Abstract
Measurement of breast tissue estradiol levels could provide a powerful method to predict the risk of developing breast cancer but obtaining sufficient amounts of tissue from women is difficult from a practical standpoint. Assessment of aromatase in ductal lavage fluid or fine needle aspirates from breast might provide a surrogate marker for tissue estrogen levels but highly sensitive methods would be required. These considerations prompted us to develop an ultra-sensitive, “nested” PCR assay for aromatase which is up to one million fold more sensitive than standard PCR methods. We initially validated this assay using multiple tissues from the aromatase transgenic mouse and found that coefficients of variation for measurement of replicate samples averaged less than 5%. We demonstrated a 60-fold enhancement in aromatase message in the transgenic versus the wild type mouse breast but surprisingly, levels in the transgenic animals were highly variable, ranging from 0.4 to 27 relative units. The variability of aromatase expression in the transgenic breast did not correlate with the degree of breast development and did not appear to relate to hormonal manipulation of the MMTV promoter but probably related to lack of exhaustive inbreeding and mixed zygocity of transgenic animals. Extensive validation in mouse tissues provided confidence regarding the assay in human tissues, since nearly identical methods were used. The human assay was sufficiently sensitive to detect aromatase in a single human JAR (choriocarcinoma) cell, in all breast biopsies measured, and in 7/23 ductal lavage fluids.
Keywords: Aromatase, Breast tissue, Ductal lavage, Estrogen, Fine needle aspirate (FNA), Nested PCR
Introduction
The concentrations of estradiol (E2) in breast tissue from post-menopausal women are similar to those from pre-menopausal patients even though plasma E2 levels are 50-fold higher prior to the menopause [1–17]. To explain this finding, several investigators have postulated that post-menopausal breast tissue converts androgens to estrogens in situ via the aromatase enzyme in amounts sufficient to maintain high tissue E2 levels [5, 7, 17]. Both animal and human studies have demonstrated aromatase enzyme activity in breast tissue as assessed by in vitro radiometric, titriated water formation assays [5, 18–23]. Our prior data in nude mice demonstrated that in situ production of estrogen in aromatase transfected MCF-7 cells exceeds the amount taken up from plasma [24]. In women, in vivo isotopic kinetic measurements have directly quantitated local estrogen synthesis in breast and determined the relative proportion of estrogen made in the breast versus uptake from plasma [17]. Specifically, the studies of Reed et al. and Miller et al demonstrated that local synthesis accounts for 30–100% of the estrogen in the postmenopausal breast whereas uptake from plasma contributes to a lesser extent [25–28].
The in vivo assays in women require administration of radioisotopes and are technically demanding. The more commonly used tritiated water formation in vitro assays lack sensitivity and are capable of detecting aromatase in only two thirds of breast cancers [5]. Taken together, these results suggest the need for a readily applicable, highly sensitive, in vitro assay suitable for the routine assessment of aromatase in human breast samples.
A substantial rationale exists to measure estrogen levels in breast tissue. Plasma E2 levels predict the development of breast cancer over the ensuing 5–10 year period in healthy post-menopausal women [29]. It is plausible then to believe that breast tissue estrogen levels might correlate with risk to an even greater extent. Measurement of breast tissue estrogen levels has been hindered in the past by the need to obtain biopsies from normal women in order to assess E2 levels. This difficulty has also limited the study of pre-malignant histologic changes and the study of biomarkers in breast tissue. To circumvent this problem, major recent emphasis has been directed toward the development of methods to obtain and study nipple aspirates, ductal lavage fluid, and FNAs (fine needle aspirates) [30]. These techniques are more acceptable to patients than excisional biopsies but yield only a small amount of tissue and consequently require highly sensitive and precise methods of analysis of selected biomarkers.
These considerations stimulated us to develop an ultra-sensitive, “nested”, Q-PCR assay suitable for measurement of aromatase with high precision in fine needle aspirates from breast and in nipple aspirate and ductal lavage fluids. We reasoned that an initial PCR amplification of a larger segment of aromatase cDNA followed by a second amplification of a “nested” internal cDNA would markedly enhance sensitivity and not compromise specificity or precision. An initial 15 cycles of amplification provide 32,000-fold higher sensitivity (and 20 cycles a 1,000,000-fold increase) than with the standard, one-amplification method. The “nested” second amplification ensures a high degree of specificity.
To systematically validate this approach, we initially utilized tissues from the aromatase over-expressing transgenic mouse to determine sensitivity, precision, and specificity of the assay [31, 32]. We also used these animals to demonstrate the ability to quantitate differences between tissues containing the aromatase transgene promoter and those not. After this validation, we then chose analogous human primers to measure aromatase in human JAR cells (a choriocarcinoma cell line) [33], ductal lavage fluid and in breast tissue itself. The “nested” method is sufficiently sensitive to measure aromatase message in one human JAR cell and in all breast biopsies. Even in highly diluted ductal lavage samples, this method was capable of detecting aromatase in 7/23 samples. From the data reported in this manuscript, we conclude that a nested Q-PCR method is valid and useful for quantitating aromatase message in human samples with low aromatase levels.
