Abstract
The NH2-terminal domains of membrane-bound sterol regulatory element-binding proteins (SREBPs) are released into the cytosol by regulated intramembrane proteolysis, after which they enter the nucleus to activate genes encoding lipid biosynthetic enzymes. Intramembrane proteolysis is catalyzed by Site-2 protease (S2P), a hydrophobic zinc metalloprotease that cleaves SREBPs at a membrane-embedded leucine-cysteine bond. In the current study, we use domain-swapping methods to localize the residues within the SREBP-2 membrane-spanning segment that are required for cleavage by S2P. The studies reveal a requirement for an asparagine-proline sequence in the middle third of the transmembrane segment. We propose a model in which the asparagine-proline sequence serves as an NH2-terminal cap for a portion of the transmembrane α-helix of SREBP, allowing the remainder of the α-helix to unwind partially to expose the peptide bond for cleavage by S2P.
Keywords: membrane proteins, cholesterol metabolism, proteolysis
Regulated intramembrane proteolysis has been recognized recently as a mechanism that controls numerous responses in animals and bacteria (1). The hallmark event is the controlled cleavage of a transmembrane protein within the membrane-spanning segment to release an intracellular fragment. In animal cells, this fragment moves to the nucleus, where it regulates gene transcription. The mechanism by which intramembrane cleavage takes place and the mechanisms for its regulation are only partially understood.
Our laboratory has been studying the role of regulated intramembrane proteolysis in the processing of sterol regulatory element-binding proteins (SREBPs), membrane-bound transcription factors that control lipid synthesis and uptake in animal cells (2). The SREBPs are synthesized as polypeptide chains averaging 1150 amino acids in length and consisting of three domains. The NH2-terminal domain of ≈480 amino acids, a transcription factor of the basic-helix-loop-helix-leucine zipper (bHLH-Zip) family, projects into the cytosol. It is anchored to membranes by an 80-amino acid central domain composed of two membrane-spanning segments separated by a short hydrophilic loop of 31 amino acids that projects into the lumen of the endoplasmic reticulum (ER). The COOH-terminal domain of ≈590 amino acids protrudes into the cytosol, where it binds SREBP cleavage-activating protein (SCAP), a polytopic membrane protein (3). After synthesis in the ER, the SCAP/SREBP complex moves to the Golgi apparatus, where a membrane-bound serine protease designated Site-1 protease (S1P) cleaves a leucine-serine bond within the luminal loop of the SREBP, separating the two membrane-spanning segments (4). The NH2-terminal domain of SREBP remains attached to the membrane through the first transmembrane segment. This segment is then cleaved by Site-2 protease (S2P), which hydrolyzes a leucine-cysteine bond that is located three residues into the hydrophobic segment (5). The NH2-terminal domain then leaves the membrane with three hydrophobic amino acids at its COOH terminus. It translocates to the nucleus, where it activates more than 20 genes encoding enzymes of cholesterol and fatty acid synthesis as well as the low density lipoprotein receptor (6, 7). When sterols build up in cells, the SREBP/SCAP complex fails to exit the ER, and it never reaches S1P (8, 9). As a result, the NH2-terminal domains of the SREBPs are no longer released into the nucleus, and transcription of the target genes declines. This mechanism allows cholesterol to inhibit its own synthesis and uptake, thereby preventing cholesterol overaccumulation in cells.
The human gene encoding S2P was cloned by complementation in M19 cells, a line of Chinese hamster ovary cells that is auxotrophic for cholesterol because of a failure to carry out Site-2 cleavage of SREBPs (10). The S2P gene encodes a hydrophobic protein of 519 amino acids that contains the sequence HEIGH, which conforms to the HExxH consensus (where x is any amino acid) found in a large family of zinc metalloproteases (11). In the classic members of this family, such as thermolysin, the two histidines help to coordinate the zinc atom, and the glutamic acid activates a water molecule, allowing it to make a nucleophilic attack on a peptide bond. Most zinc metalloproteases contain a remote residue (histidine, tyrosine, glutamic, or aspartic acid) that forms an additional coordination site for the zinc (11). In S2P, this function is believed to be filled by an aspartic acid that is part of an LDG sequence that is located 300 amino acids to the COOH-terminal side of the HEIGH motif (12). Mutation of either of the two histidines or the glutamic acid of the HEIGH sequence or of the aspartic acid of the LDG sequence abolishes the ability of an S2P cDNA to restore SREBP cleavage in the mutant M19 cells (12).
In contrast to the classic zinc metalloproteases, S2P is distinguished by its extreme hydrophobicity. Indeed, the HEIGH and LDG sequences are both embedded in long hydrophobic segments (12). The orientation of S2P in membranes, as studied by a combination of protease protection and glycosylation site mapping, revealed that much of the protein is embedded in the membrane (12). The protein contains only three long hydrophilic segments. One of these contains a stretch of 23 contiguous serine residues, and another is cysteine-rich. All three of the hydrophilic segments are oriented toward the lumen of the ER and post-ER organelles.
