Abstract
The ROCK-I serine/threonine protein kinase mediates the effects of RhoA to promote the formation of actin stress fibres and integrin-based focal adhesions. ROCK-I phosphorylates the unconventional G-protein RhoE on multiple N- and C-terminal sites. These phosphorylation events stabilise RhoE, which functions to antagonise RhoA-induced stress fibre assembly. Here, we provide a molecular explanation for multi-site phosphorylation of RhoE from the crystal structure of RhoE in complex with the ROCK-I kinase domain. RhoE interacts with the C-lobe αG helix of ROCK-I by means of a novel binding site remote from its effector region, positioning its N and C termini proximal to the ROCK-I catalytic site. Disruption of the ROCK-I:RhoE interface abolishes RhoE phosphorylation, but has no effect on the ability of RhoE to disassemble stress fibres. In contrast, mutation of the RhoE effector region attenuates RhoE-mediated disruption of the actin cytoskeleton, indicating that RhoE exerts its inhibitory effects on ROCK-I through protein(s) binding to its effector region. We propose that ROCK-I phosphorylation of RhoE forms part of a feedback loop to regulate RhoA signalling.
Keywords: G proteins, multi-site phosphorylation, RhoE, ROCK-I, stress fibres
Introduction
Rho family G proteins, the functions of which are mediated by a variety of effector molecules, are critical regulators of cell motility, polarity, adhesion, cytoskeletal reorganisation, proliferation and apoptosis (Jaffe and Hall, 2005; Ridley, 2006). The related serine/threonine protein kinases ROCK-I (ROKβ/p160ROCK) and ROCK-II (ROKα/Rho-kinase) (Kimura et al, 1996; Leung et al, 1996; Amano et al, 1997, 2000; Riento and Ridley, 2003) were the first Rho effectors to be discovered, being initially characterised for their roles in mediating the formation of RhoA-induced stress fibres and focal adhesions. ROCK kinases elicit these effects through the phosphorylation of myosin light chain (MLC) and the MLC phosphatase subunit MYPT1, thereby enhancing acto-myosin contractility (Amano et al, 1996; Kimura et al, 1996).
RhoA-induced stress fibre and focal adhesion formation are inhibited by two atypical members of the Rho family, Rnd1 and RhoE (also termed Rnd3) (Guasch et al, 1998; Nobes et al, 1998; Nobes and Hall, 1999). The direct molecular targets of RhoE responsible for exerting these effects are unknown, but appear to be mediated through RhoE-induced antagonism of RhoA activation of ROCK-I-mediated signalling. This notion is supported by studies showing that RhoE's capacity to promote stress fibre disassembly and increased cell motility is associated with decreased MYPT1 phosphorylation, resembling the effects of small molecule inhibitors of ROCK kinases (Guasch et al, 1998; Riento et al, 2003). There is evidence that RhoE-mediated inhibition of RhoA signalling occurs either through inactivation of RhoA and/or regulation of the RhoA effector ROCK. RhoE, similarly to Rnd1, stimulates p190 RhoGAP to reduce intracellular levels of RhoA-GTP (Wennerberg et al, 2003). However, RhoE also binds ROCK-I directly, and is phosphorylated by the kinase on multiple sites within its N and C termini, increasing RhoE stability and altering its cellular localisation (Riento et al, 2005a). Growth factor-induced ROCK-I-mediated phosphorylation of RhoE Ser11 correlates with stress fibre disassembly, and its dephosphorylation coincides with their reappearance (Riento et al, 2005a).
In contrast to the canonical mode of G-protein activity and regulation, RhoE and the related Rnd1 and Rnd2 G proteins are defective as GTPases even in the presence of RhoGAPs (Foster et al, 1996; Guasch et al, 1998; Nobes et al, 1998; Nobes and Hall, 1999; Riento et al, 2005b; Chardin, 2006). Moreover, as Rnd1, Rnd2 and RhoE bind GTP 100-fold more tightly than GDP, in vivo they exist constitutively in the GTP-bound form (Foster et al, 1996). Regulation of these G proteins is exerted at the level of gene expression (Hansen et al, 2000; Riento et al, 2003; Ongusaha et al, 2006), phosphorylation and controlled protein degradation (Riento et al, 2005a).
To understand the molecular basis for ROCK-I-catalysed multi-site phosphorylation of RhoE, we have determined the crystal structure of RhoE in complex with the kinase/dimerisation domains of ROCK-I. RhoE interacts with the C-terminal lobe of the ROCK-I kinase domain, presenting its N and C termini in proximity to the kinase catalytic site facilitating their phosphorylation. The ROCK-I interaction sites of RhoE are remote from a so-called ‘effector' region, which mediates all small G-protein/effector interactions thus far characterised. The ROCK-I:RhoE structure therefore has implications for understanding how G proteins recognise proteins independent of their effector/switch regions. Significantly, although mutants of RhoE that disrupt ROCK-I binding are as effective as wild-type RhoE in inducing loss of stress fibres, mutations within the effector region of RhoE profoundly inhibit this response. These data indicate that RhoE mediates an indirect inhibitory effect on ROCK-I through effector site-mediated interactions with regulators of RhoA signalling.
Results
ROCK-I kinase domain forms a 1:1 complex with RhoE
A region of ROCK-I (residues 1–420) incorporating both the kinase (residues 70–404) and the N-terminal dimerisation domains (residues 1–69) (Doran et al, 2004; Jacobs et al, 2006) was defined earlier to be necessary and sufficient for direct RhoE interactions (Riento et al, 2003). Significantly, this region of ROCK-I is incapable of binding RhoA (Riento et al, 2003). To obtain protein boundaries optimised for crystallisation, guided by tryptic analysis and the published ROCK-I structure, we removed an additional 14 residues from the ROCK-I C terminus (ROCK-I1−406). This shorter ROCK-I fragment interacted with RhoE indistinguishably from ROCK-I1−420 (Supplementary Figure S1A), and microinjection of both ROCK-I proteins induced stress fibres in Swiss 3T3 cells (Supplementary Figure S1B). We co-crystallised ROCK-I1−406 in complex with RhoE-GTP/Mg2+ (residues 1–200) in the presence of AMP-PNP/Mg2+. The complex crystallised in space group P6522, and the structure was determined at 3.7 Å resolution. Diffraction data quality was compromised by a long cell axis and radiation damage. Moreover, only a few crystals diffracted to an intrinsic resolution limit of 3.7 Å. The structure was solved by molecular replacement using the previously determined structures of the ROCK-I kinase domain (Jacobs et al, 2006) and RhoE (Garavini et al, 2002) (Table I and Materials and methods). The asymmetric unit comprises a dimer of ROCK-I, with each kinase subunit engaging an individual RhoE molecule (Figure 1). Examination of molecular packing within the unit cell indicated that this inter-molecular interaction corresponds to the only significant ROCK-I:RhoE contact, and therefore represents the biologically relevant ROCK-I:RhoE interface (Supplementary Figure S2A). Application of strict NCS restraints during refinement, combined with an 80% solvent content, resulted in electron density maps of exceptional quality (Supplementary Figure S3A). Although not present in the individual search models, nucleotide density was discernible in every protein molecule (Supplementary Figure S3C and D), and these ligands were refined with full occupancy. In RhoE, electron density is well defined for residues Gln20 to Lys200, whereas in ROCK-I residues 6–404 are similarly well resolved. Calculation of simulated annealing OMIT maps indicated that the refined structure was free of model bias (Supplementary Figure S3B).
Table 1.