Materials and methods
Source of mouse samples and collection procedures
A total of 55 female aromatase transgenic mice of BALB/c genetic background [31, 32] ranging in age from 3 to 6 months with mixed zygocity (largely hetero and some homozygous animals) were utilized for PCR studies. The breast tissues from these mice were classified into four groups (0 minimal development; 1 moderate development; 2 average development; 3 marked development) according to the level of ductal and gland proliferation estimated by subjective examination of whole mounts. As controls, 29 wild-type female mice were similarly classified and used for comparison. Samples were also collected from mouse ovary, uterus, kidney, liver, and adrenal.
The third and fourth right mammary glands as well as the left third glands were excised from non-castrate female mice at ages 3–6 months. The outside one third of the glands were dissected from the remaining two thirds and discarded. The remaining two thirds of the glands (80–100 mg) were dissected free, placed into clean RNase-free tubes, immediately immersed in liquid nitrogen, and then transferred into a −70° Revco ultra-cold freezer for storage. At the same time, 80–100 mg of mouse liver, kidney, adrenal, uterus, and ovary were obtained and similarly processed.
Human tissues
Human JAR cells, a choriocarcinoma cell line, were obtained from American Tissue Type Culture (ATCC) and have been used in our laboratory for more than 15 years [33]. Additional samples were obtained from women and included 23 ductal lavage samples, 10 breast cancer specimens, and 3 breast tissues obtained during reduction mammoplasty. Ductal lavage fluid was obtained by standard techniques at the time of surgery from women undergoing breast lump excision for various clinical indications [30]. Breast biopsies were obtained as part of a human tissue bank program at the University of Virginia or from reduction mammoplasties performed in the plastic surgical department of Hôpital Rothschild (Paris) and processed in INSERM Unit 673, Hôpital Saint Antoine. At both institutions, samples were quickly frozen using methods designed to preserve RNA. Ethical guidelines from both institutions were followed.
RNA extraction
Total RNA was extracted from frozen tissue specimens by using an Aurum Kit (Bio-Rad) for total RNA extraction according to the manufacturer’s instructions. Extracted RNA was dissolved in TE buffer and stored at −70°C in the ultra-cold Revco. The quantity of RNA extracted was determined by spectrophotometric determination. The quality of extracted RNA was assessed by electrophoresis through 1.2% agarose gel and staining with ethidium bromide. The 18s and 28s RNA bands were visualized under ultraviolet light. Additionally we confirmed the quality of extracted RNA by calculating the A260/A280 ratio = (adequate = 1.7–2.1).
JAR cell dilution study
JAR cells were grown until there were 1 × 108 to 1 × 109 cells present. These were then collected, diluted to a concentration of 1 × 108 cells, and subjected to the RNA extraction procedure. This method was chosen to obviate the need to extract RNA from small numbers of cells. The RNA was then diluted with the medium used for constructing standards to a content representing 100,000, 10,000, 1000, 100, 10, and 1 cell. PCR amplifications were then preformed and the results mathematically reduced to the amount of RNA per cell. The amplicons were confirmed to be specific for human aromatase by melt curves.
Design of primers and probes
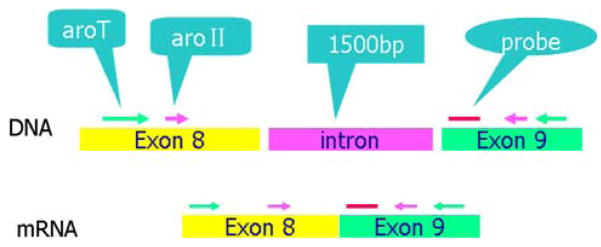
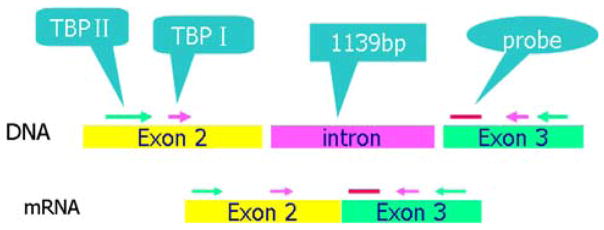
Mouse and human primers and probes were chosen from inspection of the cDNA sequence of the aromatase and TBP genes from the Nucleotide sequence database in GenBank (Figs. 1 and 2). We then performed a BLAST search using the whole cDNA sequence to determine the location of exon–exon junctions and designed primers by specifying a target region corresponding to the junctions. The specificity of the primer pair enclosed sequences were confirmed by BLAST search and the absence of polymorphisms in these regions were verified. To avoid detection of contaminating genomic DNA, the primers were located in exons and not in introns. The sequence of mouse aromatase mRNA is from GenBank (D00659, Mus musculus CYP mRNA for aromatase p450, complete cds).
Fig. 1.