Proteins that resemble S2P have now been identified in all eukaryotic species whose genomes have been studied, with the exception of Saccharomyces cerevisiae. Similar proteins are encoded by the genomes of eubacteria (both Gram-positive and Gram-negative) and archaea (1, 13, 14). All of these proteins share the hydrophobicity profile of S2P, and they all contain HExxH and φDG sequences (where φ is leucine or phenylalanine) embedded in long hydrophobic segments. Two of the bacterial proteases have been implicated in the cleavage of membrane-bound proteins. One of these, SpoIVFB, cleaves a membrane-embedded transcription factor (pro-σK) during sporulation in Bacillus subtilis (14). The other, designated Eep from Enterococcus faecalis, cleaves a transmembrane protein in the middle of a membrane-spanning sequence (1, 15, 16).
The mechanism by which S2P and its relatives cleave membrane-spanning sequences is not clear. Most membrane-spanning sequences are folded as α-helices. Peptide bonds within α-helices are poorly accessible to proteases because of the steric hindrance provided by the framework of hydrogen bonds that stabilize the helix (17). This has led to the suggestion that the helices must unfold at least partially before the protease can act (1).
A second unanswered question relates to the mechanism by which these proteases choose their targets. The target sequence for S2P was studied by measurement of Site-2 cleavage in cells transfected with cDNAs encoding mutant versions of SREBP-2 (5). Cleavage was shown to require all or part of the sequence DRSR, which immediately precedes the first membrane-spanning segment of SREBPs. Replacement of the DRSR sequence with unrelated amino acids markedly reduced the cleavage by S2P. Surprisingly, S2P does not cleave at the DRSR sequence. Rather, it cleaves a Leu-Cys bond that is located three residues to the COOH-terminal side of the DRSR (5). Cleavage by S2P does not require either the leucine or cysteine at these positions; replacement of either residue with alanine does not interfere with cleavage. Even more remarkably, cleavage was not abolished when any of the other residues of the membrane-spanning segment was replaced singly with alanine (5).
In the current studies, we use a domain-swapping approach to identify the requirements for cleavage of SREBP-2 by S2P. The results reveal a requirement for at least one member of the asparagine-proline (NP) sequence that is found in the middle third of the first membrane-spanning segment of SREBP-2. This NP sequence is conserved in all SREBPs from all species identified so far, including worms, flies, and humans. We propose a model in which the NP sequence acts as a cap that permits a partial unfolding of the NH2-terminal part of the transmembrane α-helix, allowing attack by S2P.
Materials and Methods
Materials.
M19 cells are a mutant line of CHO-K1 cells auxotrophic for cholesterol and unsaturated fatty acids (18), attributable to a deletion in the S2P gene (10). We obtained Lipofectamine from GIBCO/BRL and N-biotinylaminoethyl methanethiosulfonate from Toronto Research Chemical (Downsview, ON, Canada). Other reagents were obtained from sources as described (5, 10, 19).
Recombinant Plasmids.
The following plasmids were described in the indicated reference: pTK-HSV-SREBP2, encoding herpes simplex virus (HSV) epitope-tagged human SREBP-2 driven by the herpes simplex virus thymidine kinase (TK) promoter (19); pCMV-SCAP(D443N), encoding hamster SCAP(D443N) driven by the cytomegalovirus (CMV) enhancer/promoter (3); pCMV-Myc-S2P, encoding Myc epitope-tagged human S2P driven by the CMV enhancer/promoter (10); pTK-HSV-8His-ACBP/BP2 (5), encoding HSV and histidine-tagged acyl-CoA-binding protein/SREBP-2 fusion protein driven by the herpes simplex virus TK promoter; and pVAI, encoding the adenovirus-associated I RNA gene, which enhances translation of transfected cDNAs (20).
To facilitate creation of mutants in the first transmembrane domain of human SREBP-2, we used the single-stranded, oligonucleotide-mediated mutagenesis kit from Bio-Rad (Muta-gene Phagemid In Vitro Mutagenesis, Version-2) to mutate codons 470 and 471 of SREBP-2 to synonymous codons that constitute an AgeI restriction site. We also mutated codons 506–508 of SREBP-2 (5′-GCCCACGAC-3′), which created a BsiWI site (5′-GCGTACGAC-3′), replacing histidine-507 with tyrosine. This plasmid, designated pTK-HSV-SREBP2(AgeI/BsiWI), produced a protein that behaved identically to the protein produced by pTK-HSV-SREBP2 when transfected into M19 cells and HEK 293 cells and is thus designated as wild-type in the figure legends. pTK-HSV-SREBP2(AgeI/BsiWI) allows ready excision and cassette replacement of DNA encoding amino acids 470–508 of SREBP-2. Sense and antisense oligonucleotides containing the desired mutations or chimeric GPP130 sequence (21) along with Age I/BsiWI overhangs were annealed and ligated into Age I/BsiWI-digested pTK-HSV-SREBP2(AgeI/BsiWI).