Data collection and refinement statistics
| ROCK1:RhoE complex | |||
|---|---|---|---|
| Data collection statistics | |||
| Beamline | ID29 ESRF | ||
| Wavelength (Å) | 1.000 | ||
| Space group | P6522 | ||
| Unit cell (Å) | a=152.48 | ||
| b=152.48 | |||
| c=531.27 | |||
| Overall | Inner shell | Outer shell | |
| Resolution (Å) | 131–3.70 | 131–11.7 | 3.90–3.70 |
| Observed reflections | 150 886 | 4549 | 22 440 |
| Unique reflections | 39 411 | 1326 | 5707 |
| Redundancy | 3.8 | 3.4 | 3.9 |
| Completeness (%) | 98.9 | 91.8 | 99.9 |
| Rmerge | 0.189 | 0.096 | 0.420 |
| 〈I/σI〉 | 8.4 | 20.7 | 3.0 |
| Refinement statistics | |||
| Reflections in test set | 809 | ||
| Rcryst | 0.237 | ||
| Rfree | 0.270 | ||
| Number of protein atoms | 9131 | ||
| Number of ligand atoms | 62 | ||
| Wilson B (Å2) | 75.0 | ||
| 〈B〉 protein (Å2) | 81.7 | ||
| 〈B〉 ligand (Å2) | 108.8 | ||
| RMSD from ideal geometry | |||
| Bond length (Å) | 0.008 | ||
| Bond angles (deg) | 1.306 | ||
| Ramachandran statistic (preferred/ allowed/outliers) | 885 (78%)/ 76 (15%)/ 79 (7%) | ||
| Values in brackets are for the highest resolution shell. All measured data were included in structure refinement. | |||
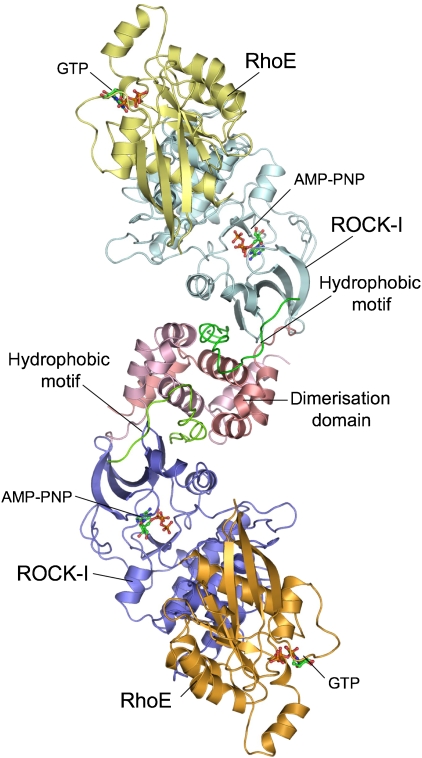
Figure 1.
Overview of ROCK-I:RhoE complex. Each subunit of the ROCK-I dimer binds one molecule of RhoE, forming a dyad-symmetrical molecule. The complex dimerises through the dimerisation domain (pink) of ROCK-I. Structural figures were created using PYMOL (www.pymol.org).
RhoE comprises a canonical GTP-binding domain with extended N and C termini relative to most other Rho family members (Figure 1). In the crystal structures of isolated RhoE, the N- and C-terminal regions (19 and 43 residues, respectively) were absent (Fiegen et al, 2002; Garavini et al, 2002). However, in our crystallised complex, the N-terminal residues of RhoE were included. Importantly, Ser7 and Ser11 of RhoE are sites of ROCK-I phosphorylation (Riento et al, 2005a).
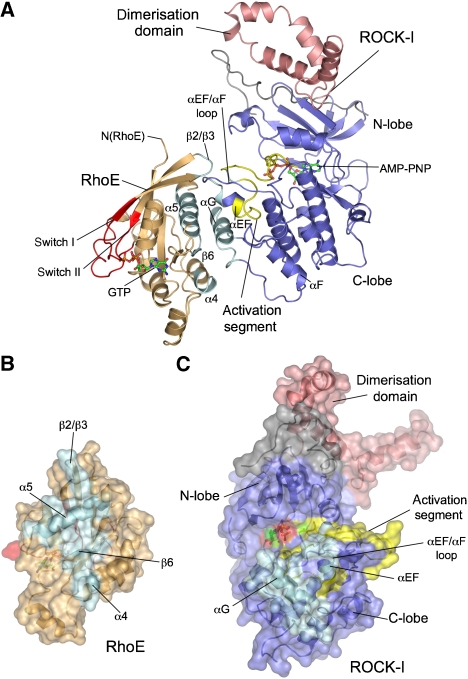
The N-terminal α-helical dimerisation domain of ROCK-I organises two kinase domains in a head-to-head configuration (Figure 1) (Jacobs et al, 2006; Yamaguchi et al, 2006). Accounting for its requirement for catalytic activity (Leung et al, 1996), the dimerisation domain incorporates and stabilises the hydrophobic motif located C-terminal to the kinase domain, a site of regulatory phosphorylation in diverse AGC family kinases (Pearl and Barford, 2002; Gold et al, 2006; Jacobs et al, 2006; Yamaguchi et al, 2006). Similarly to the isolated ROCK-I and -II structures (Jacobs et al, 2006; Yamaguchi et al, 2006), the kinase domain of ROCK-I in complex with RhoE adopts a conformation consistent with that of a catalytically active kinase (Johnson et al, 1996; Nolen et al, 2004) (Figure 2A).
Figure 2.
RhoE interacts through its α4–β6–α5 module with the C-lobe of the kinase. (A) View of the ROCK-I:RhoE complex, with secondary structural elements at the protein interface coloured cyan. Remainder of ROCK-I in blue and pink (dimerisation domain) and RhoE in orange with switch I and II coloured red. Surface representation of RhoE (B) and ROCK-I (C) rotated in opposite directions to (A) to reveal subunit interface. Colour coding as in (A).
RhoE interacts with the C-lobe of ROCK-I independent of its switch regions
The most striking feature of the ROCK-I:RhoE interface is its complete independence of the switch I/effector and switch II regions of RhoE, a mode of G-protein–protein recognition that contrasts with all previously known G-protein–effector complexes (Vetter and Wittinghofer, 2001). Regions of RhoE that interact with ROCK-I lie on the opposite surface of the molecule to the switch regions (Figure 2A). Interactions between RhoE and ROCK-I are dominated by the α4-β6-α5 module of RhoE that engages with the C-lobe of the kinase, centred on its αG helix. The kinase's N-lobe and dimerisation domain do not participate in RhoE interactions. Residues of RhoE and ROCK-I mediating these interactions are invariant in mammalian forms of both proteins (data not shown). The α4-β6-α5 module of RhoE defines a shallow channel, contacting the kinase domain such that the kinase αG helix is aligned parallel to β6, and antiparallel to the α4 and α5 helices of RhoE (Figure 2A). The protein–protein interface is further augmented by contacts between the tip of the β2/β3 hairpin of RhoE and the αEF/αF loop, preceding αEF helix, and neighbouring activation segment of the kinase. In total, around 2500 Å2 solvent accessible surface area is buried at the protein interface (Figure 2B and C).