The locations of the genomic DNA and cDNA regions chosen for the first amplification, the second “nested” amplification, and the probe used for real time PCR quantification. Primer for mouse aromatase
Fig. 2.
The locations of the genomic DNA and cDNA regions chosen for the first amplification, the second “nested” amplification, and the probe used for real time PCR quantification. Primer for mouse TBP
The mouse primer pairs chosen included:
| AroT: outside primers for initial amplification | |
| sense primer | 5′–agc atg cgg tac cag cct gt–3′ pos 1087 |
| anti-sense primer | 5′–tca tca tca cca tgg cga tgt–3′ pos 1342 |
| Aro II: inside (“nested”) primers for amplification | |
| sense primer | 5′–tga aca tcg gaa gaa tgc ac–3′ pos 1187 |
| anti-sense primer | 5′–caa agc caa aag gct gaa ag–3′ pos 1297 |
| probe | 5′–FAM ccc taa gcc caa tga att tac cct t–TAMRA–3′ |
The sequence of mouse TBP mRNA is from GenBank (BC016476, Mus musculus TATA box binding protein mRNA; cDNA clone MGC:5877 IMAGE:3584341, complete cds; BC050136). The primers for mouse TBP included:
| TBP II: outside primers for initial amplification | |
| sense primer | 5′–ggg aga atc atg gac cag aa–3′ pos 10 |
| anti-sense primer | 5′–tgt gtg ggt tgc tga gat gt–3′ pos 265 |
| TBP I: inside (“nested”) primers for nested amplification | |
| sense primer | 5′–gga cca gaa caa cag cct tc–3′ pos 3 |
| anti-sense primer | 5′–ccg taa ggc atc att gga ct–3′ pos 104 |
| probe | 5′–FAM cat gac tcc tgg aat tcc cat ctt t–TAMRA–3′ |
The human primer pairs chosen included:
Outside aromatase primers for initial amplification
| aromatase: h-aroΠ(p515–681 of cds, 564–730 of full length, amplicon is 167 bps) | |
| sense primer | 5′–tgg aca ggt tgg agg agg tg–3′, Tm 63.5 |
| anti-sense primer | 5′–gag agc ttg cca tgc atc aa–3′, Tm 62.5 |
Inside primers for nested amplification
| Aromatase h-aroШ(p552–644 of cds) | |
| sense primer | 5′–tgt gga cgt gtt gac cct tct–3′, Tm 62.57 |
| anti-sense primer | 5′–acc acg ata gca ctt tcg tcc a–3′, Tm 62.67 |
(Probe not listed since SYBR green method is used)
Outside TBP primers for initial amplification
Initial TBP amplification
| Tbp3a (p8–173 of cds) | |
| sense primer | 5′–aga aca aca gcc tgc cac ct–3′ Tm 62.2 |
| anti-sense primer | 5′–tgc ctt tgt tgc tct tcc aa–3′ Tm 61.85 |
Inside primers for nested amplification
Inside “nested” TBP:
| Tbp3b (p10–132 of cds) | |
| sense primer | 5′–aac aac agc ctg cc acct ta–3′ Tm 60.69 |
| anti-sense primer | 5′–ctg aat agg ctg tgg ggt ca–3′ Tm 61.05 |
(Probe not listed since SYBR green method is used)
PCR amplification
For mouse aromatase determination
Measurement of aromatase mRNA from extracted RNA from mouse tissues were performed by using nested quantitative Q-PCR with the TaqMan Probe Method. The kit for PCR was iScriptTM One-Step RT-PCR Kit (Bio-Rad) using the manufacturer’s instructions. The initial amplification utilized the outside primers (see primer lists) to amplify the chosen longer segment of DNA. The second amplification involved primers chosen to replicate a “nested” segment within the larger amplified segment. Components of the PCR reaction medium are provided in the appendix online.
For human aromatase determination
Measurement of aromatase from human breast samples was performed by nested quantitative Q-PCR in a similar fashion. The first amplification was carried out by using the SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity kit (Invitrogen), and the nested Q-PCR by SYBR Green PCR Master Mix kit (Applied Biosystems) according to the manufacturer’s instructions. The initial amplification used the outside human aromatase primers (see primer list) to amplify the chosen longer segment of DNA. The second amplification utilized the primers chosen to duplicate the “nested” segment within the longer amplified segment. Components of the PCR reaction medium are provided in the appendix online.
Analysis and reporting of results
The relative level of expression of aromatase mRNA is related to the amount of TBP mRNA which serves as a normalizing gene. Relative aromatase mRNA expression was calculated using the following formula:
Note: In the above formula “Ct” indicates threshold values and “N” indicates relative quantity of mRNA expression.
Performance characteristics
The performance of this PCR amplification system was characterized with respect to efficiency, reproducibility, and specificity. For each PCR run, a standard curve was constructed from five serial dilutions of a stock of extracted RNA from a pool of mouse ovarian tissue as a source of aromatase. The expression of aromatase mRNA in these serial dilutions was quantitated by real-time qRT-PCR. The standard curve was plotted as the threshold cycle against the log dilution.
