Abstract
The voltage-dependent anion channel (VDAC) mediates trafficking of small molecules and ions across the eukaryotic outer mitochondrial membrane. VDAC also interacts with anti-apoptotic proteins from the Bcl-2 family and this interaction inhibits release of apoptogenic proteins from the mitochondrion. We present the NMR solution structure of recombinant human VDAC-1 reconstituted in detergent micelles. It forms a 19-stranded β-barrel with the first and last strand parallel. The hydrophobic outside perimeter of the barrel is covered by detergent molecules in a belt-like fashion. In the presence of cholesterol recombinant VDAC-1 can form voltage-gated channels in phospholipid bilayers similar to the native protein. NMR measurements revealed the binding sites of VDAC-1 for the Bcl-2 protein Bcl-xL, for β-NADH and for cholesterol. Bcl-xL interacts with the VDAC barrel laterally at strands 17 and 18.
The integral membrane protein VDAC forms the primary path for diffusion of metabolites between the mitochondrial intermembrane space and the cytosol (1, 2). VDAC is conserved across eukaryotes, with about 30% sequence identity between yeast and human. The three isoforms VDAC-1, VDAC-2 and VDAC-3 found in humans are 68% to 75% pairwise identical. All three isoforms allow the exchange of metabolites through the membrane, but have distinct physiological roles and expression profiles (3, 4).
Numerous reports have suggested that VDAC-1 is involved in mitochondrial apoptosis (5-7). Apoptotic signals lead to the formation of a mitochondrial exit channel that allows the release of apoptogenic proteins, which in turn cause cell death by activating executioner caspase or through other mechanisms (8-10). Functional studies indicate that VDAC-1 closure leads to the opening of the mitochondrial exit channel (11). The anti-apoptotic protein Bcl-xL opens the VDAC-1 channel for trafficking of metabolites and thus inhibits the release of apoptogenic proteins (12). Direct interaction between VDAC-1 and Bcl-xL has been demonstrated (11, 13).
Insights into VDAC organization have come from biochemical and biophysical studies (14, 15) and low resolution electron microscopy data showing that VDAC-1 is a cylindrical channel with a diameter of 20–30 Å (16, 17). Electrophysiological experiments reveal that at low membrane potentials of 10 mV VDAC is in the open state, but it switches to the closed state at increased membrane potentials of about 30 mV (18). Thus, Donnan or diffusion potentials across the outer mitochondrial membrane may regulate small-molecule passage (19). In a lipid bilayer, recombinant VDAC can form stable open states and feature voltage-dependent transitions (20).
Here we present the three-dimensional solution structure of human VDAC-1 reconstituted in detergent micelles as determined by high-resolution nuclear magnetic resonance (NMR). We have assayed the binding of VDAC-1 with its natural ligands β-NADH and cholesterol and with the protein Bcl-xL by NMR and identify the location of these interaction sites on the structure of VDAC-1.
For the structure determination of VDAC-1 with NMR spectroscopy, bacterially expressed human VDAC-1 was refolded into lauryldimethylamine oxide (LDAO) detergent micelles (21). The channel exhibits a well-dispersed 2D [15N,1H]-TROSY spectrum indicating the presence of extensive β-sheet secondary structure (Fig. S1). High-field triple-resonance TROSY-type experiments and selective labeling enabled sequence-specific resonance assignment of 80% of the protein backbone including the Cβ resonances (Figs. 1, S2, S3). 19 β-strands are formed within residues 25–283 and a short α-helix is located at the N-terminus containing residues 6–10, as indicated by Cα and Cβ secondary chemical shifts (Fig. S4). To complement the backbone assignments, specifically isotope-labelled samples were prepared to assign methyl groups of Ile, Leu and Val residues (22) (Fig. S5). These side-chain assignments were essential for defining the overall structure of the protein and the location of the N-terminal helix with respect to the channel.
Figure 1.