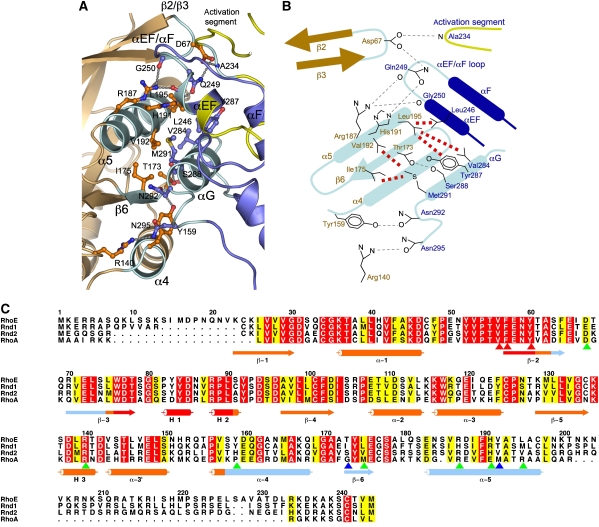
Interactions at the ROCK-I:RhoE interface are dominated by the α5 helix of RhoE and the αEF/αF loop and αG helix of ROCK-I (Figure 2A). As noted by Fiegen et al (2002), the α5 helix of RhoE is notably richer in nonpolar residues when compared with other small G proteins, and the contacts between α5 and the ROCK-I αG helix are entirely hydrophobic (Figure 3A and B). In ROCK-I, a cluster of exposed residues extending a hydrophobic ridge on the αG helix (Val284, Tyr287 and Met291), together with the side chain of Leu246 of the αEF helix, creates a contiguous hydrophobic surface to engage complementary nonpolar residues (Val192 and Leu195) of the RhoE α5 helix. Additional contacts contributed by the α5 helix include the imidazole side chain of His191 donating a hydrogen bond to the amide side chain of Gln249 (αEF helix) and, promoted through a shift of ∼5 Å, the guanidinium side chain of Arg187 of α5 donates hydrogen bonds to main-chain carbonyls at the C terminus of the αEF helix. The β6 strand of RhoE contacts the kinase αG helix through hydrogen bonds between Thr173 (β6) and Ser288 (αG), whereas the side-chain hydroxyl group of Tyr159 at the N terminus of α4 of RhoE donates a hydrogen bond to the amide side chain of Asn292 at the C terminus of αG. Finally, from the β2/β3 loop of RhoE, the Asp67 side chain accepts hydrogen bonds from the amide side chain of Gln249 (αEF/αF loop) and main-chain amide of Ala234 of the activation segment, the only contact between RhoE and the kinase activation segment. Gln249 becomes available for interactions with both Asp67 (β2/β3 loop) and His191 (α5 helix) of RhoE following the conformational shift of the αEF/αF loop (see below).
Figure 3.
Details of ROCK-I:RhoE interface. (A) View of interface formed by the α4–β6–α5 module of RhoE to the αG and αEF helices, αEF/αF loop and activation segment of ROCK-I. Colour scheme as in Figure 2. (B) Schematic of interface shown in (A). Hydrogen bonds and van der Waals interactions are indicated by grey and red dashed lines, respectively. ROCK-I and RhoE residue labels are in blue and orange. (C) Multiple sequence alignment of RhoE, Rnd1, Rnd2 and RhoA. Residues contacting ROCK-I are indicated by blue and green arrowheads, with blue arrowheads showing mutated residues (Thr173, Val192). Red arrowheads denote residues of the effector region required for RhoE function. Figure was produced using ALSCRIPT (Barton, 1993).
Notably, only RhoE, but not Rnd1 or Rnd2, interacts with ROCK-I (Riento et al, 2005a). Residues of RhoE that contact ROCK-I are poorly conserved in Rnd1 and Rnd2, explaining their inability to bind ROCK-I (Figure 3C, blue and green arrowheads). ROCK-II phosphorylates RhoE only weakly, and direct interactions between ROCK-II and RhoE could not be detected (Riento et al, 2005a). The regions of ROCK-I that interact with RhoE are highly conserved in ROCK-II, with only a conservative substitution of an Asp for Asn292 (see below). The side chain of Asn292, which contacts Tyr159 of α4 (Figure 3A and B), is also within 3–4 Å of the main-chain carbonyl groups of Tyr174 and Glu176 of RhoE (β6), and it is therefore possible that the repulsive electrostatic interaction conferred by the negative charge of an Asp side chain would destabilise interactions between the two proteins.
ROCK-I:RhoE interactions are facilitated through localised conformational changes of both proteins
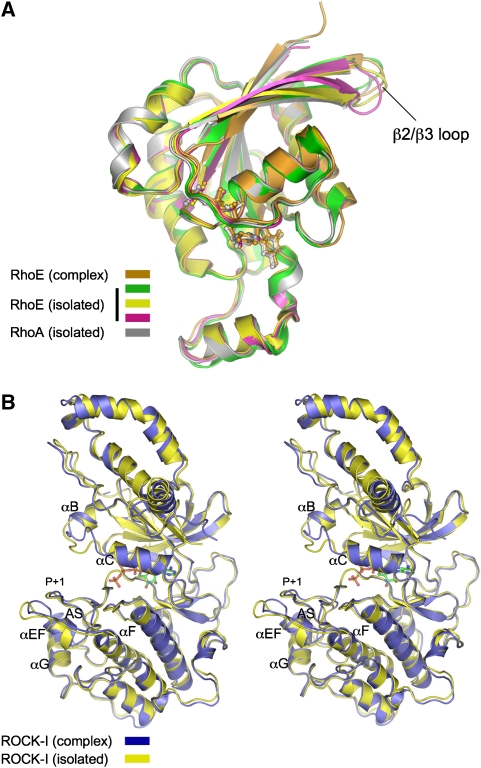
Specific individual structural elements of ROCK-I and RhoE that participate at the interface of the complex differ in conformation to their isolated counterparts (Figure 4). Although the overall conformation of RhoE in association with ROCK-I is virtually identical to that in isolation, notably in the vicinity of the GTP-binding site (overall RMSD of 0.6 Å between equivalent Cα atoms to RhoE of PDB code 1m7b), the tip of the β2/β3 loop, which contacts the αEF/αF loop of ROCK-I, is shifted relative to all isolated RhoE structures (Figure 4A). Conformational changes of ROCK-I are mainly confined to the N terminus of the αEF/αF loop, together with accompanying small shifts of the P+1 loop of the activation segment (Figure 4B). The αEF/αF loop shifts ∼5 Å relative to the isolated ROCK-I structure (Jacobs et al, 2006), with the new position well resolved in the electron density maps. Facilitating its interaction with RhoE, the αEF/αF loop encompasses a four-residue insertion (Gly251–Tyr254) unique to ROCKs, which protrudes from the kinase domain, buttressing the overlying activation segment N-terminal to the P+1 loop (Jacobs et al, 2006). Another difference between the two ROCK-I structures involves the αB/αC loop of the N-lobe (Figure 4B). The conformation of this loop in the ROCK-I:RhoE complex is virtually identical to its counterparts in the related kinase ROCK-II (Yamaguchi et al, 2006) (data not shown). However, in the isolated ROCK-I structure (Jacobs et al, 2006), its participation in crystal packing interactions most likely accounts for its non-canonical conformation.
Figure 4.
Conformational changes of RhoE and ROCK-I on forming a complex are confined to the ROCK-I:RhoE interface. (A) RhoE with comparison to isolated RhoE (Fiegen et al, 2002; Garavini et al, 2002) and RhoA structures (Ihara et al, 1998) and (B) stereoview of ROCK-I, isolated ROCK-I from Jacobs et al (2006).