Efficiency
The reaction efficiency was calculated by the following formula:
Reproducibility
Twenty-four replicate mouse samples were assayed by Q-PCR and the coefficient of variation (CV) determined. Testing of linearity involved setting up the standard curve and calculating correlation coefficients (r2) between dose and response.
Specificity
Specificity was demonstrated by agarose gel electrophoresis with DNA ladder markers to demonstrate the appropriate molecular size and by sequencing the amplicon.
Accuracy
In order to assure the accuracy of results obtained by nested Q-PCR, internal and external controls were included in all assays. In the Q-PCR assay, the level of expression of the CYP19 gene was normalized to an internal control “housekeeping” gene TBP. Because TBP is a relatively consistently expressed, it is suitable for use as an internal standard to correct for internal variation caused by a range of parameters in real-time RT-PCR assays. A stock of RNA extracted from mixed ovarian tissues of 10 MMTV-Transgenic mice was used as the external control and used in each assay for quality control.
Results
Mouse aromatase determinations
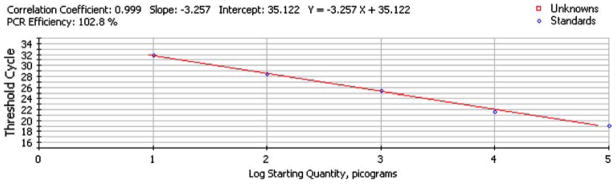
The linearity of response for serial dilutions of an ovarian tissue pool are shown on Fig. 3. Descriptive statistics indicate that the correlation coefficient was 0.99, slope −3.257×, and efficiency 102.8%. The reproducibility of the nested Q-PCR for measurement of aromatase in ovarian samples was 1.3% (online Fig. IA) and for TBP 1.22% (online Fig. IB). Six separate ovarian samples were then each run in two separate assay runs and the results are shown in Table 1.
Fig. 3.
Mouse aromatase standard curve obtained by nested Q-PCR
Table 1.
Reproducibility between two lots of measurement in six mammary gland samples from mouse
| Sample No. | Aromatase mRNA (1000 × N)
|
|
|---|---|---|
| Lot 1 | Lot 2 | |
| 1561 | 1.99 | 2.29 |
| 1486 | 0.25 | 0.43 |
| 1522 | 2.65 | 3.4 |
| 1524 | 1 | 1.25 |
| 1487 | 0.55 | 0.6 |
| 1491 | 3.17 | 2.76 |
| Mean ± SEM | 1.60 ± 0.48 | 1.79 ± 0.49 |
Note: N = aromatase mRNA/TBP mRNA, see above “Materials and methods” about Analysis and reporting of results
We then conducted another reproducibility assay to include both the measurement of aromatase and the housekeeping gene (TBP). This involved mixing the RNA from the ovaries of 10 mice and running on four separate occasions (Table 2) with the results expressed as the ratio of aromatase to TBP times 1000. This experiment, which yielded a CV of 2.47%, reflected the reproducibility of the entire nested assay corrected for the quantity of the TBP housekeeping gene.
Table 2.
Repeatability between different four lots of measurement with the same sample
| Lot No. | 1 | 2 | 3 | 4 | CV (%) |
|---|---|---|---|---|---|
| Aromatase mRNA (1000× N) | 1,319 | 1,414 | 1,275 | 1,319 | 2.47 |
To determine specificity, the amplicons for mouse mRNA obtained by RT-PCR were sequenced with the following results:
aromatase amplicon sequence (sense chain):
aattcgccctttgaacatcggaagaatgcacaggctcgagtacttccctaagcccaatgaatttacccttgaaaactttgagaagaatgttccctacaggtactttcagccttttggctttgaagggcgaattcc
Sequence of aromatase from GenBank(NM007810):
ctaacatcat tctgaacatcggaagaatgc acaggctcga gtacttccct aagcccaatg aatttaccct tgaaaacttt gagaagaatg ttccctacaggtactttcag ccttttggctttgggccccg
TBP Amplicon sequence (antisense chain):
aattcgcccttccgtaaggcatcattggactaaagatgggaattccaggagtcatggcgccctgtggggaggccaagccctgagcataaggtggaaggctgttgttctggtccaagggcgaatt
Sequence of TBP from GenBank (BC016476):
aagaaaggga gaatcatgga ccagaacaac agccttccac cttatgctca gggcttggcc tccccacagg gcgccatgac tcctggaatt 301 cccatcttta gtccaatgat gccttacggcacaggactta
According to the above data, it is clear that amplicons are specific to mouse aromatase mRNA and to mouse TBP mRNA.
Having validated the assay for mouse tissue, we then determined the relative levels of aromatase in MMTV aromatase transgenic mice (i.e., breast specific expression) and in the breast tissues from wild type mice. Data are shown in online Table I and summarized here. The mean of the results for the aromatase transfected mouse mammary tissue were 60-fold higher (Mean ± SEM, 4.17 ± 0.56) than in the wild type mammary glands(-Mean ± SEM, 0.07 ± 0.033) and this represented a highly statistically significant difference (P < 0.0001).
