the Affymetrix GeneChip Mapping 10K 2.0 Array to search for regions of shared genotypes among the affected siblings. Sample processing for this part of the study was carried out at the Microarray Facility in Tübingen, Germany. For Family 2, SNP genotyping was performed on Affymetrix GeneChip Human Mapping 50K Hind 240 SNP microarrays (Affymetrix, Santa Clara, CA). We allowed SNPs that could not be scored to be included in the regions of interest. An average call rate >95% was obtained.
Screening of BBS1, BBS2, BBS4, MKKS, and BBS10 by DHPLC did not reveal causative mutations in the five patients. We therefore performed SNP genotyping in order to identify other BBS loci. Since we had no information of consanguinity in Family 1, we searched for regions of shared genotypes covering more than 5 Mb among the four affected sibs and different from the unaffected sib. We identified three major regions: Two regions at 6q15 (23 Mb) and 12q32 (34 Mb) did not contain any known BBS gene while a region spanning 11 Mb at 2q31 contained the BBS5 locus (see the online Table I at http://www.interscience.wiley.com/jpages/1552-4825/suppmat/index.html). Mutational analysis of BBS5 revealed a homozygous nucleotide change, c.214G>A (p.Gly72Ser) in exon 4, identified in all four affected siblings in Family 1 but not in the unaffected sib. Both parents were carriers. The mutation was absent in 202 ethnically matched control chromosomes. Sequence alignment showed the change to be localized within a conserved region (Fig. 1).
Fig. 1.
Evolutionary conservation of BBS5 surrounding novel missense mutation sites showing local alignment of amino acid sequence. A: c.214G>A (p.Gly72Ser). B: c.574A>G (p.Thr183Ala). Hs, Homo sapiens; M, Mus musculus; Gg, Gallus gallus; Md, Monodelfis domesticus; Rn, Rattus norvegicus; Bt, Bos Taurus; Cf, Canis familiaris; Pt, Pan troglodytes.
SNP genotyping of the patient in Family 2 lead to the identification of nine homozygous regions including two regions containing a known BBS gene. One locus spanning 19.6 Mb at 1p32 contained TRIM32 (BBS11) while the other locus spanning 16.6 Mb at 2p31 contained BBS5 (see the online Table I at http://www.interscience.wiley.com/jpages/1552-4825/suppmat/index.html). Sequence analysis of TRIM32 did not reveal any causative nucleotide changes in the patient. However, direct screening of the entire BBS5 gene identified a novel single base pair change, c.547A>G, in exon 7 in the homozygous state predicted to result in a non-conserved amino acid change, p.Thr183Ala (Fig. 1). The mutation was absent in 108 ethnically matched control chromosomes. Furthermore, it was not detected among 60 BBS patients primarily of Northern European origin (authors' unpublished data, manuscript submitted). No family members were available for testing.
We report here on two novel missense mutations in BBS5. Both mutations are localized within conserved regions of the gene, are present in the homozygous state in the patients, and are absent in control chromosomes. In silico analysis predicts the mutations to affect protein function and in Family 1 the mutation segregates with the phenotype. Most of the known mutations in BBS5 are localized within either of two putative domains, called DM16 (Fig. 2); as for the mutations reported here, p.Gly72Ser is localized in the first domain while p.Thr183Ala is localized in the second DM16 domain of BBS5. DM16 is a domain of unknown function and evolutionary conserved among many species [Li et al., 2004]. The fact that the mutations reported here are located in the DM16 domains supports their pathogenecity. We cannot rule out though, the existence of another mutation in linkage disequilibrium with these missense mutations. However, all coding regions plus 20 base pairs in the flanking regions were sequenced. Our findings represent the first missense mutations detected in the homozygous state in BBS5 patients. Thus, these BBS5 missense mutations might be useful for functional studies (Table II).
Fig. 2.
Diagram of the BBS5 protein. Origin of exons is shown as boxes. The positions of the previously reported sequence variations and those reported here are indicated with reference to the exon where mutations occurred. Protein alterations are indicated with the one-letter abbreviations. Though the mutations mainly are truncating mutations and thereby affecting other parts than the DM16 domains, the missense mutations are all localized within the two domains. p.Asn184Ser (N184S) and p.Arg207His (R207H) are of uncertain pathogenecity due to their detection in the heterozygous state in BBS patients.