RhoE as a substrate of ROCK-I
The interaction of RhoE with ROCK-I positions RhoE to present both its N and C termini in close proximity to the kinase catalytic site cleft (Figure 5A). The visible N and C termini of RhoE (residues 20 and 200, respectively) are located some 20 Å from the kinase phosphoacceptor-binding site. Model building studies, extending the polypeptide chain N-terminal from Gln20, and with reference to the position of the GSK3β peptide bound to PKBβ (Yang et al, 2002), indicate that both N-terminal phosphorylation sites, Ser7 and Ser11, are within reach of the catalytic site (Figure 5A). Similarly, the five phosphorylation sites within the C terminus are also all capable of accessing the catalytic site. Although contiguous electron density bridging Gln20 of RhoE with the kinase catalytic cleft is not observed, electron density (maps contoured at 3σ) corresponding to the main-chain of the phosphoacceptor and adjacent P−1 and P−2 residues of a peptide substrate modelled on GSK3β bound to PKBβ (Yang et al, 2002) is visible in molecule A of the ROCK-I:RhoE complex (Figure 5B). This electron density is consistent with partial, and possibly alternate, occupancy of one or both phosphoacceptor residues (Ser7 and Ser11) at the kinase catalytic cleft. The relatively poorly resolved electron density at the catalytic site of ROCK-I is not due to a disrupted catalytic site or hindered access, as the structure of ROCK-I appears compatible with catalysis and protein substrate recognition. Supporting our conclusions, in a crystal structure of ROCK-I with RhoE lacking its N terminus, which we determined at the same resolution in the same space group, no electron density is observed at the phosphoacceptor site (data not shown).
Figure 5.
RhoE phosphorylation by ROCK-I. (A) View of the ROCK-I:RhoE dimer with a substrate peptide based on GSK3β bound to PKB (magenta) (Yang et al, 2002), modelled into the catalytic cleft of ROCK-I. The distance between the visible N and C termini of RhoE to the phosphoacceptor site in ROCK-I is 20 Å. Colour scheme as in Figure 2. (B) Details of the catalytic site of ROCK-I, Fo–Fc electron density (contoured at 3σ in green) overlapping a GSK3β peptide bound to PKB modelled into the ROCK-I catalytic site.
Substrates of ROCK-I typically comprise basic residues at P−2 and/or P−3 (Amano et al, 1996; Kawano et al, 1999; Hagerty et al, 2007), and all seven RhoE phosphorylation sites conform to this consensus sequence (Supplementary Figure S4A). However, relative to ROCK-I phosphorylation sites in MYPT1 and ZIPK (Amano et al, 1996; Kawano et al, 1999; Hagerty et al, 2007) (Supplementary Figure S4B), RhoE phosphorylation sites comprise fewer basic residues, potentially attenuating their affinity for the ROCK-I catalytic site. The notion that optimised phosphorylation sites of ROCK-I may require numerous N-terminal basic residues is supported by analogy to the basophilic protein kinases PKA and PKB, the substrates of which contain multiple basic residues at sites P−2 to P−5 (Kennelly and Krebs, 1991; Alessi et al, 1996), and which, in common with ROCK-I, feature acidic residues lining the N-terminal region of their substrate-binding clefts.
Kinase G helix is a common module for substrate and inhibitor recognition
The αG helix is a relatively exposed structural feature of the C-lobe of protein kinases, and together with the proximal αEF helix and αEF/αF loop, protrudes from the main body of the molecule. In our ROCK-I:RhoE structure, the αG helix dominates interactions with RhoE, inserting into the shallow channel created by the RhoE α4–β6–α5 module, facilitating a mode of kinase–protein interaction also observed in other complexes (Supplementary Figure S5). The structure of the ROCK-I:RhoE complex is strikingly reminiscent of the protein substrate complex formed between the RNA-dependent protein kinase PKR and its substrate, eIF2α (Dar et al, 2005) (Supplementary Figure S5A). As observed in our ROCK-I:RhoE complex, eIF2α bound to PKR is disposed such that its phosphoacceptor site is accessible to the kinase catalytic site, although the site of eIF2α phosphorylated by PKR is not resolved within the kinase catalytic site (Dar et al, 2005). In CDK2, the αG helix, together with the C-terminal segment of αEF, mediates the major site of interaction with its protein phosphatase KAP, positioning the phosphatase catalytic site optimally to contact and dephosphorylate pThr160 of the CDK2 activation segment (Song et al, 2001) (Supplementary Figure S5B). Lastly, in the PKA holoenzyme complex, the cAMP-binding domain CBD-A of the regulatory RIα subunit contacts both the αG and αEF helices of the catalytic subunit to engage its inhibitor segment at the catalytic site cleft in a mode reminiscent of a peptide substrate (Kim et al, 2005) (Supplementary Figure S5C). Notably, a common feature of these complexes is that protein interactions facilitated through a secondary site of the kinase—the αG and αEF helices—optimise contacts with the primary site of the activation segment, important for substrate recognition (Dar et al, 2005), pseudo-substrate inhibition by regulatory subunits (Kim et al, 2005) and phosphatase catalysed dephosphorylation of the activation segment (Song et al, 2001).
RhoE binds to ROCK-I through a distinctive mechanism
In contrast to other G-protein:effector complexes, interactions between RhoE and the kinase domain of ROCK-I are independent of switches I and II (Vetter and Wittinghofer, 2001), which are consequently accessible for potential interactions with other domains of ROCK-I, possibly with sites within the coiled-coil region, and/or with other proteins. Interestingly, the involvement of the α4–β6–α5 module of a G protein as observed in the ROCK-I:RhoE complex, is reminiscent of how Ran contacts the acidic loop of importin/karyopherinβ (Chook and Blobel, 1999; Vetter et al, 1999). In the karyopherinβ:Ran complex (Chook and Blobel, 1999), a portion of the acidic loop, a 60-residue insertion within HEAT repeat 7, adopts a 310 helical conformation lying antiparallel to the β6 strand of Ran within its α4–β6–α5 channel. However, karyopherinβ also interacts with the effector region of Ran, and not exclusively through the α4–β6–α5 channel.
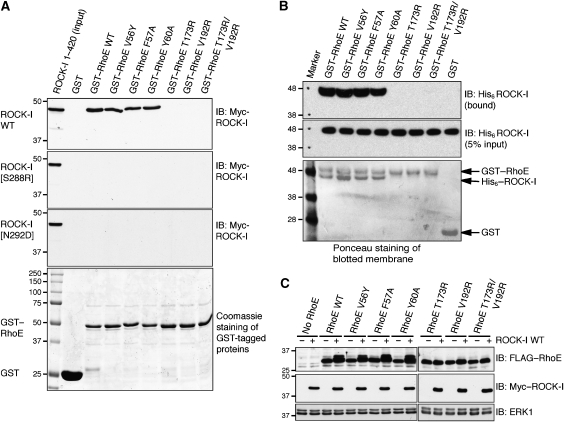
Mutagenesis of the ROCK-I:RhoE interface prevents binding and phosphorylation of RhoE by ROCK-I
To test whether disruption of the ROCK-I:RhoE interface affects RhoE interaction with ROCK-I, RhoE amino acids Thr173 and Val192 of the β6 strand and α5 helix, respectively, were mutated to arginines. Although recombinant glutathione-S-transferase (GST)–RhoEWT interacted with ROCK-I1−420 from COS-7 cell lysates, GST–RhoET173R and RhoEV192R, as well as the double mutant RhoET173R/V192R, were unable to pull down ROCK-I1−420 (Figure 6A) or to interact with purified ROCK-I1−406 in vitro (Figure 6B), confirming the importance of these residues for the interaction of RhoE with ROCK-I. The interaction between RhoEWT and ROCK-I1−420 occurred in the absence of additional AMP-PNP. Furthermore, in contrast to RhoEWT, these mutants were not phosphorylated when co-expressed with ROCK-I1−420 in COS-7 cells (Figure 6C). However, mutation of effector site residues (Val56, Phe57 and Tyr60) did not inhibit binding to ROCK-I1−420 (Figure 6A and B) or ROCK-I phosphorylation of RhoE in cells (Figure 6C), consistent with the lack of involvement of these effector residues in ROCK-I interactions. Mutation of Ser288 to Arg and Asn292 to Asp in the ROCK-I αG helix prevented interaction with wild-type RhoE (Figure 6A) and these mutants were unable to phosphorylate RhoE in cells (Supplementary Figure S6).