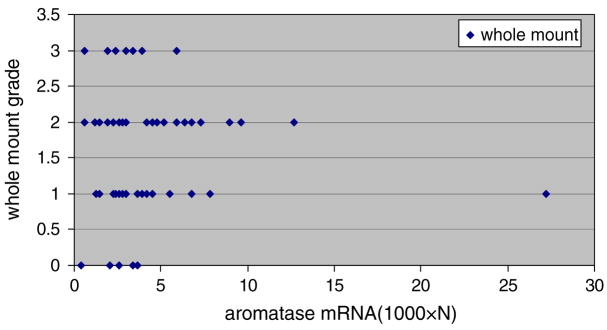
We were impressed that the range of levels of aromatase in breast tissue from transgenic animals was substantial (i.e., range 0.4–27) and that the degree of breast development in transgenic animals also varied (See Fig. 4). This raised the question whether the amount of aromatase transgene expression correlated with the degree of breast development. As shown in Fig. 4 and online Table II, there appeared to be no such correlation.
Fig. 4.
Aromatase activity for each level of breast differentiation classified subjectively from whole mounts of the breast on a scale of 0–3 (see “Materials and methods” for definitions of grades 0–3)
Since there was a 60-fold difference in aromatase expression (i.e., 0.4–27) in the breast tissue of transgenic animals, we postulated that this might reflect an influence of circulating hormone levels on the activity of the MMTV promoter. For this reason we treated animals with testosterone propionate (0.25 mg per day IM), (a hormone known to stimulate MMTV promoter activity) or corn oil as a control for 5 days and assessed its effect on aromatase expression. There were no significant differences in mammary mRNA (1000 × N) between pre-treatment and post-treatment in the testosterone propionate group (aromatase activity, Mean ± SEM: 4.77 ± 0.90 in pre-treatment and 2.95 ± 0.79 in post-treatment, respectively, P = NS). Pre-treatment and post-treatment levels in the corn oil group were 3.25 ± 0.84 and 4.58 ± 1.50, respectively, P = NS as shown in online Table III. These results suggested that the variation in aromatase expression in breast tissues of the transgenic mice probably reflected subtle genetic differences in animals that were not exhaustively inbred and maintained as a heterozygous colony since male homozygous animals develop Leydig cell tumors and are sub- or infertile [34].
To determine the tissue specificity of the aromatase promoter and whether the transgene was over-expressed in tissues other than breast, we then compared aromatase message levels in various tissues of transgenic and wild type animals. Surprisingly, (Table 3) the transgene was over-expressed in each tissue except ovary and adrenal although not to the same extent as in the breast tissue.
Table 3.
Comparison of aromatase in various tissues between wild-type and MMTV-transgenic mice
| Tissue type | Wild-type mouse (n = 5) Mean ± SEM (1000 × N) | MMTV-transgenic mouse (n = 5) Mean ± SEM (1000 × N) | P value |
|---|---|---|---|
| Ovary | 489.60 ± 158.8 | 264.60 ± 61.0 | 0.223 |
| Uterus | 0.14 ± 0.058 | 21.80 ± 4.8 | 0.002 |
| Adrenal | 2.30 ± 1.11 | 1.31 ± 0.43 | 0.445 |
| Liver | 0.25 ± 0.15 | 5.52 ± 2.93 | 0.111 |
| Kidney | 0.40 ± 0.35 | 0.97 ± 0.19 | 0.196 |
| Mammary | 0.03 ± 0.009 | 5.02 ± 2.10 | 0.046 |
Human aromatase determinations
To demonstrate the validity of the human reactions, we examined the PCR amplification and melt curves for human aromatase which are shown in online Figs. 2 and 3. To assess the level of sensitivity, we utilized the human JAR cell line which is known to contain substantial levels of aromatase and made serial dilutions of RNA extracted from a large pool of cells (see “Materials and methods”). In this human cell model system, we detected aromatase in cells diluted down to the amount of RNA contained in one cell (Table 4).
Table 4.
JAR cells and respective aromatase activity (Ct)
| Cell number | Ct |
|---|---|
| 100,000 cells | 13.1 |
| 10,000 cells | 16.7 |
| 1000 cells | 20.4 |
| 100 cells | 24.1 |
| 10 cells | 27.5 |
| 1 cell | 31.7 |
Human tissue
We assayed aromatase message in 23 ductal lavage samples, 10 breast biopsies and in 3 reduction mammoplasty samples. We detected aromatase in 7 of 23 ductal lavage samples and in an all breast biopsies and reduction mammoplasty specimens. Ten breast cancer samples were assayed with a mean value of 90 and a range of 20–120 (1000 × N). Three benign breast reduction mammoplasty specimens were assayed with levels of 0.008, 0.002, and 2.09. Finally, 23 ductal lavage specimens were assayed with 7 of 23 containing measurable aromatase activity ranging from 4.8 to 143.6 (Table 5).