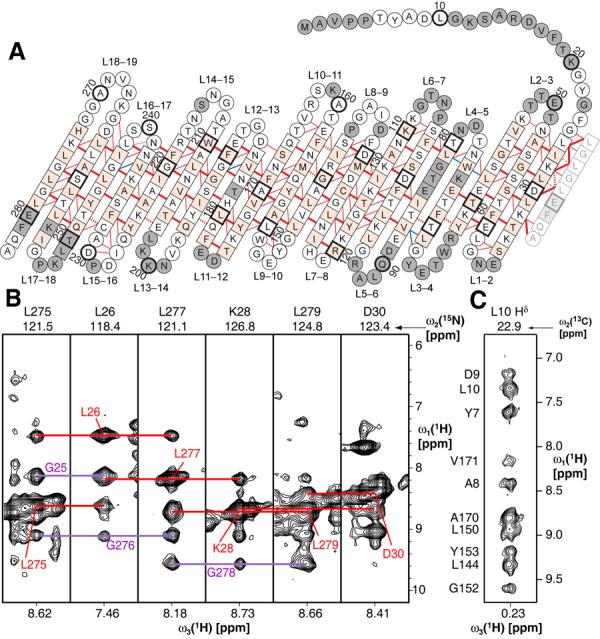
Architecture of VDAC-1. (A) The amino acid sequence of VDAC-1 in one-letter code is arranged according to the secondary and tertiary structure. Amino acids in squares denote β-sheet secondary structure as identified by secondary chemical shifts, all other amino acids are in circles. Red and blue lines denote experimentally observed NOE contacts between two amide protons and NOE contacts involving side chain atoms, respectively. Bold lines indicate strong NOEs typically observed between hydrogen bonded residues in β-sheets. For clarity of the presentation, not all observed NOEs are shown. The 19th strand is duplicated at the right next to strand 1 to allow indicating the barrel-closure NOEs. The side chains of white and orange residues point towards the inside and outside of the barrel, respectively. Dashed lines show probable contacts between protons with degenerate 1H chemical shifts. Grey residues could not be assigned so far. Every tenth amino acid is marked with a heavy outline, and corresponding residue numbers are indicated. (B) Strips from a 3D [1H,1H]-NOESY-15N-TROSY defining the barrel closure between parallel strands 1 and 19. Red lines show the interstrand contacts for the depicted residues, whereas the violet lines indicate the NOE contacts for the respective opposite residues. (C) Strip from a 3D [1H,1H]-NOESY-13C-HMQC taken at the position of a methyl group of Leu 10. The assignments of the individual NOE signals are indicated on the left and exemplify the NOEs defining the location of the N-terminal helix in the barrel.
For the acquisition of long-range structural information, nuclear Overhauser effect spectroscopy (NOESY) was used to establish spatial correlations between nuclear spins (23). To obtain adequate signal, it was necessary to record these NOESY experiments on a fully deuterated background, including the use of perdeuterated LDAO molecules. From the cross-peaks observed in 3- and 4-dimensional NOESY experiments, a network of more than 600 NOE contacts were identified that confirmed the 19-stranded β-sheet topology for VDAC-1 in LDAO micelles and established the relative orientation of the strands (Fig. 1, S6, S7, Table S1). Strands 1–19 form an antiparallel β-sheet, which is closed to a β-barrel by parallel pairing of strands 19 and 1.
32 discrete β-barrel membrane protein structures have so far been solved, all from prokaryotic organisms and all consisting of even numbers of β-strands with antiparallel orientation (24) (Fig. S8). The eukaryotic VDAC-1 thus represents a new class of a β-barrel membrane protein fold. Whereas N- and C-terminal ends are aligned next to each other in even-stranded barrels, they protrude from opposite sides of the VDAC-1 barrel. However, the N-terminus of VDAC reaches through the channel to return to the same side of the barrel as the C-terminus. In previous predictions of the VDAC topology the number of β-strands ranged from 12–19 with one corresponding to the topology determined here (25).