Figure 6.
Disruption of ROCK-I:RhoE interaction and effects on RhoE/ROCK-I responses. (A) The indicated wild-type (WT) and mutant GST–RhoE proteins on glutathione beads were incubated with cell lysates from COS-7 cells transfected with pCAG-myc-ROCK-I1−420 (wild-type and indicated mutants). Proteins that bound to GST–RhoE were resolved by SDS–PAGE and western blotted with anti-myc antibody to identify myc–ROCK-I. The lower panel (Coomassie staining of polyacrylamide gel) shows that equal amounts of each GST–RhoE mutant were used in the binding assay. (B) GST–RhoE was incubated with His-ROCK-I purified from baculovirus-infected insect cells. ROCK-I bound to glutathione agarose immobilised RhoE was identified by western blotting with anti-His antibody. (C) ROCK-I phosphorylation of RhoE in cells. COS-7 cells were co-transfected with the indicated FLAG–RhoE and myc–ROCK-I1−420 expression vectors. After 24 h, cells were lysed and cell lysates were western blotted with anti-FLAG antibody to detect RhoE. ROCK-I-phosphorylated RhoE migrates more slowly. ERK1 panels indicate equivalent loading in each lane.
ROCK-I:RhoE interaction is not required for RhoE-induced loss of stress fibres
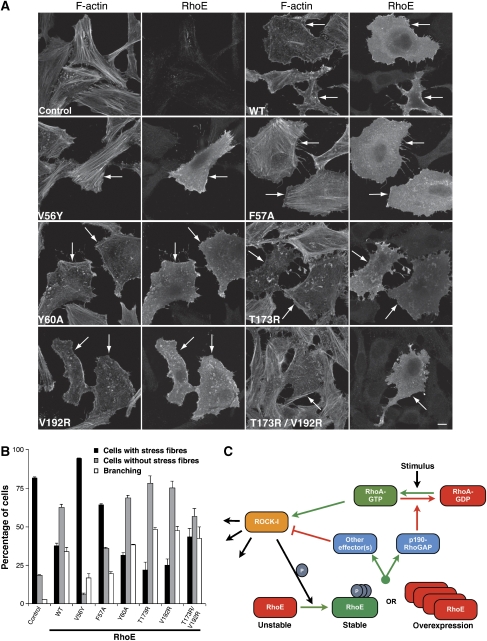
To investigate whether RhoE interaction with ROCK-I is necessary for its ability to alter actin organisation in cells, RhoE mutants were expressed in HeLa and Swiss 3T3 cells. Significantly, the RhoE mutants RhoET173R and RhoEV192R, as well as the double mutant RhoET173R/V192R, induced similar responses to wild-type RhoE in both cell types (Figure 7A and B; Supplementary Figure S7A). All of these mutants induced a strong reduction of stress fibres in the majority of HeLa cells, and a rounded phenotype with branched protrusions (branching) in 30–40% of cells (Figure 7A and B). Cells with a branched phenotype also lacked stress fibres, and this phenotype was generally induced at higher levels of RhoE. In contrast, RhoE effector domain mutants RhoEV56Y and RhoEF57A were defective in their ability to stimulate loss of stress fibres and cell rounding, and in some cells increased stress fibre levels, suggesting that they could function as dominant-negative RhoE mutants (Figure 7A and B). Interestingly, the RhoEY60A mutant had a similar phenotype to wild-type RhoE, indicating that this amino acid (equivalent to Tyr40 in Ras and Rac) is not necessary for RhoE-induced actin reorganisation. Similarly, Tyr40 in Rac is not required for changes to the actin cytoskeleton (Lamarche et al, 1996). The localisation of RhoE was not detectably affected by the different mutations (Figure 7A). These results indicate that binding of a protein to the RhoE effector region is important for RhoE-induced loss of stress fibres, rather than direct binding to ROCK-I, which is independent of the effector site.
Figure 7.
Effect of RhoE mutants on the actin cytoskeleton. HeLa cells were transfected with the indicated FLAG–RhoE wild-type (WT) and mutant constructs, or with empty vector (control). After 24 h, cells were fixed and stained for actin filaments and with anti-FLAG antibody. (A) Representative images of transfected cells. Left panels of each pair of images show actin filaments (F-actin); cells expressing transfected FLAG epitope-tagged RhoE are indicated with arrows, and were identified by staining with anti-FLAG antibody (right panels of each pair, labelled ‘RhoE'). Note that cells expressing transfected RhoE have much fewer stress fibres than surrounding untransfected cells, except for cells expressing V56Y and F57A mutants. (B) Cells expressing transfected RhoE were scored for loss of stress fibres (compared with surrounding untransfected cells) and for rounded, ‘branching' phenotype; 100 FLAG antibody-stained cells were counted on each of three coverslips; mean % of cells in each category ±s.e.m. is shown. Note that the ‘branching' phenotype is a stronger response to RhoE than loss of stress fibres. (C) Model for RhoE and ROCK-I regulation. RhoE exerts negative feedback regulation of ROCK-I. Stabilised RhoE (through RhoA-activated ROCK-I phosphorylation or RhoE over-expression) antagonises RhoA-stimulation of ROCK-I through either activation of p190 RhoGAP or through regulation of an alternative RhoA effector.
It is possible that other ROCK-I substrates employ a similar interface as RhoE to bind the kinase. To test this possibility, we expressed ROCK-I αG helix mutants in Swiss 3T3 cells. Both ROCK-IS288R (1–420) and ROCK-IN292D (1–420) induce stress fibre bundling to form stellate arrays of stress fibres, similar to wild-type ROCK-I (1–420) (Supplementary Figure S7B). The αG helix is therefore not essential for ROCK-I binding to substrates required for acto-myosin contractility, such as MYPT1 and MLC.
Discussion
The ROCK-I:RhoE structure provides insights into mechanisms of multi-site phosphorylation of an individual substrate by a specific protein kinase. Consistent with the crystal structure, mutations of RhoE within β6 or α5 that comprise the ROCK-I-binding site abolished interactions with ROCK-I in vitro. Furthermore, the same mutations, and those within the αG helix of ROCK-I, abrogated ROCK-I-catalysed RhoE phosphorylation in vivo. Interestingly, one of the ROCK-I mutations defective for RhoE binding, a substitution of Asp for Asn292, converts the RhoE-binding site of ROCK-I to that of ROCK-II, demonstrating that this single amino-acid difference is sufficient to prevent ROCK-II interactions with, and subsequent ROCK-II-mediated phosphorylation of RhoE. These findings indicate that the formation of a complex between ROCK-I and RhoE involving the kinase αG helix, representing a secondary site of interaction, is a prerequisite for ROCK-I-catalysed phosphorylation of RhoE. This suggests that the ROCK-I phosphorylation sites within the N and C termini of RhoE are only presented to the kinase catalytic site in the context of an interaction between the two proteins mediated by the αG helix. We predict therefore that each of the seven RhoE phosphorylation sites individually is poorly optimised for ROCK-I phosphorylation. Within the context of the crystal structure, the two ROCK-I phosphorylation sites present in RhoE (Ser7 and Ser11) are likely interchanging at the catalytic (primary binding) site, with resultant low substrate occupancy and consequently disordered and poorly resolved electron density. Conceivably, where a protein substrate is presented to the catalytic site of its cognate kinase through interactions to a secondary site, the context of the phosphoacceptor site deviates from that required to optimise phosphorylation of a short peptide sequence or where no secondary site contributes to the phosphorylation reaction. Studies by Haystead and colleagues have indicated that peptides modelled on ROCK-I phosphorylation sites of MYPT1 and ZIPK are efficiently phosphorylated by the kinase (Hagerty et al, 2007). In contrast to RhoE phosphorylation sites, these optimised substrates feature multiple basic residues N-terminal to the phosphoacceptor site (Supplementary Figure S4B).