Table II.
Mutations Reported in BBS5
| BBS families | c.DNA | Predicted effect | Exon | State | Origin | Reference |
|---|---|---|---|---|---|---|
| 1 | c.123delA | p.Gly42GlnfsX11 | 2 | Ho | Tunisia | Smaoui et al. 2006 |
| 1 | 263_271indelGCTCTTA1 | Indel-1 fs X1 | 3 | Ho | Turkey | Li et al. 2004 |
| 1 | c.176G>A | p.Trp59X | 3 | Ho | Kurdish | Li et al. 2004 |
| 1 | c.214G>A | p.Gly72Ser | 4 | Ho | Somalia | This study |
| 1 | c.181T>A/G | p.Leu142X | 6 | Ho | Saudi Arabia | Li et al. 2004 |
| 1 | IVS6+3A>G1 | fsX in exon 71 | 7 | Ho | New Foundland | Li et al. 2004 |
| 1 | c.547G>A | p.Thr183Ala | 7 | Ho | Sri Lanka | This study |
| 2 | c.551A>G | p.Asn184Ser2 | 7 | He | Caucasian | Li et al. 2004 |
| 2 | c.620G>A | p.Arg207His2 | 8 | He | Caucasian | Li et al. 2004 |
| 1 | Deletion in intron 8→3′UTR | Exon 9–12 spliced out | 9–12 | Ho | Turkey | Nishimura et al. 2005 |
For cDNA numbering +1 corresponds to the A of the first ATG translation initiation codon, except for 1 where the mutations are reported as in the reference paper; 2 uncertain pathogeneicity; nucleotide numbers are derived from GenBank, RefSeq cDNA accession numbers: NM_152384.2 for BBS5.
Only five other point mutations, two indels and one large deletion have been previously reported in BBS5 (Table II) [Li et al., 2004; Nishimura et al., 2005; Smaoui et al., 2006]. Most of these were detected in the homozygous state with the exception of the two previously reported missense variants (p.Asn184Ser and p.Arg207His). The previously published mutations are identified in patients from Africa and the Middle East, and in a single patient from New Foundland [Li et al., 2004; Smaoui et al., 2006].
In conclusion, we report two novel missense mutations in BBS5. Several pieces of information support that the mutations are pathogenic. Both patients are non-European. Screening of 60 patients from Northern Europe revealed no mutations in BBS5. These data might have implications for the mutational screening strategy of BBS5.
ELECTRONIC DATABASES
Online Inheritance of Man (OMIM)—BBS5:# 603650.
GenBank RefSeq cDNA accession number: NM_152384.2.
Acknowledgments
This work was supported by grants from the Danish Eye Research Foundation, the Danish Eye Protection Society, the AP Moller Foundation, and the National Institutes of Health (VCS and DYN). The authors acknowledge the Africa American Diabetes Mellitus (AADM) network of investigators for African control DNA samples. AADM was supported by grants obtained from multiple institutes at NIH (NCMHD, NHGRI and NIDDK). We also acknowledge the role of Dr. Duncan Ngare in the collection of the Kenyan samples in collaboration with Dr. Charles Rotimi VCS is an investigator of the Howard Hughes Medical Institute.