Table 5.
Aromatase mRNA in 7 of 23 ductal lavage samples from human breast
| Sample No. | 1000 × N |
|---|---|
| 6288R | 67.0 |
| 5171L | 10.3 |
| 6170L | 4.8 |
| 5711L | 143.6 |
| 5381R | 62.5 |
| 6288L | 64.6 |
| 5171R | 14.0 |
Discussion
This study validated the use of a “nested” Q-PCR assay which is sufficiently sensitive to detect aromatase in a single JAR cell [33], in human breast biopsies, and in one third of ductal lavage samples (i.e., 7/23 samples). Without the initial amplification step, which increases sensitivity by 32,000–1,000,000-fold, it would not have been possible to detect aromatase in the ductal lavage samples. The nested nature of the assay does not impede precision which approximated a CV of 5%. Specificity was maintained as demonstrated by the sequencing of the products of the Q-PCR reaction. We conclude that this assay will provide a powerful new tool to assess aromatase expression in ductal lavage samples, nipple aspirate fluid, and fine needle aspirates from women [30]. In the future, this method should provide a means to assess the role of aromatase in contributing to breast density and to examine the utility of aromatase message as a biomarker for breast cancer risk.
During the process of validation of the nested Q-PCR assay, we learned much about the expression of aromatase in a transgenic mouse model. Prior data suggested only a three- to fourfold enhancement of aromatase in the breast tissue of this transgenic animals based on conventional PCR [31, 32]. However, recent studies from Tekmal’s group also show that using more sensitive quantitative real PCR the differences in aromatase levels between wild type and transgenic animals were more than 50-fold at the RNA levels and protein levels were more than tenfold [35].
It was surprising to us that the degree of over-expression of aromatase in breast tissue from transgenic animals varied substantially as did the degree of ductal development (Fig. 3) and E2 levels (unpublished data E. Cavalieri and E. Rogan, Eppley Cancer Center). We considered the possibility that this might reflect the action of endogenous hormones on MMTV expression. This did not appear to be the case since administration of testosterone propionate did not change the level of aromatase expression. We now consider it likely that the variation reflects subtle differences in the genetic backgrounds of the animals which may not be sufficiently inbred at this point in their breeding career. In addition, the zygocity of the colony is mixed since homozygous males are subfertile or infertile due to increased circulating estrogen levels and these animals form Leydig cells tumors [35].
Use of mouse tissues allowed us to fully validate the nested Q-PCR assay using multiple samples and various parameters to study. This is not practical with valuable human tissues. However, we believe that the validation in the mouse tissues provides sufficient validation for human tissues, since the only difference was the precise human primers used.
With this highly sensitive assay, a number of human samples may now be assayed. We believe that fine needle aspirates will be the best means to assess breast tissue aromatase levels. Nipple aspirate and ductal lavage fluid provide an assessment of shed cells into the ductal lumina but not necessary a reflection of breast tissue itself. In addition, this method should facilitate using very small amounts of tissue obtained by laser capture dissection from frozen sections. With the availability of this assay, we now plan to systematically exam these possibilities.
Several weaknesses are apparent in the examination of our data. We did not have cell counts, protein level determinations, or DNA levels in the ductal lavage samples so that one could correlate aromatase expression with cellular composition of the fluids. We also did not assay nipple aspirates nor FNA samples to directly demonstrate feasibility of measuring aromatase in these samples. However, one would expect FNAs and nipple aspirates to contain substantially more cells then ductal lavage fluids. According, it is likely that our assay will be sufficiently sensitive to detect aromatase in these samples. Future studies are planned to examine these samples directly. We have not correlated aromatase message with enzyme levels in this study and will need to do this in the future. Finally, measurements of DNA in each sample to provide a denominator for aromatase will be preferable to use of TBP and this is also planned in future studies.
Acknowledgments
This study was funded by the Department of Defense Centers of Excellence Grant DAMD17-03-1-0229 and NIH/NCI grants (CA 75018, P30 CA 54174 (RRT)).