To create plasmids that encode proteins suitable for cysteine panning, we used PCR on the template pTK-HSV-8His-ACBP/SREBP2 to generate a NotI site at the 5′ end of the HSV tags and an XmaI site at the 3′ end of the acyl-CoA binding protein (ACBP) coding region. The PCR fragment NotI-HSV-8His-ACBP-XmaI was double-digested with NotI/XmaI and was ligated into pTK-HSV-SREBP2(AgeI/BsiWI) that had been previously digested with NotI/AgeI. (The NotI site is located 5′ to the initiator methionine in all plasmids.) This plasmid was used to produce ACBP fusion proteins whose cleavage could be studied by cysteine panning. All constructs were extensively sequenced for confirmation.
Transient Transfection of Cells.
On day 0, M19 cells were set up at a density of 7 × 105 cells per 60-mm dish in medium A [a 1:1 mixture of Ham's F-12 medium and DMEM containing 100 unit/ml of penicillin, 100 μg/ml streptomycin sulfate, 5% (vol/vol) FCS, 5 μg/ml cholesterol, 1 mM sodium mevalonate, and 20 μM sodium oleate]. On day 1, cells were transfected with the indicated amount of DNA by using 3 μl of Lipofectamine per 1 μg of DNA in serum-free medium (final volume, 2 ml) according to the manufacturer's protocol. After incubation for 3 h at 37°C in a 8–9% CO2 incubator, the existing 2 ml of medium in each dish was supplemented with 2 ml of medium to give the following final concentrations: 5% FCS containing 1 μg/ml of 25-hydroxycholesterol and 10 μg/ml of cholesterol added in 0.2% ethanol. After incubation for 20 h at 37°C, the cells received N-acetyl-leucinal-leucinal-norleucinal to a final concentration of 25 μg/ml. After a further incubation for 2 h at 37°C, the cells were harvested and fractionated into nuclear extract and membrane fractions as described (19).
Immunoblot Analysis.
Immunoblot analysis was carried out after 8% SDS/PAGE as described (19) by using the SuperSignal CL-HRP Substrate System (Pierce). HSV-SREBP-2 was detected with 0.2 μg/ml monoclonal IgG-HSV-Tag (Novagen) and was visualized with a 1:5,000 dilution of peroxidase-conjugated, affinity-purified donkey anti-mouse IgG (Jackson ImmunoResearch). Gels were exposed at room temperature to X-Omat Blue XB-1 film (Kodak) for the indicated time.
Cysteine Panning.
Monolayers of HEK293 cells were set up on day 0 at a density of 4 × 105 cells per 60-mm dish. On day 2, the cells were transfected as described (5) using the MBS Transfection Kit (Stratagene) with 4 μg of pTK-HSV-8His-ACBP/BP2, 0.5 μg of pCMV-SCAP(D443N), 0.5 μg of pVAI, and 0.25 μg of pCMV-Myc-S2P. The cells were incubated for 20 h at 37°C in the absence of sterols, after which the cells were harvested, and cytosol was prepared as described (5). To identify the cleavage site, the cytosolic fractions were subjected to Ni2+ chromatography to purify the cleaved His-tagged ACBP/SREBP-2 proteins as described (5). Proteins eluted from the Ni-NTA column (Qiagen, Chatsworth, CA) were then derivatized with 0.5 mM N-biotinylaminoethyl methanethiosulfonate. This reagent was substituted for the previous reagent (maleimide propionyl biotin) (5) because it produced faster and more efficient labeling. After repeat purification on the Ni-NTA column, the eluted proteins were precipitated with streptavidin-agarose (Prozyme, San Leandro, CA) and were centrifuged. Supernatant and pellet fractions were resolved by SDS/PAGE and were analyzed by immunoblotting as described above. Exposed film was quantified by using a Hewlett–Packard ScanJet 5100C scanner and the PC version of the nih image program available over the internet from Scion (www.scioncorp.com).
Results
As a first step in localizing the residues in the first transmembrane segment of SREBP-2 that are required for Site-2 cleavage, we replaced this segment with the membrane-spanning sequence from another membrane protein that is not expected to be cleaved by S2P. A search of protein databases revealed a Golgi-specific single-pass transmembrane protein, designated GPP130, that is oriented like the intermediate fragment of SREBP-2: i.e., with its NH2 terminus in the cytosol (type II membrane orientation). Moreover, the length of the hydrophobic segment is precisely the same in the two proteins (Fig. 1). In making the chimeric constructs, we retained the DRSR sequence from SREBP-2 that was shown previously to be required for Site-2 cleavage. To test for S2P-dependent cleavage of the chimeric proteins, we adopted a standard protocol designed to assure that all observed cleavage was carried out by S2P. The cDNA encoding the epitope-tagged chimeric protein was introduced into M19 cells, the mutant line of CHO cells that does not express S2P. Transfections were carried out in the absence or presence of a cDNA encoding SCAP(D443N), a mutant version of SCAP that is resistant to suppression by sterols. We also transfected a cDNA encoding S2P tagged with a Myc epitope. The cells were incubated in the presence of sterols, which suppresses the activity of endogenous SCAP and makes all SREBP cleavage dependent on the presence of the mutant SCAP (3, 22). Cell membranes and nuclei were isolated, were subjected to SDS/PAGE, and were blotted with an antibody against the HSV epitope tag on the SREBP-2.