Based on the experimental constraints, the three-dimensional structure for VDAC-1 in LDAO micelles was calculated (Fig. 2, Table S1). The 19 β-strands form the wall of an open barrel with the strands being tilted by about 45° with respect to the barrel main axis. Alternating between the two ends of the barrel, adjacent strands are connected by loops with lengths of 2–10 residues. The height of the barrel including the loops is approximately 30 Å and a circular conformer of VDAC-1 has an open diameter of about 25 Å, corresponding nicely to electron microscopy data (16, 17).
Figure 2.
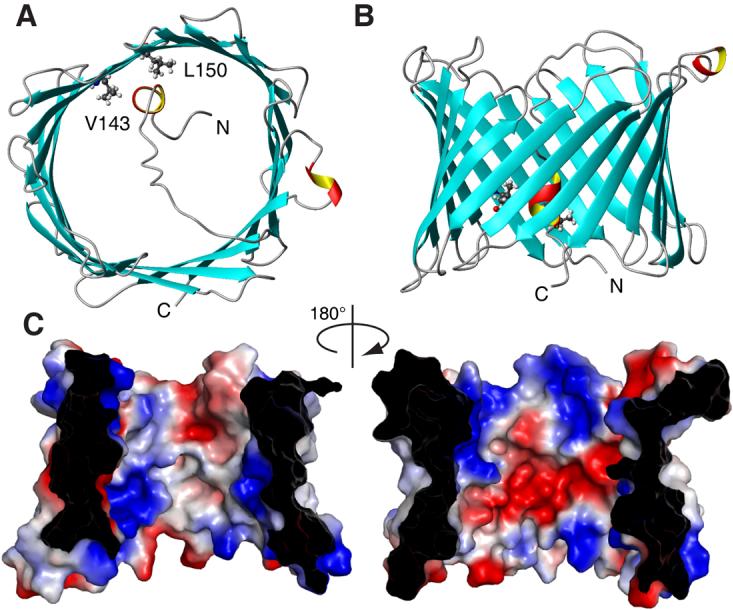
NMR solution structure of VDAC-1 in LDAO micelles. (A) and (B) Top- and side-view, respectively, of the conformer closest to the mean of the conformational ensemble in ribbon representation. β-sheets are shown blue and α-helical secondary structure in red and yellow. N- and C-termini and residues L150 and V143 are indicated. (C) Van der Waals surface of VDAC-1. The surface is colored according to the surface potential, calculated using vacuum electrostatics in the program Pymol. Blue indicates positive charge and red negative charge.
Consistent with the function of VDAC-1 as a wide diffusion pore, no tertiary contacts were observed between residues across the barrel diameter. This is reflected within the calculated bundle of 20 structural conformers, among which the β-barrel adopts differently circular and oval shapes (Fig. S9). It is unclear, whether this variation of the barrel shape represents actual molecular motions. The relative orientations of the β-strands with respect to each other are well defined by the observed NOEs and thus identical in all members of the conformational ensemble.
The N-terminal tail of VDAC-1, consisting of residues 1–23 is not part of the barrel wall but is located inside the channel. NOE contacts connect residues 7, 8, 9 and 10 with several residues around a small hydrophobic patch formed by residues Val 143 and Leu 150 on the inside of the barrel wall (Figs. 1, 2). The observation that the N-terminal 23 residues are structurally not involved in the main barrel architecture is consistent with experiments showing that a deletion mutant with the N-terminal part missing properly targets to the mitochondrial outer membrane (26). Since the N-terminal region is involved in voltage gating (27), this segment might adopt different conformations depending on the external conditions.
The overall theoretical charge of VDAC-1 at neutral pH is +3, since 29 negative charges from Asp and Glu residues oppose 32 positive charges from Arg and Lys residues. These charges are located predominantly in the loops and in the interior, water-accessible wall of the channel, where they cluster to form one negative and two positive patches (Fig. 2). The VDAC channel is known to be open to both anions and cations, with a 2:1 preference for anions (28, 29) and these two features of the channel could thus be correlated.