References
- 1.Santen RJ, Demers L, Ohorodnik S, et al. Superiority of gas chromatography/tandem mass spectrometry assay (GC/MS/MS) for estradiol for monitoring of aromatase inhibitor therapy. Steroids. 2007;72(8):666–671. doi: 10.1016/j.steroids.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Wang S, Paris F, Sultan CS, et al. Recombinant cell ultrasensitive bioassay for measurement of estrogens in post-menopausal women. J Clin Endocrinol Metab. 2005;90(3):1407–1413. doi: 10.1210/jc.2004-0766. [DOI] [PubMed] [Google Scholar]
- 3.Geisler J, Haynes B, Anker G, et al. Treatment with high-dose estrogen (diethylstilbestrol) significantly decreases plasma estrogen and androgen levels but does not influence in vivo aromatization in postmenopausal breast cancer patients. J Steroid Biochem Mol Biol. 2005;96(5):415–422. doi: 10.1016/j.jsbmb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Geisler J. Breast cancer tissue estrogens and their manipulation with aromatase inhibitors and inactivators. J Steroid Biochem Mol Biol. 2003;86(3–5):245–253. doi: 10.1016/s0960-0760(03)00364-9. [DOI] [PubMed] [Google Scholar]
- 5.Lipton A, Santner SJ, Santen RJ, et al. Aromatase activity in primary and metastatic human breast cancer. Cancer. 1987;59(4):779–782. doi: 10.1002/1097-0142(19870215)59:4<779::aid-cncr2820590419>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 6.van Landeghem AA, Poortman J, Nabuurs M, et al. Endogenous concentration and subcellular distribution of estrogens in normal and malignant human breast tissue. Cancer Res. 1985;45(6):2900–2906. [PubMed] [Google Scholar]
- 7.Vermeulen A, Deslypere JP, Paridaens R, et al. Aromatase, 17 beta-hydroxysteroid dehydrogenase and intratissular sex hormone concentrations in cancerous and normal glandular breast tissue in postmenopausal women. Eur J Cancer Clin Oncol. 1986;22(4):515–525. doi: 10.1016/0277-5379(86)90121-5. [DOI] [PubMed] [Google Scholar]
- 8.Fishman J, Nisselbaum JS, Menendez-Botet CJ, Schwartz MK. Estrone and estradiol content in human breast tumors: relationship to estradiol receptors. J Steroid Biochem. 1977;8(8):893–896. doi: 10.1016/0022-4731(77)90100-5. [DOI] [PubMed] [Google Scholar]
- 9.Pasqualini JR, Chetrite G, Blacker C, et al. Concentrations of estrone, estradiol, and estrone sulfate and evaluation of sulfatase and aromatase activities in pre- and postmenopausal breast cancer patients. J Clin Endocrinol Metab. 1996;81(4):1460–1464. doi: 10.1210/jcem.81.4.8636351. [DOI] [PubMed] [Google Scholar]
- 10.Thorsen T, Tangen M, Stoa KF. Concentration of endogenous oestradiol as related to oestradiol receptor sites in breast tumor cytosol. Eur J Cancer Clin Oncol. 1982;18(4):333–337. doi: 10.1016/0277-5379(82)90002-5. [DOI] [PubMed] [Google Scholar]
- 11.Mistry P, Griffiths K, Maynard PV. Endogenous C19-steroids and oestradiol levels in human primary breast tumour tissues and their correlation with androgen and oestrogen receptors. J Steroid Biochem. 1986;24(6):1117–1125. doi: 10.1016/0022-4731(86)90372-9. [DOI] [PubMed] [Google Scholar]
- 12.Edery M, Goussard J, Dehennin L, et al. Eur J Cancer. 1. Vol. 17. (Oxford); 1981. Endogenous oestradiol-17beta concentration in breast tumours determined by mass fragmentography and by radioimmunoassay: relationship to receptor content; pp. 115–120. [DOI] [PubMed] [Google Scholar]
- 13.Recchione C, Venturelli E, Manzari A, et al. Testosterone, dihydrotestosterone and oestradiol levels in postmenopausal breast cancer tissues. J Steroid Biochem Mol Biol. 1995;52(6):541–546. doi: 10.1016/0960-0760(95)00017-t. [DOI] [PubMed] [Google Scholar]
- 14.Millington DS. Determination of hormonal steroid concentrations in biological extracts by high resolution mass fragmentography. J Steroid Biochem. 1975;6(3–4):239–245. doi: 10.1016/0022-4731(75)90139-9. [DOI] [PubMed] [Google Scholar]
- 15.Bonney RC, Reed MJ, Davidson K, Beranek PA, James VH. The relationship between 17 beta-hydroxysteroid dehydrogenase activity and oestrogen concentrations in human breast tumours and in normal breast tissue. Clin Endocrinol. 1983;19(6):727–739. doi: 10.1111/j.1365-2265.1983.tb00051.x. [DOI] [PubMed] [Google Scholar]
- 16.Reed MJ, Aherne GW, Ghilchik MW, Patel S, Chakraborty J. Concentrations of oestrone and 4-hydroxyandrostenedione in malignant and normal breast tissues. Int J Cancer. 1991;49(4):562–565. doi: 10.1002/ijc.2910490415. [DOI] [PubMed] [Google Scholar]
- 17.Reed MJ. The role of aromatase in breast tumors. Breast Cancer Res Treat. 1994;30(1):7–17. doi: 10.1007/BF00682737. [DOI] [PubMed] [Google Scholar]
- 18.Peaker M, Taylor E. Oestrogen production by the goat mammary gland: transient aromatase activity during late pregnancy. J Endocrinol. 1990;125(1):R1–R3. doi: 10.1677/joe.0.125r001. [DOI] [PubMed] [Google Scholar]
- 19.Sourdaine P, Parker MG, Telford J, Miller WR. Analysis of the aromatase cytochrome P450 gene in human breast cancers. J Mol Endocrinol. 1994;13(3):331–337. doi: 10.1677/jme.0.0130331. [DOI] [PubMed] [Google Scholar]
- 20.Pauley RJ, Santner SJ, Tait LR, Bright RK, Santen RJ. Regulated CYP19 aromatase transcription in breast stromal fibroblasts. J Clin Endocrinol Metab. 2000;85(2):837–846. doi: 10.1210/jcem.85.2.6345. [DOI] [PubMed] [Google Scholar]
- 21.Santen RJ, Martel J, Hoagland M, et al. Demonstration of aromatase activity and its regulation in breast tumor and benign breast fibroblasts. Breast Cancer Res Treat. 1998;49:S93–S99. doi: 10.1023/a:1006081729828. [DOI] [PubMed] [Google Scholar]
- 22.Santner SJ, Pauley RJ, Tait L, Kaseta J, Santen RJ. Aromatase activity and expression in breast cancer and benign breast tissue stromal cells. J Clin Endocrinol Metab. 1997;82(1):200–208. doi: 10.1210/jcem.82.1.3672. [DOI] [PubMed] [Google Scholar]
- 23.Miller WR. Aromatase activity in breast tissue. J Steroid Biochem Mol Biol. 1991;39(5B):783–790. doi: 10.1016/0960-0760(91)90026-2. [DOI] [PubMed] [Google Scholar]
- 24.Yue W, Wang JP, Hamilton CJ, Demers LM, Santen RJ. In situ aromatization enhances breast tumor estradiol levels and cellular proliferation. Cancer Res. 1998;58(5):927–932. [PubMed] [Google Scholar]
- 25.Larionov AA, Berstein LM, Miller WR. Local uptake and synthesis of oestrone in normal and malignant postmenopausal breast tissues. J Steroid Biochem Mol Biol. 2002;81(1):57–64. doi: 10.1016/s0960-0760(02)00047-x. [DOI] [PubMed] [Google Scholar]
- 26.Miller WR, Howie F, Mason I. Aromatase and breast cancer (Review 45 refs Russian) Vopr Onkol. 2001;47(2):182–186. [PubMed] [Google Scholar]
- 27.James VH, McNeill JM, Lai LC, Newton CJ, Ghilchik MW, Reed MJ. Aromatase activity in normal breast and breast tumor tissues: in vivo and in vitro studies. Steroids. 1987;50(1–3):269–279. doi: 10.1016/0039-128x(83)90077-6. [DOI] [PubMed] [Google Scholar]
- 28.Reed MJ, Owen AM, Lai LC, et al. In situ oestrone synthesis in normal breast and breast tumour tissues: effect of treatment with 4-hydroxyandrostenedione. Int J Cancer. 1989;44(2):233–237. doi: 10.1002/ijc.2910440208. [DOI] [PubMed] [Google Scholar]
- 29.Key T, Appleby P, Barnes I, Reeves G, Endogenous Hormones Breast Cancer Collaborative Group. Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. J Natl Cancer Inst. 2002;94(8):606–616. doi: 10.1093/jnci/94.8.606. [DOI] [PubMed] [Google Scholar]
- 30.Sidawy MK, Stoler MH, Frable WJ, et al. Interobserver variability in the classification of proliferative breast lesions by fine-needle aspiration: results of the Papanicolaou Society of Cytopathology Study. Diagn Cytopathol. 1998;18(2):150–165. doi: 10.1002/(sici)1097-0339(199802)18:2<150::aid-dc12>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 31.Kirma N, Mandava U, Wuichet K, Tekmal RR. The effects of aromatase overexpression on mammary growth and gene expression in the aromatase x transforming growth factor alpha double transgenic mice. J Steroid Biochem Mol Biol. 2001;78(5):419–426. doi: 10.1016/s0960-0760(01)00121-2. [DOI] [PubMed] [Google Scholar]
- 32.Mandava U, Kirma N, Tekmal RR. Aromatase overexpression transgenic mice model: cell type specific expression and use of letrozole to abrogate mammary hyperplasia without affecting normal physiology. J Steroid Biochem Mol Biol. 2001;79(1–5):27–34. doi: 10.1016/s0960-0760(01)00133-9. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Chen S. Induction of aromatase gene expression in human placental choriocarcinoma (JAR) cells by phorbol esters. Biochim Biophys Acta. 1994;1218(1):48–54. doi: 10.1016/0167-4781(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 34.Fowler KA, Gill K, Kirma N, Dillehay DL, Tekmal RR. Overexpression of aromatase leads to development of testicular leydig cell tumors: an in vivo model for hormone-mediated TesticularCancer. Am J Pathol. 2000;156(1):347–353. doi: 10.1016/S0002-9440(10)64736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tekmal RR, Nair HB, Perla RP, Kirma N. HER-2/neu x aromatase double transgenic mice model: the effects of aromatase overexpression on mammary tumorigenesis. J Steroid Biochem Mol Biol. 2007;106:111–118. doi: 10.1016/j.jsbmb.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]