Phosphorylation of suboptimal sites by a kinase presented with a substrate bound through a secondary binding site is reminiscent of phosphorylation in trans of non-consensus sites within activation segments (Oliver et al, 2007; Wu et al, 2008). As mutations of the αG helix of ROCK-I do not prevent MYPT1 phosphorylation as inferred from their ability to induce stress fibre formation and cell contraction, and that MYPT1 is also a ROCK-II substrate, it is likely that MYPT1 does not utilise a secondary ROCK-I-binding site centred on the αG helix.
The structure of the ROCK-I:RhoE complex provides a molecular framework for testing the role of direct RhoE:ROCK-I interactions in mediating RhoE-dependent stress fibre disassembly. Significantly, those mutations of RhoE that disrupt ROCK-I interactions have no effect on its ability to disrupt actin stress fibres in vivo. In contrast, however, mutations within the effector region of RhoE prevent loss of stress fibres without affecting ROCK-I-mediated RhoE phosphorylation. These data clearly demonstrate that the ability of RhoE to induce loss of stress fibres is independent of its capacity to bind ROCK-I directly, suggesting that RhoE either negatively regulates ROCKs through upstream effectors, or interacts with other proteins to induce loss of stress fibres. The finding that high-level expression of RhoE inhibits a constitutively active form of ROCK-I (Riento et al, 2003) is likely explained by substrate-level inhibition.
A potential candidate for a RhoE target is p190 RhoGAP. In an earlier study, RhoE was found to stimulate p190 RhoGAP activity, reducing cellular RhoA-GTP. An effector mutant of RhoE (Thr55Ala) defective for p190 RhoGAP binding, was unable to induce stress fibre loss (Wennerberg et al, 2003). However, in contrast to these findings, overexpression of RhoE in Swiss 3T3 cells did not reduce RhoA-GTP levels (Villalonga et al, 2004), indicating that an alternative RhoE effector could be responsible for the inhibition of ROCK-I signalling. One possibility is Socius, which binds to both Rnd1 and RhoE and induces loss of stress fibres (Katoh et al, 2002).
Our results indicate that the primary function of the ROCK-I:RhoE interaction is to allow RhoE phosphorylation on multiple sites. ROCK-I-catalysed phosphorylation of RhoE stabilises the protein and correlates with disruption of actin stress fibres (Riento et al, 2005a). Stabilisation of RhoE through ROCK-I activation might enable efficient stimulation of a RhoE effector resulting in a negative feedback loop in vivo that then inhibits ROCK-I. Under conditions of RhoE overexpression, such as in the p53-mediated DNA damage response, or in transfection and microinjection experiments, RhoE levels and stability are presumably sufficient to activate a RhoE effector and induce loss of stress fibres without phosphorylation by ROCK-I (Figure 7C).
In conclusion, the structure of a complex of RhoE bound to ROCK-I, together with mutagenesis studies, has revealed that the main function of the ROCK-I:RhoE interaction is to mediate multi-site phosphorylation of RhoE. Multi-site phosphorylation is characterised by suboptimal phosphoacceptor sites engaged at the primary catalytic site with affinity and specificity for the protein substrate augmented by a remote secondary interaction site.
Materials and methods
Cloning and mutagenesis
pCAG-myc-ROCK-I1−420 and pCMV-FLAG-RhoE have been described earlier (Riento et al, 2003). ROCK-I1−406 was cloned with an N-terminal His tag into the pFastBac system for the generation of baculovirus, and RhoE1−200 were cloned into the pET28 vector for bacterial expression. GST-tagged versions of RhoE generated using the pGEX6 and pGEX4 vectors (GE Healthcare). Mutagenesis on ROCK-I and RhoE was performed using the Quikchange system (Stratagene).
Protein expression, purification and crystallisation
His-tagged ROCK-I1−406 was produced using baculovirus-mediated insect cell expression in Sf9 cells, with 72 h of infection at an MOI of 2. A cell pellet from 2.5 l cells was lysed in lysis buffer A (300 mM NaCl, 50 mM Tris (pH 8.0), 15 mM imidazole, Complete protease inhibitor) and after centrifugation (40 min, 20 000 g) was applied to 15 ml TALON resin. The His-tagged protein was eluted using lysis buffer supplemented with 150 mM imidazole at pH 7, and the His tag was cleaved using 100 μg His-TEV protease overnight during dialysis into buffer A (100 mM NaCl, 25 mM Tris (pH 7.5) and 1 mM DTT). ROCK-I was further purified by anion exchange and gel filtration in buffer B (200 mM NaCl, 20 mM Tris (pH 7.5) and 1 mM DTT). RhoE1−200 was expressed in Escherichia coli strain BL21 with an N-terminal His6 tag, and purified according to Garavini et al (2002).
RhoE was mixed with ROCK-I in a 3:1 molar ratio, 5 mM AMP-PNP and 10 mM MgCl2 was added, the mixture was concentrated to 6 mg/ml, and used in crystallisation screening. Hexagonal plate crystals were obtained from 0.6 to 1 M Na/K tartrate, 0.1 M MES (pH 5–6) and 0.2 M LiSO4. Prior to data collection, crystals were soaked briefly in mother liquor containing 20% (v/v) ethylene glycol, and frozen.
Data collection, structure solution and refinement
Diffraction data for the ROCK-I:RhoE complex were collected at the ESRF beamline ID29. The crystals displayed a hexagonal lattice (point group P622) and diffracted to a maximum of 3.7 Å with only few crystals showing useable diffraction. Owing to a long cell axis (c=532 Å), oscillation angles of 0.25° were collected. Indexing of different crystals, where possible, revealed large deviations in the long cell axis, eliminating the possibility of merging datasets from different crystals. Furthermore, crystals suffered from radiation damage during data collection. The structure was solved by molecular replacement in PHASER (McCoy et al, 2007), using the structure of ROCK-I (pdb-id 2etr) and the structure of RhoE (1gwn) as search models. Despite the large unit cell, PHASER identified just two copies of both ROCK-I and RhoE, and unambiguously determined the space group as P6522. Inspection of the packing and the resulting map verified the solution; this determines a Matthews coefficient of 6.3 (80.5% solvent content), possibly explaining the observed intrinsic resolution limit of the crystals. Molecular replacement statistics are listed in Supplementary Figure S2B. Calculations of electron density maps using phases based on the ROCK-I model alone, revealed additional electron density adjacent to the ROCK-I αG helix consistent with the position of RhoE determined using molecular replacement (Supplementary Figure S2C).