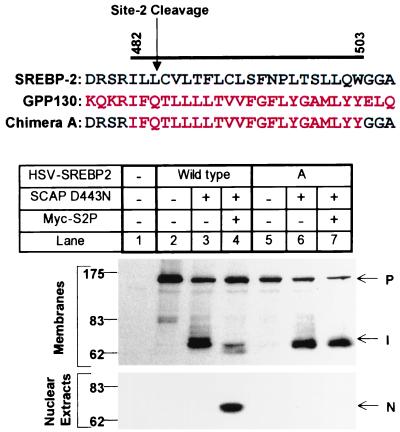
Figure 1.
Replacement of the first transmembrane domain of SREBP-2 with that of GPP130 abolishes Site-2 cleavage. The DNA sequence encoding the first transmembrane domain of SREBP-2 (designated wild-type) was replaced with DNA encoding the membrane-spanning segment of GPP130 (designated chimera A). The bar denotes the first transmembrane domain with relevant amino acids numbered above. Black and red letters represent SREBP-2 and GPP130 sequences, respectively. M19 cells were set up on day 0 and were transfected on day 1 with a plasmid encoding wild-type or chimeric HSV-tagged SREBP-2 (3 μg/dish) together with pCMV-SCAP(D443N) (0.5 μg/dish) with or without pCMV-Myc-S2P (1 μg/dish) as indicated. The total amount of DNA was adjusted to 4.5 μg/dish by addition of pcDNA3 empty vector. After incubation at 37°C with sterols for 20 h, the cells were harvested and fractionated as described in Materials and Methods. Aliquots of nuclear extracts or membranes (30 μg protein) were subjected to SDS/PAGE and immunoblot analysis as described in Materials and Methods. Filters were exposed for 15 sec. P, I, and N denote the precursor, intermediate, and nuclear forms of HSV-tagged SREBP-2, respectively.
As shown in Fig. 1, when we transfected HSV-tagged wild-type SREBP-2 into the M19 cells in the absence of SCAP(D443N) and Myc-S2P, we failed to observe the NH2-terminal fragment in the nuclear extracts (lane 2). When we cotransfected SCAP(D443N), the SREBP-2 was cleaved by S1P, generating the NH2-terminal intermediate fragment of ≈70 kDa that remained membrane-bound (Fig. 1, lane 3). When we added the cDNA encoding S2P, the membrane-bound intermediate was cleaved, and the mature NH2-terminal fragment appeared in nuclear extracts (Fig. 1, lane 4). When we transfected the cDNA encoding the SREBP/GPP130 chimera, the protein was cleaved by S1P, generating the membrane-bound intermediate in a reaction that depended on SCAP(D443N) (Fig. 1, lanes 5 and 6). When S2P was co-expressed, the intermediate failed to be cleaved, and no nuclear form was seen (Fig. 1, lane 7). These data indicate that the GPP130 transmembrane sequence lacks one or more residues that are required for Site-2 cleavage.
To narrow down the residues in SREBP-2 required for Site-2 cleavage, we systematically replaced portions of the GPP130 sequence with sequences from SREBP-2 (Fig. 2). In making these chimeras, we encountered a problem with the glutamine at the third intramembrane position of GPP130 (see Fig. 1). This glutamine did not cause a problem when it was present in the context of the complete GPP130 transmembrane segment. However, when we began to swap portions of the SREBP-2 sequence into the GPP130 sequence, we noted that some of the constructs failed to insert into the membrane properly as indicated by the finding of the uncleaved full-length protein in nuclear extracts. This problem was resolved when we changed the glutamine of GPP130 to phenylalanine, and so the experiments of Fig. 2 were performed with a GPP130 sequence containing this substitution.
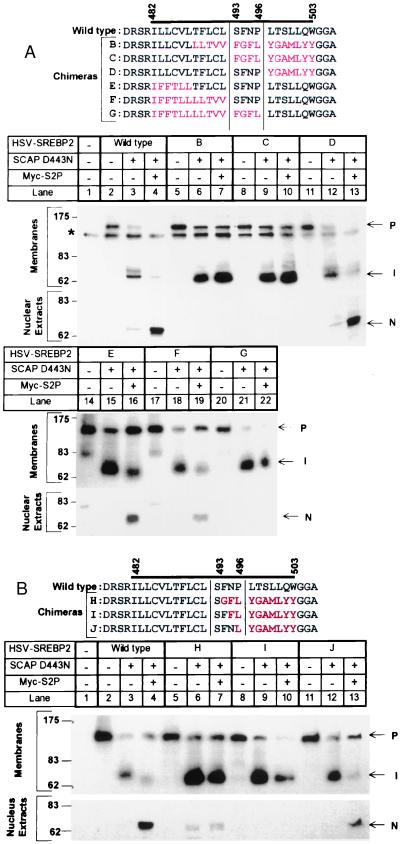
Figure 2.