The surface of VDAC-1 that is in contact with the detergent micelle was determined with the help of the detergent 16-DSA that has a paramagnetic spin-label attached to its hydrophobic tip, quenching all NMR resonances of residues in close contact with the micelle interior (30). The data show that the detergent micelle covers the entire periphery of the VDAC-1 barrel like a belt, corresponding to the position of hydrophobic residues on the outside of the barrel (Figs. 3, S10). On the inner wall only two hydrophobic residues are located, the above mentioned L150 and V143 (Fig. 3C). The protein–micelle complex in aqueous solution has thus a topology that would be consistent with the assumption that the protein adopts a similar fold in a lipid bilayer as in the LDAO micelle. The data obtained in the micellar state can not address which opening of the channel faces into the mitochondria and which faces into the cytosol. Previous studies attempting to determine this orientation have reached different conclusions (15, 31). Experiments on Neurospora crassa VDAC suggested a different barrel topology and proposed a membrane-inserted N-terminal helix (32), which is inconsistent with the structure presented here. The VDAC-1 solution structure will stimulate revisiting the large body of previous biochemical data and the design of new experiments to resolve this issue.
Figure 3.
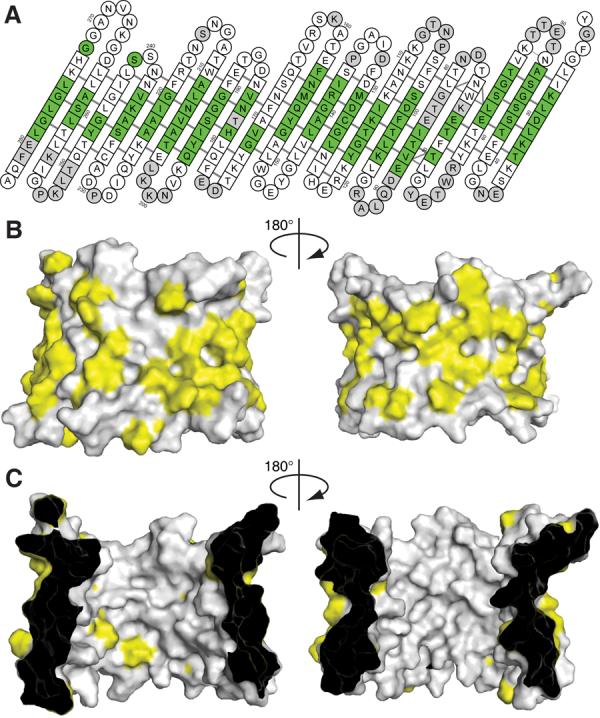
Hydrophobic surface of VDAC-1. (A) Result of a titration with the spin-labelled detergent 16-DSA. Residues with a relaxation enhancement ε > 20 s−1mM−1 are green (30). These residues are in close contact to the hydrophobic interior of the micelle. Residues with ε ≤ 20 s−1mM−1 are white. Grey residues are unassigned. Residues 1–21 have been omitted, no interaction with the spin label was observed for these. (B) and (C) Surface plot of the outer and inner surface of VDAC-1, respectively, with the side chains of the hydrophobic residues Leu, Val, Ile, Met, Phe, Trp shown in yellow and all other residues in white.
The possibility of a multimeric state of VDAC is a matter of ongoing debate. There is evidence to support the formation of dimers, trimers and tetramers, possibly in dynamic equilibrium with monomers (13). Since the entire perimeter of VDAC-1 is in contact with the micelle-immersed spin labels (Fig. 3A), our data excludes formation of stable oligomers in the LDAO micelles, however a dynamic inter-channel interaction cannot be excluded. A possible interaction site is located on strands 4 and 5 where the resonances of several residues are unresolved presumably due to exchange broadening (Fig. 1). This potential contact may be due to the high protein/detergent ratio used here and may not exist in mitochondrial membranes.
In its native environment in the outer mitochondrial membrane, VDAC has several posttranslational modifications such as phosphorylation and acetylation (33). In an attempt to determine whether these affect function, we compare the gating parameters of our unmodified recombinant protein with those of native VDAC carrying posttranslational modifications. To achieve functional properties similar to wild-type, recombinant VDAC-1 had to be reconstituted into planar membranes in the presence of cholesterol and the detergent triton X100, which were both also present in the reference measurements with native VDAC (34, 35). Under these conditions, the refolded VDAC-1 exhibited similar functional behavior to native VDAC from eukaryotic sources as measured by the single-channel conductance, the effective valence of the voltage sensor, the energy difference between open and closed states, and ion selectivity (Fig. S11, Table 1). While the present data does not directly address the structure of VDAC-1 in lipid bilayers, it is clear that our recombinantly produced VDAC-1 polypeptide can be induced to adopt a structure with very similar functional properties as native wild-type VDAC and that the gating properties of VDAC are thus not substantially affected by the post-translational modifications in native VDAC or by the C-terminal His-tag present in our preparations.
Table 1.
Functional properties of VDAC Voltage gating in lipid bilayers.
Since cholesterol was necessary to obtain wild-type like function of recombinant VDAC-1 and since mammalian VDAC has tightly-bound cholesterol (35), we used chemical shift mapping to examine the effect of cholesterol on recombinant VDAC-1 in LDAO micelles (Fig. S12). The overall structure of VDAC-1 in LDAO micelles is unchanged by cholesterol concentrations up to 1:5:400 for VDAC-1:cholesterol:LDAO. The chemical shift changes induced by cholesterol revealed two distinct interaction sites located at β-strands 7+8, and 11 (Fig. 4A). Cholesterol is dissolved in the hydrophobic phase of the micelle and thus approaches the residues at these interaction sites from the outside of the barrel.
Figure 4.
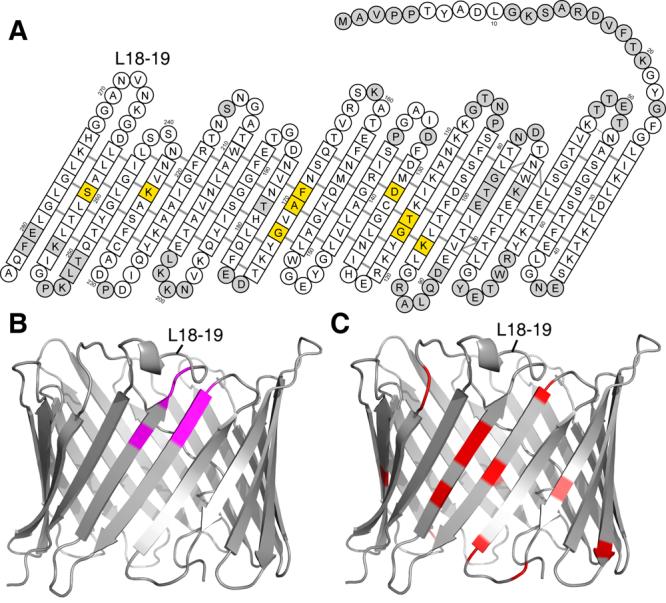
Interactions of VDAC-1. (A) Residues with significant chemical shift changes (Δδ(HN) > 0.5 ppm) caused by cholesterol binding are shown in yellow (Fig. S12). The amino acids of VDAC-1 are shown as in Fig. 1A. (B) Amide resonances of VDAC-1 with significant chemical shift changes (Fig. S13) caused by β-NADH are labeled magenta in this ribbon representation, all other residues are grey. (C). Residues involved in Bcl-xL binding (13) are marked red in this ribbon representation, all other residues are grey. In all three panels, the loop connecting strands 18 and 19 is indicated for orientation purpose.
Weak interactions with VDAC have also been reported for some of its water-soluble natural ligands, such as ATP (13). NMR chemical shift mapping indicates that neither ATP nor β-NAD do interact with a specific site of VDAC-1 (Fig. S13). However, for the interaction with β-NADH, a distinct interaction surface was observed at strands 17 and 18 involving residues G242, L243, I244, A261, L263 and D264 (Figs. S13, 4B). Our results are consistent with a study on native VDAC, which showed that β-NADH, but not NAD, favors the closure of VDAC and predicted that the NADH binding site would comprise the partial Walker B motif SALLD at residues 260–264 (36).
We have mapped the binding site of the native binding partner Bcl-xL onto reconstituted VDAC-1 (Figs. 4C, S14). Bcl-xL binds to VDAC-1 at strands 17 and 18. Interestingly, the loops adjacent to the Bcl-xL binding site contain several basic residues (Lysines 266, 252, 200, and 201) so that this face of the molecule has a positive electrostatic surface. Bcl-xL, with an estimated pI of 4.9 is negatively charged at neutral pH and the interaction between VDAC-1 and Bcl-xL may thus contain a substantial electrostatic component. Some residues affected by Bcl-xL binding are not located closely to this binding site. These effects could originate from allosteric changes or from a second Bcl-xL binding site at the lumen opening.
Supplementary Material
Acknowledgments
This work was supported by the NIH roadmap grant GM075879. Purchase, operation and maintenance of instruments used was supported by grants GM066360, GM47467 and EB 002026. S.H. was supported by the Swiss National Science Foundation. Initial research on this project was supported by the Ludwig Foundation for Cancer Research. We thank Vlado Gelev from FBReagents.com, Cambridge, MA, for custom synthesis of the 2H-LDAO, Soumya Samanta for help with the electrophysiological measurements, and Dominique Frueh, Alexander Koglin, Zhen-Yu J. Sun, Kirill Oxenoid and Simon Jenni for technical help and valuable discussions. The atomic coordinates of VDAC-1 in LDAO micelles have been deposited at the PDB with code 2k4t.
Footnotes
Publisher's Disclaimer: This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. Their manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.”
References and Notes
- 1.Hodge T, Colombini M. J. Membr. Biol. 1997;157:271. doi: 10.1007/s002329900235. [DOI] [PubMed] [Google Scholar]
- 2.Rostovtseva T, Colombini M. Biophys. J. 1997;72:1954. doi: 10.1016/S0006-3495(97)78841-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sampson MJ, et al. J. Biol. Chem. 2001;276:39206. doi: 10.1074/jbc.M104724200. [DOI] [PubMed] [Google Scholar]
- 4.Hinsch KD, et al. J. Biol. Chem. 2004;279:15281. doi: 10.1074/jbc.M313433200. [DOI] [PubMed] [Google Scholar]
- 5.Vander Heiden MG, Thompson CB. Nat. Cell Biol. 1999;1:E209. doi: 10.1038/70237. [DOI] [PubMed] [Google Scholar]
- 6.Brustovetsky N, Dubinsky JM, Antonsson B, Jemmerson R. J. Neurochem. 2003;84:196. doi: 10.1046/j.1471-4159.2003.01545.x. [DOI] [PubMed] [Google Scholar]
- 7.Rostovtseva TK, et al. J. Biol. Chem. 2004;279:13575. doi: 10.1074/jbc.M310593200. [DOI] [PubMed] [Google Scholar]
- 8.Desagher S, Martinou JC. Trends Cell Biol. 2000;10:369. doi: 10.1016/s0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]
- 9.Danial NN, Korsmeyer SJ. Cell. 2004;116:205. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 10.Kroemer G, Galluzzi L, Brenner C. Physiol. Rev. 2007;87:99. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 11.Shimizu S, Narita M, Tsujimoto Y. Nature. 1999;399:483. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- 12.Vander Heiden MG, et al. J. Biol. Chem. 2001;276:19414. doi: 10.1074/jbc.M101590200. [DOI] [PubMed] [Google Scholar]
- 13.Malia TJ, Wagner G. Biochemistry. 2007;46:514. doi: 10.1021/bi061577h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas L, Blachly-Dyson E, Colombini M, Forte M. Proc. Natl. Acad. Sci. USA. 1993;90:5446. doi: 10.1073/pnas.90.12.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanley S, Dias JA, Darcangelis D, Mannella CA. J. Biol. Chem. 1995;270:16694. doi: 10.1074/jbc.270.28.16694. [DOI] [PubMed] [Google Scholar]
- 16.Mannella CA. J. Cell Biol. 1982;94:680. doi: 10.1083/jcb.94.3.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo XW, Smith PR, Mannella CA. J. Struct. Biol. 1995;114:41. doi: 10.1006/jsbi.1995.1004. [DOI] [PubMed] [Google Scholar]
- 18.Colombini M. Nature. 1979;279:643. doi: 10.1038/279643a0. [DOI] [PubMed] [Google Scholar]
- 19.Colombini M. Mol. Cell. Biochem. 2004;256:107. doi: 10.1023/b:mcbi.0000009862.17396.8d. [DOI] [PubMed] [Google Scholar]
- 20.Koppel DA, et al. J. Biol. Chem. 1998;273:13794. doi: 10.1074/jbc.273.22.13794. [DOI] [PubMed] [Google Scholar]
- 21.Information on materials and methods is available on Science Online.
- 22.Rosen MK, et al. J. Mol. Biol. 1996;263:627. doi: 10.1006/jmbi.1996.0603. [DOI] [PubMed] [Google Scholar]
- 23.Wüthrich K. NMR of Proteins and Nucleic Acids. Wiley; New York: 1986. [Google Scholar]
- 24.White SH. Membrane proteins of known structure. 2008 April; http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html.
- 25.Forte M, Guy HR, Mannella CA. J. Bioenerg. Biomembr. 1987;19:341. doi: 10.1007/BF00768537. [DOI] [PubMed] [Google Scholar]
- 26.De Pinto V, et al. ChemBioChem. 2007;8:744. doi: 10.1002/cbic.200700009. [DOI] [PubMed] [Google Scholar]
- 27.Song J, Midson C, Blachly-Dyson E, Forte M, Colombini M. Biophys. J. 1998;74:2926. doi: 10.1016/S0006-3495(98)78000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schein SJ, Colombini M, Finkelstein A. J. Membr. Biol. 1976;30:99. doi: 10.1007/BF01869662. [DOI] [PubMed] [Google Scholar]
- 29.Komarov AG, Graham BH, Craigen WJ, Colombini M. Biophys. J. 2004;86:152. doi: 10.1016/S0006-3495(04)74093-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilty C, Wider G, Fernández C, Wüthrich K. ChemBioChem. 2004;5:467. doi: 10.1002/cbic.200300815. [DOI] [PubMed] [Google Scholar]
- 31.De Pinto V, Prezioso G, Thinnes F, Link TA, Palmieri F. Biochemistry. 1991;30:10191. doi: 10.1021/bi00106a017. [DOI] [PubMed] [Google Scholar]
- 32.Song JM, Midson C, Blachly-Dyson E, Forte M, Colombini M. J. Biol. Chem. 1998;273:24406. doi: 10.1074/jbc.273.38.24406. [DOI] [PubMed] [Google Scholar]
- 33.Olsen JV, et al. Cell. 2006;127:635. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 34.Colombini M. J. Membr. Biol. 1989;111:103. doi: 10.1007/BF01871775. [DOI] [PubMed] [Google Scholar]
- 35.De Pinto V, Benz R, Palmieri F. Eur. J. Biochem. 1989;183:179. doi: 10.1111/j.1432-1033.1989.tb14911.x. [DOI] [PubMed] [Google Scholar]
- 36.Zizi M, Forte M, Blachly-Dyson E, Colombini M. J. Biol. Chem. 1994;269:1614. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