The structure was refined with PHENIX (Adams et al, 2002), initially using simulated annealing to overcome model bias. Non-crystallographic symmetry restraints were applied on chains of ROCK-I and RhoE individually. The maps are of exceptional quality for the given resolution, and several loops that were initially removed could be rebuilt, and additional residues lacking in the search models could be included in subsequent rounds of model building using COOT (Emsley and Cowtan, 2004). Electron density was present for AMP-PNP/Mg2+ in the kinase domain and GTP/Mg2+ bound to RhoE, and both ligands were refined with full occupancy (Supplementary Figure S2C and S2D). The models of ROCK-I lack interpretable electron density for residues 1–5 and 372–377 (molecule A) or 372–374 (molecule C). RhoE residues 1–19 were missing in the electron density (see Results). Complete data collection and refinement statistics can be found in Table I.
COS-7 cell transfections and GST–RhoE pull downs
COS-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS). For transfection with constructs encoding myc epitope-tagged ROCK-I and/or FLAG epitope-tagged RhoE, cells were washed with 5 ml of cold electroporation buffer (120 mM KCl, 10 mM K2PO4·KH2PO4 (pH 7.6), 25 mM HEPES (pH 7.6), 2 mM MgCl2 and 0.5% Ficoll). The buffer was removed and cells were resuspended in 250 μl of cold electroporation buffer and electroporated at 250 V and 960 mF with 5 μg of DNA. Cells were then incubated for 24 h prior to lysis.
Recombinant RhoE wild-type and RhoE point mutants were expressed as GST fusion proteins in E. coli and purified as described earlier (Riento et al, 2003). Recombinant GST was used as a control. Transfected COS-7 cells were washed and lysed in lysis buffer (1% Triton X-100, 20 mM Tris–HCl (pH 8), 130 mM NaCl, 10 mM NaF, 1% aprotonin, 10 mg/ml leupeptin, 1 mM DTT, 0.1 mM Na3VO4 and 1 mM PMSF). Insoluble material was removed by centrifugation and the clarified cell lysates were incubated with rotation for 2 h at 4°C with the recombinant GST fusion proteins on glutathione-sepharose beads (30 μl). Beads were washed extensively with lysis buffer and then the proteins were eluted in Laemmli sample buffer. Western blotting to indicate the presence of bound ROCK-I was performed as described in Garg et al (2008).
In vitro pull-down experiments
GST-tagged RhoE proteins were expressed in E. coli BL21 cells as for crystallisation. GST–RhoE was purified by affinity purification (GSH sepharose 4B; GE Healthcare) in lysis buffer B (270 mM sucrose, 150 mM NaCl, 50 mM Tris (pH 8), 1 mM EGTA, 10 mM MgCl2 and 10 mM β-mercaptoethanol). The resin was washed extensively, and GST-tagged RhoE proteins were eluted with lysis buffer B plus 30 mM glutathione.
For pull-down experiments, 30 μg of GST–RhoE was incubated with 30 μl GSH-sepharose 4B equilibrated in pull-down buffer (PDB: 150 mM NaCl, 50 mM Tris (pH 7.6), 0.1 % NP-40 and 5 mM DTT) supplemented with 1 mg/ml bovine serum albumin (BSA). After 1 h incubation at 4°C with agitation, the resin was washed 4 × with PDB, and incubated with PDB+BSA with 0.1 mg/ml His6-ROCK-I (purified for crystallisation without cleavage of the His6 tag) overnight at 4°C. The resin was then washed six times with PDB, mixed with SDS sample buffer and resolved on SDS–PAGE. Western blotting with αHis-HRP (Sigma-Aldrich) was used to reveal RhoE-bound ROCK-I.
Analysis of actin cytoskeleton
HeLa and Swiss 3T3 cells were grown in DMEM containing 10% FCS. For transfection, 6 × 105 HeLa cells were seeded onto 60-mm Petri dishes and plasmids encoding FLAG epitope-tagged RhoE wild-type or point mutants (2.5 μg) were transfected in serum-free OptiMEM medium using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Fresh serum-containing medium was added after 4 h. After transfection, cells were incubated for 24 h. For microinjection, Swiss 3T3 cells were seeded on coverslips at 3 × 104 cells/ml. Cells were microinjected with 100 ng/μl of FLAG–RhoE or myc–ROCK-I expression vectors. After microinjection, cells were incubated for 6 h to allow expression of proteins. HeLa and Swiss 3T3 cells were then fixed with 3.7% formaldehyde solution, permeabilised with 0.2% Triton X-100 in PBS and stained for actin filaments (F-actin) with 0.1 μg/ml TRITC-labeled phalloidin (Sigma-Aldrich) and for RhoE or ROCK-I expression with rabbit anti-FLAG antibody (Sigma) or mouse anti-myc (9E10) antibody (Santa Cruz Biotechnology), followed by fluorescein isothiocyanate-conjugated goat anti-mouse or goat–anti-rabbit antibodies (Jackson Immunoresearch). Coverslips were mounted in Dako mounting fluid (Dako Cytometrics) and images were generated with a Zeiss LSM Exciter or LSM510 confocal microscope using a × 40/1.3 NA objective. To quantify RhoE-induced loss of stress fibres and acquisition of a rounded ‘branching' phenotype in HeLa cells, 10 random images of cells were generated by confocal microscopy from each of three coverslips, and FLAG antibody-stained cells were scored for loss of stress fibres (compared with surrounding uninjected cells) and for ‘branching' phenotype. Note that cells with the ‘branching' phenotype were not included in the ‘no stress fibre' category.