Site-2 cleavage of SREBP-2/GPP130 chimeric proteins in transfected M19 cells. The sequence flanking the first transmembrane domain of wild-type SREBP-2 and various GPP130 chimeric substitutions is shown. The bar denotes the putative first transmembrane domain. Black and red letters represent amino acids derived from SREBP-2 and GPP130, respectively. M19 cells were set up, were transfected with the indicated plasmids, were incubated for 20 h at 37°C with sterols, and were harvested for immunoblot analysis as described in the legend to Fig. 1. Filters were exposed to film for 30 sec in A and 1 min in B. P, I, and N denote the precursor, intermediate, and nuclear forms of HSV-tagged SREBP-2, respectively. The asterisk (*) denotes a cross-reactive band that is present in mock-transfected cells.
As shown in Fig. 2A, Site-2 cleavage was not reconstituted when we restored the SREBP-2 sequence at the NH2-terminal end of the membrane-spanning region, which includes the actual cleavage site (Fig. 2A, chimera C, lane 10). However, cleavage returned when we extended the SREBP-2 sequence to include four additional residues, SFNP (Fig. 2A, chimera D, lane 13). Cleavage also took place when the entire NH2-terminal half of the membrane-spanning domain was derived from GPP130, so long as the SFNP sequence from SREBP-2 was retained (Fig. 2A, chimeras E and F, lanes 16 and 19). Cleavage was abolished when we extended the GPP130 sequence by four residues so that it replaced the SFNP sequence (compare Fig. 2A, chimera G, lane 22, with chimera F, lane 19). This experiment pointed to the SFNP sequence as containing some element essential for Site-2 cleavage.
To determine which residues within the SFNP sequence are sufficient for cleavage, we substituted these residues one-by-one into the GPP130 sequence (Fig. 2B). Restoration of the serine or phenylalanine failed to restore cleavage (Fig. 2B, lanes 7 and 10). Cleavage was partially rescued when we restored the asparagine (Fig. 2B, SFNL, lane 13), but the level of cleavage was still below that observed when the sequence contained both the asparagine and proline (SFNP, lane 4). These data implicate the asparagine and proline as the crucial residues within the SFNP sequence necessary for cleavage by S2P.
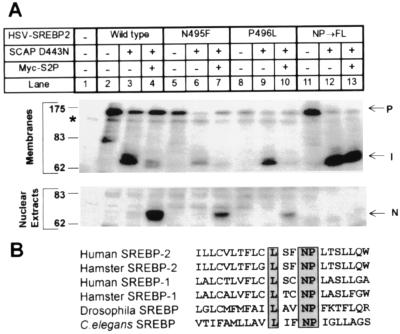
To explore the roles of the asparagine and proline in the context of the wild-type SREBP-2 sequence, we replaced asparagine-495 and proline-496 of SREBP-2 with the corresponding residues of GPP130 (Fig. 3A). When either the asparagine or the proline was replaced individually, Site-2 cleavage was diminished, but not abolished (compare Fig. 3A, lanes 7 and 10 with lane 4). When both residues were replaced, cleavage no longer occurred (Fig. 3A, lane 13). Fig. 3B shows that the asparagine-proline (NP) sequence is conserved in all SREBPs that are available to date, including those of Drosophila melanogaster and Caenorhabditis elegans.
Figure 3.
NP sequence in the first transmembrane domain of SREBP-2 is required for cleavage at Site-2. (A) M19 cells were set up, were transfected with the indicated plasmids, were incubated for 20 h at 37°C with sterols, and were harvested for immunoblot analysis as described in the legend to Fig. 1. Filters were exposed to film for 30 sec. P, I, and N denote the precursor, intermediate, and nuclear forms of HSV-tagged SREBP-2, respectively. The asterisk (*) denotes a cross-reactive band that is present in mock-transfected cells. (B) Sequence alignment of the first transmembrane domain of SREBPs from various species. Completely conserved amino acids are boxed. The sequences of human SREBP-1 (28), human SREBP-2 (29), hamster SREBP-1 (30), hamster SREBP-2 (31), and Drosophila SREBP (32) are published in the indicated references. The C. elegans sequence is predicted from the sequence of a cosmid that was obtained from the European Molecular Biology Laboratory Database (ID code AL031635.1).
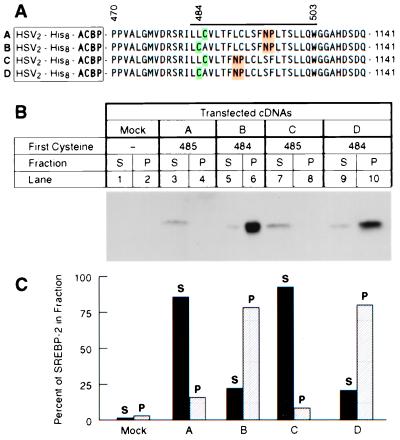
We next asked whether Site-2 cleavage would occur if the NP sequence were moved to another location within the SREBP-2 transmembrane domain and whether such movement would affect the location of the cleaved peptide bond. To localize the cleavage site, we used the cysteine panning technique that we previously described (5). For this purpose, we replaced the DNA sequence encoding the NH2-terminal domain of SREBP-2 with a sequence encoding rat acyl-CoA binding protein (ACBP), a small protein of 87 amino acids that lacks cysteine residues (23). We also modified the first membrane-spanning domain in this construct by inserting a cysteine codon either at position 484 or 485 (see Fig. 4). We have shown previously that this fusion protein is recognized by the Site-1 and Site-2 proteases and that the ACBP is released from the membrane into the cytosol in a sterol-regulated fashion (5). To determine whether the cleaved cytosolic fragment contains a cysteine, we derivatize the fragment with a biotin reagent that attaches to sulfhydryls of cysteine residues. The protein is then incubated with streptavidin-agarose beads. If the cytosolic fragment contains a cysteine, it adheres to the beads and is found in the pellet fraction (P in Fig. 4), where it can be detected by immunoblotting. Otherwise, it remains in the supernatant (S in Fig. 4). When the chimeric protein contained the wild-type SREBP-2 transmembrane sequence with a cysteine at position 485, the cytosolic fragment remained in the supernatant, indicating that it lacked a cysteine (Fig. 4B, construct A, lane 3). When the cysteine was moved to position 484, the cytosolic fragment was found mostly in the pellet (Fig. 4B, construct B, lane 6). This indicates that S2P is cleaving between positions 484 and 485, which is its normal site of cleavage. We then moved the NP sequence five positions closer to the NH2 terminus (Fig. 4B, constructs C and D). Again the cysteine-485 fragment remained in the supernatant (Fig. 4B, lane 7), and the cysteine-484 fragment was in the pellet (Fig. 4B, lane 10). These data indicate that movement of the NP sequence within the transmembrane domain does not eliminate cleavage, nor does it change the site of cleavage.
Figure 4.
Movement of NP sequence fails to alter site of cleavage by Site-2 protease as measured by cysteine panning. (A) Sequences of His8-tagged ACBP/SREBP-2 chimeric proteins in which positions of cysteine (green) and asparagine-proline (NP) (yellow) sequences have been moved. The bar above the sequences denotes the first transmembrane domain. Proline-470 is the first amino acid derived from SREBP-2. The first cysteine in the chimeric proteins is placed at a position corresponding to residue 484 or 485 of the SREBP-2 sequence. The NP sequence is highlighted in yellow. (B) Immunoblots of the cleaved proteins after derivatization by N-biotinylaminoethyl methanethiosulfonate and precipitation by streptavidin-agarose. Monolayers of HEK-293 cells were transfected with 4 μg of the indicated plasmid together with 0.5 μg of pCMV-SCAP(D443N), 0.5 μg pVAI, and 0.25 μg of pCMV-Myc-S2P. After incubation at 37°C for 20 h without sterols, the cells were harvested, cytosol was prepared, and the cleaved NH2-terminal domains in the cytosolic fraction were purified by Ni-NTA chromatography, were treated with N-biotinylaminoethyl methanethiosulfonate, and were precipitated with streptavidin-agarose as described in Materials and Methods. Equal aliquots (50% of total volume of the streptavidin-agarose supernatant (S) and pellet (P) fractions) were subjected to SDS/PAGE and were immunoblotted with IgG-HSV-Tag. The filter was exposed to film for 20 sec at room temperature. (C) Quantification of the bands in A as determined by densitometry as described in Materials and Methods. Each bar represents the percentage of total ACBP/SREBP-2 found in the supernatant (closed bars) or pellet (hatched bars). The results shown are representative of two experiments.
Discussion
The current data establish that the NP sequence, which is located deep within the first membrane-spanning segment of all known SREBPs (Fig. 3B), is required for cleavage of this segment by S2P. Cleavage occurs at a Leu-Cys bond that is 11 residues to the NH2-terminal side of the NP sequence and 3 residues to the COOH-terminal side of the DRSR sequence that was shown previously to be required for cleavage. The requirement for both residues of the NP sequence is not absolute: cleavage takes place at a reduced but detectable rate when either the asparagine or the proline is eliminated, but not when both are eliminated.
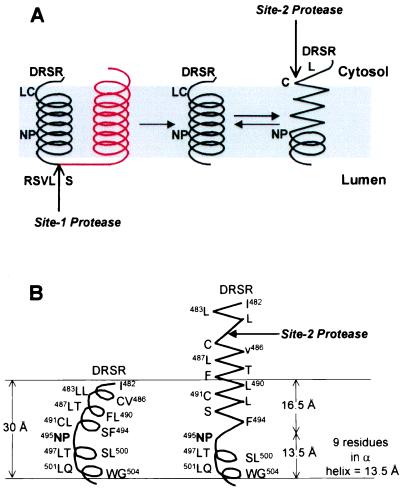
The mechanism by which the NP sequence facilitates S2P cleavage is not known. These residues might interact directly with S2P, but this seems somewhat unlikely because the NP sequence is remote from the actual cleavage site and because the NP sequence can be moved by five positions without affecting either the extent or the site of cleavage (Fig. 4). The NP sequence could be required for interaction with another protein that facilitates cleavage by S2P, but such a protein has not been identified in an extensive search for mutants that block S2P cleavage and cause cholesterol auxotrophy (22, 24). We favor an alternative theory: namely, that the NP sequence is required in order for the transmembrane segment of SREBP-2 to undergo a conformational change that exposes the peptide bond to cleavage by S2P. This model is illustrated in Fig. 5.
Figure 5.
Proposed model for the effect of NP on the conformation of the first transmembrane domain of SREBP-2. (A) The first (black) and second (red) transmembrane domains of SREBP-2 are shown. The NP motif, S1P cleavage site (RSVL↑S), S2P cleavage site (L↓C), and the DRSR sequence immediately adjacent to the first transmembrane domain are shown. After cleavage at Site-1, the first transmembrane domain separates from the second transmembrane domain. This allows the NH2-terminal portion of the transmembrane α-helix to unwind to form a more extended structure, thereby exposing the Leu-Cys (LC) bond to the cytosolic face. The unwinding of the helix requires the NP sequence, which caps the unwinding, allowing the COOH-terminal portion of the transmembrane domain to maintain a helical conformation. (B) Proposed dimensions of the first transmembrane domain in relation to the membrane thickness. The transmembrane domain at the left is presented as a kinked α-helix. The spacing between every turn of the helix is assumed to be 1.5 Å (33), and the thickness of the hydrocarbon region of the bilayer is assumed to be 30 Å (34). On the right, the NH2-terminal end of the transmembrane domain is shown in an extended conformation followed by an α-helix that is capped by the NP motif.
In the model of Fig. 5, we envision that the two closely spaced transmembrane segments of SREBP-2 exist as paired α-helices. The first helix is metastable because of the presence of the NP sequence, which has the propensity to cap α-helices at their NH2 termini, forming a boundary between the helix and more extended structures (25). The NP sequence is not required for cleavage by S1P (Fig. 1), which indicates that the NP motif does not play a role in the insertion of SREBP-2 into the membrane, nor in the interaction of the protein with SCAP, nor in the SCAP-dependent transport to the Golgi complex in which cleavage takes place. After cleavage by S1P, the two helices separate, as demonstrated previously by experiments in which an antibody against the COOH terminus of SREBP-2 failed to bring down the NH2-terminal intermediate in a co-precipitation assay (26). We envision that this separation allows the first helix to unwind between boundaries formed by the DRSR and NP sequences. This unwinding would allow the Leu-484/Cys-485 bond to assume an extended structure, pushing it up to the surface of the membrane, where it is now accessible to S2P. In the absence of the NP sequence, the transmembrane α-helix would not unwind, and cleavage by S2P would not occur.
A precedent for proteolytic cleavage of an unfolded structure at the end of a membrane-spanning sequence was established recently in structural studies of bacterial signal peptidase in complex with a substrate inhibitor (17). The region of the membrane-spanning sequence that is cleaved by the enzyme was shown to form an extended structure, rendering the scissile bond accessible to the enzyme, in this case a membrane-embedded serine protease (17, 27).
The model of Fig. 5 is speculative at present. The strongest support, albeit indirect, comes from previous studies by Richardson and Richardson (25), who surveyed known protein structures to determine whether there was a preference for specific amino acids at the ends of α-helices. Remarkably, they found a 3.5-fold enrichment for asparagine at the NH2-terminal end of the helix and a 2.6-fold enrichment for proline at the adjacent position. Thus, the NP sequence would be favored to form the NH2-terminal cap of an α-helix as postulated in Fig. 5. One of the confounding variables of regulated intramembrane proteolysis is the inaccessibility of water to transmembrane domains. Our model may address this issue because the theoretical dimensions shown in Fig. 5B place the scissile bond of the SREBP-2 transmembrane domain in a charged milieu, thus making it accessible to water. Confirmation of this model will require direct structural studies of SREBPs before and after cleavage by S1P.
Acknowledgments
We thank our colleague Rob Rawson for helpful discussions and critical review of the manuscript; Ken Westover for excellent technical assistance; Anna Fuller and Lisa Beatty for invaluable assistance with tissue culture; and Jeff Cormier for DNA sequencing and oligonucleotide synthesis. This work was supported by research grants from the National Institutes of Health (Grant HL20948) and the Perot Family Foundation. U.P.D. is the recipient of a Postdoctoral Fellowship for Physicians from the Howard Hughes Medical Institute.
Abbreviations
- ACBP
acyl-CoA binding protein
- CMV
cytomegalovirus
- ER
endoplasmic reticulum
- HSV
herpes simplex virus
- S1P
Site-1 protease
- SCAP
SREBP cleavage-activating protein
- SREBP
sterol regulatory element-binding protein
- TK
thymidine kinase
References
- 1.Brown M S, Ye J, Rawson R B, Goldstein J L. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 2.Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hua X, Nohturfft A, Goldstein J L, Brown M S. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 4.Sakai J, Rawson R B, Espenshade P J, Cheng D, Seegmiller A C, Goldstein J L, Brown M S. Mol Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- 5.Duncan E A, Davé U P, Sakai J, Goldstein J L, Brown M S. J Biol Chem. 1998;273:17801–17809. doi: 10.1074/jbc.273.28.17801. [DOI] [PubMed] [Google Scholar]
- 6.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 7.Horton J D, Shimomura I. Curr Opin Lipidol. 1999;10:143–150. doi: 10.1097/00041433-199904000-00008. [DOI] [PubMed] [Google Scholar]
- 8.Nohturfft A, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1998;95:12848–12853. doi: 10.1073/pnas.95.22.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeBose-Boyd R A, Brown M S, Li W-P, Nohturfft A, Goldstein J L, Espenshade P J. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- 10.Rawson R B, Zelenski N G, Nijhawan D, Ye J, Sakai J, Hasan M T, Chang T-Y, Brown M S, Goldstein J L. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 11.Rawlings N D, Barrett A J. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 12.Zelenski N G, Rawson R B, Brown M S, Goldstein J L. J Biol Chem. 1999;274:21973–21980. doi: 10.1074/jbc.274.31.21973. [DOI] [PubMed] [Google Scholar]
- 13.Lewis A P, Thomas P J. Protein Sci. 1999;8:439–442. doi: 10.1110/ps.8.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudner D Z, Fawcett P, Losick R. Proc Natl Acad Sci USA. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An F Y, Sulavik M C, Clewell D B. J Bacteriol. 1999;181:5915–5921. doi: 10.1128/jb.181.19.5915-5921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firth N, Fink P D, Johnson L, Skurray R A. J Bacteriol. 1994;176:5871–5873. doi: 10.1128/jb.176.18.5871-5873.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paetzel M, Dalbey R E, Strynadka N C J. Nature (London) 1998;396:186–190. doi: 10.1038/24196. [DOI] [PubMed] [Google Scholar]
- 18.Hasan M T, Chang T Y. Somatic Cell Mol Genet. 1994;20:481–491. doi: 10.1007/BF02255839. [DOI] [PubMed] [Google Scholar]
- 19.Sakai J, Duncan E A, Rawson R B, Hua X, Brown M S, Goldstein J L. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 20.Akusjärvi G, Svensson C, Nygard O. Mol Cell Biol. 1987;7:549–551. doi: 10.1128/mcb.7.1.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linstedt A D, Mehta A, Suhan J, Reggio H, Hauri H-P. Mol Biol Cell. 1997;8:1073–1087. doi: 10.1091/mbc.8.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawson R B, DeBose-Boyd R, Goldstein J L, Brown M S. J Biol Chem. 1999;274:28549–28556. doi: 10.1074/jbc.274.40.28549. [DOI] [PubMed] [Google Scholar]
- 23.Mocchetti I, Einstein R, Brosius J. Proc Natl Acad Sci USA. 1986;83:7221–7225. doi: 10.1073/pnas.83.19.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawson R B, Cheng D, Brown M S, Goldstein J L. J Biol Chem. 1998;273:28261–28269. doi: 10.1074/jbc.273.43.28261. [DOI] [PubMed] [Google Scholar]
- 25.Richardson J S, Richardson D C. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- 26.Sakai J, Nohturfft A, Cheng D, Ho Y K, Brown M S, Goldstein J L. J Biol Chem. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- 27.von Heijne G. Nature (London) 1998;396:111–113. doi: 10.1038/24036. [DOI] [PubMed] [Google Scholar]
- 28.Yokoyama C, Wang X, Briggs M R, Admon A, Wu J, Hua X, Goldstein J L, Brown M S. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- 29.Hua X, Yokoyama C, Wu J, Briggs M R, Brown M S, Goldstein J L, Wang X. Proc Natl Acad Sci USA. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato R, Yang J, Wang X, Evans M J, Ho Y K, Goldstein J L, Brown M S. J Biol Chem. 1994;269:17267–17273. [PubMed] [Google Scholar]
- 31.Yang J, Sato R, Goldstein J L, Brown M S. Gene Dev. 1994;8:1910–1919. doi: 10.1101/gad.8.16.1910. [DOI] [PubMed] [Google Scholar]
- 32.Theopold U, Ekengren S, Hultmark D. Proc Natl Acad Sci USA. 1996;93:1195–1199. doi: 10.1073/pnas.93.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stryer L. Biochemistry. New York: Freeman; 1995. [Google Scholar]
- 34.Bretscher M S, Munro S. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]