References
- Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, Kim JC, Ross AJ, Eichers ER, Teslovich TM, Mah AK, Johnsen RC, Cavender JC, Lewis RA, Leroux MR, Beales PL, Katsanis N. Basal body dysfunction is a likely cause of pleiotropic Bardet–Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA. New criteria for improved diagnosis of Bardet–Biedl syndrome: Results of a population survey. J Med Genet. 1999;36:437–446. [PMC free article] [PubMed] [Google Scholar]
- Blacque OE, Leroux MR. Bardet–Biedl syndrome: An emerging pathomechanism of intracellular transport. Cell Mol Life Sci. 2006;63:2145–2161. doi: 10.1007/s00018-006-6180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang AP, Nishimura D, Searby C, Elbedour K, Carmi R, Ferguson AL, Secrist J, Braun T, Casavant T, Stone EM, Sheffield VC. Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet–Biedl syndrome (BBS3) Am J Hum Genet. 2004;75:475–484. doi: 10.1086/423903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim M. Epidemiology of retinitis pigmentosa in Denmark. Acta Ophthalmol Scand Suppl. 2002;80:1–34. doi: 10.1046/j.1395-3907.2002.00001.x. [DOI] [PubMed] [Google Scholar]
- Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, Hoskins BE, Scambler PJ, Davidson WS, Beales PL, Lupski JR. Triallelic inheritance in Bardet–Biedl syndrome, a Mendelian recessive disorder. Science. 2001;293:2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, Dutcher SK. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Mykytyn K, Nishimura DY, Searby CC, Shastri M, Yen HJ, Beck JS, Braun T, Streb LM, Cornier AS, Cox GF, Fulton AB, Carmi R, Luleci G, Chandrasekharappa SC, Collins FS, Jacobson SG, Heckenlively JR, Weleber RG, Stone EM, Sheffield VC. Identification of the gene (BBS1) most commonly involved in Bardet–Biedl syndrome, a complex human obesity syndrome. Nat Genet. 2002;31:435–438. doi: 10.1038/ng935. [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Swiderski RE, Searby CC, Berg EM, Ferguson AL, Hennekam R, Merin S, Weleber RG, Biesecker LG, Stone EM, Sheffield VC. Comparative genomics and gene expression analysis identifies BBS9, a new Bardet–Biedl syndrome gene. Am J Hum Genet. 2005;77:1021–1033. doi: 10.1086/498323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotimi CN, Dunston GM, Berg K, Akinsete O, Amoah A, Owusu S, Acheampong J, Boateng K, Oli J, Okafor G, Onyenekwe B, Osotimehin B, Abbiyesuku F, Johnson T, Fasanmade O, Furbert-Harris P, Kittles R, Vekich M, Adegoke O, Bonney G, Collins F. In search of susceptibility genes for type 2 diabetes in West Africa: The design and results of the first phase of the AADM study. Ann Epidemiol. 2001;11:51–58. doi: 10.1016/s1047-2797(00)00180-0. [DOI] [PubMed] [Google Scholar]
- Smaoui N, Chaabouni M, Sergeev YV, Kallel H, Li S, Mahfoudh N, Maazoul F, Kammoun H, Gandoura N, Bouaziz A, Nouiri E, M'Rad R, Chaabouni H, Hejtmancik JF. Screening of the eight BBS genes in Tunisian families: No evidence of triallelism. Invest Ophthalmol Vis Sci. 2006;47:3487–3495. doi: 10.1167/iovs.05-1334. [DOI] [PubMed] [Google Scholar]
- Stoetzel C, Laurier V, Davis EE, Muller J, Rix S, Badano JL, Leitch CC, Salem N, Chouery E, Corbani S, Jalk N, Vicaire S, Sarda P, Hamel C, Lacombe D, Holder M, Odent S, Holder S, Brooks AS, Elcioglu NH, Da Silva E, Rossillion B, Sigaudy S, de Ravel TJ, Lewis RA, Leheup B, Verloes A, Amati-Bonneau P, Megarbane A, Poch O, Bonneau D, Beales PL, Mandel JL, Katsanis N, Dollfus H. BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat Genet. 2006a;38:521–524. doi: 10.1038/ng1771. [DOI] [PubMed] [Google Scholar]
- Stoetzel C, Laurier V, Faivre L, Megarbane A, Perrin-Schmitt F, Verloes A, Bonneau D, Mandel JL, Cossee M, Dollfus H. BBS8 is rarely mutated in a cohort of 128 Bardet–Biedl syndrome families. J Hum Genet. 2006b;51:81–84. doi: 10.1007/s10038-005-0320-2. [DOI] [PubMed] [Google Scholar]
- Stoetzel C, Muller J, Laurier V, Davis EE, Zaghloul NA, Vicaire S, Jacquelin C, Plewniak F, Leitch CC, Sarda P, Hamel C, de Ravel TJ, Lewis RA, Friederich E, Thibault C, Danse JM, Verloes A, Bonneau D, Katsanis N, Poch O, Mandel JL, Dollfus H. Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet–Biedl syndrome. Am J Hum Genet. 2007;80:1–11. doi: 10.1086/510256. [DOI] [PMC free article] [PubMed] [Google Scholar]