Supplementary Material
Supplementary Figures
Acknowledgments
This study was funded by grants from CR-UK (DB and AJR), BBSRC (AJR) and a Beit Memorial Fellowship for Medical Research to DK. Robert Hussey is acknowledged for technical assistance. The ESRF (ID29), Grenoble, is acknowledged for crystallographic data collection. Protein coordinates deposited with PDB code: 2v55.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P (1996) Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett 399: 333–338 [DOI] [PubMed] [Google Scholar]
- Amano M, Chihara K, Kimura K, Fukata Y, Nakamura N, Matsuura Y, Kaibuchi K (1997) Formation of actin stress fibers and focal adhesions enhanced by Rho-kinase. Science 275: 1308–1311 [DOI] [PubMed] [Google Scholar]
- Amano M, Fukata Y, Kaibuchi K (2000) Regulation and functions of Rho-associated kinase. Exp Cell Res 261: 44–51 [DOI] [PubMed] [Google Scholar]
- Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K (1996) Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J Biol Chem 271: 20246–20249 [DOI] [PubMed] [Google Scholar]
- Barton GJ (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng 6: 37–40 [DOI] [PubMed] [Google Scholar]
- Chardin P (2006) Function and regulation of Rnd proteins. Nat Rev Mol Cell Biol 7: 54–62 [DOI] [PubMed] [Google Scholar]
- Chook YM, Blobel G (1999) Structure of the nuclear transport complex karyopherin-beta2-Ran × GppNHp. Nature 399: 230–237 [DOI] [PubMed] [Google Scholar]
- Dar AC, Dever TE, Sicheri F (2005) Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell 122: 887–900 [DOI] [PubMed] [Google Scholar]
- Doran JD, Liu X, Taslimi P, Saadat A, Fox T (2004) New insights into the structure–function relationships of Rho-associated kinase: a thermodynamic and hydrodynamic study of the dimer-to-monomer transition and its kinetic implications. Biochem J 384: 255–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fiegen D, Blumenstein L, Stege P, Vetter IR, Ahmadian MR (2002) Crystal structure of Rnd3/RhoE: functional implications. FEBS Lett 525: 100–104 [DOI] [PubMed] [Google Scholar]
- Foster R, Hu KQ, Lu Y, Nolan KM, Thissen J, Settleman J (1996) Identification of a novel human Rho protein with unusual properties: GTPase deficiency and in vivo farnesylation. Mol Cell Biol 16: 2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavini H, Riento K, Phelan JP, McAlister MS, Ridley AJ, Keep NH (2002) Crystal structure of the core domain of RhoE/Rnd3: a constitutively activated small G protein. Biochemistry 41: 6303–6310 [DOI] [PubMed] [Google Scholar]
- Garg R, Riento K, Keep N, Morris JD, Ridley AJ (2008) N-terminus-mediated dimerization of ROCK-I is required for RhoE binding and actin reorganization. Biochem J 411: 407–414 [DOI] [PubMed] [Google Scholar]
- Gold MG, Barford D, Komander D (2006) Lining the pockets of kinases and phosphatases. Curr Opin Struct Biol 16: 693–701 [DOI] [PubMed] [Google Scholar]
- Guasch RM, Scambler P, Jones GE, Ridley AJ (1998) RhoE regulates actin cytoskeleton organization and cell migration. Mol Cell Biol 18: 4761–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerty L, Weitzel DH, Chambers J, Fortner CN, Brush MH, Loiselle D, Hosoya H, Haystead TA (2007) ROCK1 phosphorylates and activates zipper-interacting protein kinase. J Biol Chem 282: 4884–4893 [DOI] [PubMed] [Google Scholar]
- Hansen SH, Zegers MM, Woodrow M, Rodriguez-Viciana P, Chardin P, Mostov KE, McMahon M (2000) Induced expression of Rnd3 is associated with transformation of polarized epithelial cells by the Raf-MEK-extracellular signal-regulated kinase pathway. Mol Cell Biol 20: 9364–9375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara K, Muraguchi S, Kato M, Shimizu T, Shirakawa M, Kuroda S, Kaibuchi K, Hakoshima T (1998) Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue. J Biol Chem 273: 9656–9666 [DOI] [PubMed] [Google Scholar]
- Jacobs M, Hayakawa K, Swenson L, Bellon S, Fleming M, Taslimi P, Doran J (2006) The structure of dimeric ROCK I reveals the mechanism for ligand selectivity. J Biol Chem 281: 260–268 [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Hall A (2005) Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol 21: 247–269 [DOI] [PubMed] [Google Scholar]
- Johnson LN, Noble ME, Owen DJ (1996) Active and inactive protein kinases: structural basis for regulation. Cell 85: 149–158 [DOI] [PubMed] [Google Scholar]
- Katoh H, Harada A, Mori K, Negishi M (2002) Socius is a novel Rnd GTPase-interacting protein involved in disassembly of actin stress fibers. Mol Cell Biol 22: 2952–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y, Fukata Y, Oshiro N, Amano M, Nakamura T, Ito M, Matsumura F, Inagaki M, Kaibuchi K (1999) Phosphorylation of myosin-binding subunit (MBS) of myosin phosphatase by Rho-kinase in vivo. J Cell Biol 147: 1023–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly PJ, Krebs EG (1991) Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem 266: 15555–15558 [PubMed] [Google Scholar]
- Kim C, Xuong NH, Taylor SS (2005) Crystal structure of a complex between the catalytic and regulatory (RIalpha) subunits of PKA. Science 307: 690–696 [DOI] [PubMed] [Google Scholar]
- Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K (1996) Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273: 245–248 [DOI] [PubMed] [Google Scholar]
- Lamarche N, Tapon N, Stowers L, Burbelo PD, Aspenstrom P, Bridges T, Chant J, Hall A (1996) Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell 87: 519–529 [DOI] [PubMed] [Google Scholar]
- Leung T, Chen XQ, Manser E, Lim L (1996) The p160 RhoA-binding kinase ROK alpha is a member of a kinase family and is involved in the reorganization of the cytoskeleton. Mol Cell Biol 16: 5313–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Cryst 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes CD, Hall A (1999) Rho GTPases control polarity, protrusion, and adhesion during cell movement. J Cell Biol 144: 1235–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes CD, Lauritzen I, Mattei MG, Paris S, Hall A, Chardin P (1998) A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion. J Cell Biol 141: 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B, Taylor S, Ghosh G (2004) Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell 15: 661–675 [DOI] [PubMed] [Google Scholar]
- Oliver AW, Knapp S, Pearl LH (2007) Activation segment exchange: a common mechanism of kinase autophosphorylation? Trends Biochem Sci 32: 351–356 [DOI] [PubMed] [Google Scholar]
- Ongusaha PP, Kim HG, Boswell SA, Ridley AJ, Der CJ, Dotto GP, Kim YB, Aaronson SA, Lee SW (2006) RhoE is a pro-survival p53 target gene that inhibits ROCK I-mediated apoptosis in response to genotoxic stress. Curr Biol 16: 2466–2472 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pearl LH, Barford D (2002) Regulation of protein kinases in insulin, growth factor and Wnt signalling. Curr Opin Struct Biol 12: 761–767 [DOI] [PubMed] [Google Scholar]
- Ridley AJ (2006) Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol 16: 522–529 [DOI] [PubMed] [Google Scholar]
- Riento K, Guasch RM, Garg R, Jin B, Ridley AJ (2003) RhoE binds to ROCK I and inhibits downstream signaling. Mol Cell Biol 23: 4219–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riento K, Ridley AJ (2003) Rocks: multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol 4: 446–456 [DOI] [PubMed] [Google Scholar]
- Riento K, Totty N, Villalonga P, Garg R, Guasch R, Ridley AJ (2005a) RhoE function is regulated by ROCK I-mediated phosphorylation. EMBO J 24: 1170–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riento K, Villalonga P, Garg R, Ridley A (2005b) Function and regulation of RhoE. Biochem Soc Trans 33: 649–651 [DOI] [PubMed] [Google Scholar]
- Song H, Hanlon N, Brown NR, Noble ME, Johnson LN, Barford D (2001) Phosphoprotein–protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2. Mol Cell 7: 615–626 [DOI] [PubMed] [Google Scholar]
- Vetter IR, Arndt A, Kutay U, Gorlich D, Wittinghofer A (1999) Structural view of the Ran-Importin beta interaction at 2.3 Å resolution. Cell 97: 635–646 [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A (2001) The guanine nucleotide-binding switch in three dimensions. Science 294: 1299–1304 [DOI] [PubMed] [Google Scholar]
- Villalonga P, Guasch RM, Riento K, Ridley AJ (2004) RhoE inhibits cell cycle progression and Ras-induced transformation. Mol Cell Biol 24: 7829–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerberg K, Forget MA, Ellerbroek SM, Arthur WT, Burridge K, Settleman J, Der CJ, Hansen SH (2003) Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr Biol 13: 1106–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li W, Craddock BP, Foreman KW, Mulvihill MJ, Ji QS, Miller WT, Hubbard SR (2008) Small-molecule inhibition and activation-loop trans-phosphorylation of the IGF1 receptor. EMBO J 27: 1985–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Kasa M, Amano M, Kaibuchi K, Hakoshima T (2006) Molecular mechanism for the regulation of rho-kinase by dimerization and its inhibition by fasudil. Structure 14: 589–600 [DOI] [PubMed] [Google Scholar]
- Yang J, Cron P, Good VM, Thompson V, Hemmings BA, Barford D (2002) Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat Struct Biol 9: 940–944 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures