Abstract
We demonstrate that short, medium and long-range constraints can be extracted from proton mediated, rare-spin detected correlation solid-state NMR experiments for the microcrystalline 10.4 × 2 kDa dimeric model protein Crh. Magnetization build-up curves from cross signals in NHHC and CHHC spectra deliver detailed information on side chain conformers and secondary structure for interactions between spin pairs. A large number of medium and long-range correlations can be observed in the spectra, and an analysis of the resolved signals reveals that the constraints cover the entire sequence, also including inter-monomer contacts between the two molecules forming the domain-swapped Crh dimer. Dynamic behavior is shown to have an impact on cross signals intensities, as indicated for mobile residues or regions by contacts predicted from the crystal structure, but absent in the spectra. Our work validates strategies involving proton distance measurements for large and complex proteins as the Crh dimer, and confirms the magnetization transfer properties previously described for small molecules in solid protein samples.
Electronic supplementary material
The online version of this article (doi:10.1007/s10858-008-9229-3) contains supplementary material, which is available to authorized users.
Keywords: Catabolite repression histidine-containing phosphocarrier protein (Crh), Distance constraints, MAS, 3D Protein structure, Solid-state NMR spectroscopy
Introduction
Solid state NMR (ssNMR) is rapidly developing to become complementary to solution NMR and X-ray crystallography as a method to study three-dimensional structure and dynamics in peptides and proteins. Because experiments can be conducted on insoluble and non-crystalline material, ssNMR is particularly well suited to investigate fibrous or membrane proteins at atomic resolution. Uniformly or multiply labeled protein variants enable the access to structural information regarding the complete polypeptide sequence using multidimensional ssNMR experiments. Even if structure determination has been shown to be possible using ssNMR techniques, no general protocol has been established today. Indeed, the strong one-bond couplings often hamper the measurement of distance constraints between carbon spins, and several approaches have been proposed to circumvent this problem. Extensive labeling strategies (Hong et al. 1999), removing one-bond couplings by partial labeling, have been used for the measurement of constraints and structure determination of two proteins so far (Castellani et al. 2002; Zech et al. 2005). Selective recoupling methods have been forwarded as another alternative (Böckmann et al. 2002). Indeed, it has been shown that the spin system dynamics remain sensitive to the distance of interest in chemical shift selective experiments and can be well reproduced within a quantum-mechanical multiple-spin analysis (Sonnenberg et al. 2004), making it possible to measure long-range constraints in uniformly labeled proteins. Selective transfer from carbonyl to side-chain carbons (Ladizhansky et al. 2004) or between nitrogen and carbon spins (Jaroniec et al. 2001) is another attractive approach for the measurement of long-range constraints. Recent work indicates that proton driven spin diffusion (Marulanda et al. 2005; Grommek et al. 2006) techniques are less sensitive to dipolar truncation. As an alternative to the measurement of carbon or nitrogen distances, a set of multidimensional NMR experiments for structure elucidation of proteins under MAS has been proposed, which relies on the measurement of proton distances (Heise et al. 2005). It uses proton homonuclear transfers, bracketed by 13C and/or 15N evolution times for the sake of spectral resolution. This scheme has been shown to be able to produce high resolution spectra connecting carbon or nitrogen spins through space (Lange et al. 2002, 2003), and we already applied this scheme successfully to the identification of the dimer interface of the Crh protein (Etzkorn et al. 2004). Most recent work shows that these heteronucleus-edited, proton relayed distance measurements can in principle be used for calculations of the 3D structure of solid proteins (Lange et al. 2005; Seidel et al. 2005). This approach is highly attractive, as it can be carried out on one uniformly labeled sample.
Here we explore the potential of this approach to collect local and long-range structural constraints for the larger and more complex 2 × 10.4 kDa Crh model protein in its dimeric, domain swapped form. This protein has proven to be an excellent model for solid state NMR methods developments (Carravetta et al. 2003; DePaëpe et al. 2003; Duma et al. 2003; Ernst et al. 2003; Juy et al. 2003; Lesage et al. 2003; Etzkorn et al. 2004; Giraud et al. 2004, 2005, 2006; Böckmann 2006; Böckmann et al. 2005; Lesage et al. 2006) as its solid state NMR chemical shifts and X-ray 3D structure (Böckmann et al. 2003) are available for reference. We show that a large number of proton-mediated constraints can be derived from CHHC and NHHC spectra at different mixing times. Information on side chain conformation and secondary structure, contained in intra-residue and sequential contacts, as well as long-range interactions constraining the 3D fold can be extracted from these spectra. Magnetization build-up curves confirm that the concept, established on small molecules, is valid also for proteins, contain detailed distance information and report on dynamic features of the molecule.
Materials and methods
Sample preparation
Crh was overexpressed with a C-terminal LQ(6×His) extension as described previously (Galinier et al. 1997). Uniformly [13C,15N] labeled Crh was obtained by growing bacteria in Silantes growth media. The protein was purified on Ni-NTA agarose (QUIAGEN) columns followed by anion exchange chromatography on a Resource Q column (Penin et al. 2001). Crh-containing fractions were dialyzed against 20 mM NH4HCO3. We used 20 mg/ml protein concentrations as obtained after dialysis in sitting drops of 150 μl deposited on a siliconated crystallization glass plate (Hampton). The same volume of a 20% solution of PEG 6000 and 0.02% NaN3 in 20 mM NH4HCO3 was added to the drops, over a 2 M NaCl reservoir solution. The plates were left at 4°C until a precipitate appeared (1–2 weeks). Resulting microcrystals were directly centrifuged at 2000g into 4 mm Bruker CRAMPS rotors and rotor caps were sealed. The sample used in this study contained ca. 20 mg of protein.
NMR spectroscopy
NMR experiments were performed on Bruker AVANCE DSX 500 MHz wide bore and 700 MHz standard bore spectrometers, equipped with double (1H, 13C) and triple resonance (1H, 13C, 15N) Bruker MAS probes, at spinning speeds of 11 kHz. All experiments were carried out between −10 and −5°C probe temperature. Ramped cross-polarization (Metz et al. 1994; Hediger et al. 1995) was used in all experiments to transfer proton magnetization to the 13C or 15N spins. High power proton decoupling using the SPINAL-64 decoupling scheme (Fung et al. 2000) was applied during evolution and detection periods. The relaxation delay between scans was 2.5 s. 2D CHHC correlation spectra (Lange et al. 2002) with longitudinal proton mixing using spin diffusion were recorded using a first 1H → 13C 1 ms cross polarization (CP) period followed by two short CP steps of either 125 or 150 μs. Different sets of spectra were recorded, with (1H, 1H) mixing times ranging from 50 to 400 μs. Acquisition times were 20 ms in t2 and 7.5 or 7.9 ms in t1 respectively for experiments performed on the 500 and 700 MHz spectrometer, corresponding to a total acquisition time of 39 or 46 hours. The spectral width was 350 ppm in the acquisition dimension and 90 ppm in the indirect dimension, centered in the aliphatic region. The 1H r.f. field during SPINAL-64 decoupling and CP was set to 71 kHz and 56 kHz respectively. The carbon r.f. field during CP was 34 kHz. 2D NHHC correlation spectra with longitudinal proton mixing under spindiffusion were recorded using a first 1H → 15N 900 μs CP period. Short heteronuclear CP contact times tNH and tCH were 200 and 100 μs, respectively. Experiments were conducted with (1H, 1H) mixing times from 15 to 100 μs. Acquisition times were 30 ms in t2 and 10.5 ms in t1. The total acquisition time was either 35 or 45 hours. The spectral widths in the 13C and 15N dimensions were 350 ppm and 60 ppm, respectively. The 1H decoupling power was set to 71 kHz. R.f. field strengths for 15N and 13C during CP were 38 and 34 kHz, respectively.
Results and discussion
The recorded 2D CHHC and NHHC spectra show a large number of cross signals, including intra-residue, sequential, medium-range (2 ≤ ∣j–i∣ < 5) and long-range contacts (∣j–i∣ ≥ 5). A detailed analysis of these signals allows the extraction of distance constraints, to different degrees of precision, as we will show in the following.
As only very few carbon chemical shifts are unique when considering a resolution limit at best of ±0.25 ppm (corresponding to the experimental linewidth of 0.5–1 ppm), all 1H–1H contacts in the different NHHC and CHHC spectra presented in this work were identified according to the Crh dimer crystal structure (PDB code 1mu4 (Juy et al. 2003)) and the chemical-shift assignments for microcrystalline Crh (Böckmann et al. 2003). Correspondingly, isolated cross signals were assigned if their chemical shift was within ±0.25 ppm from a unique contact predicted from the crystal structure and associated to two protons with an interatomic distance smaller than 5 Å.
Intra-residue contacts
Intra-residue contacts yield information on secondary structure, as well as on side chain conformations. The angles ϕ and χ or combinations thereof directly relate to inter-proton distances as HN–Hα, HN–Hβ, Hα–Hβ, Hα–Hγ, Hβ–Hγ, which we denote here, in compliance with common practice in solution NMR, dNα dNβ, dαβ, dαγ, dβγ (Markley et al. 1998). To evaluate the correlation between cross peak intensities and distances in detail, magnetization build-up characteristics were measured. This analysis is possible in a more quantitative manner for heteronuclei with only one proton attached. This includes the amide protons, all Hα protons but Gly; Val, Thr, Ile Hβ, and Leu Hγ. Indeed, as NHHC and CHHC correlation experiments are based on rare-spin detection, there is no chemical shift signature for protons; as a consequence, these experiments do not allow the distinction between protons located on the same carbon or nitrogen. For a CH2 group for example, it is likely that both protons are affected in a different manner by the polarization transfer from an amide proton, and the detected signal will be the sum of two distinct transfers.
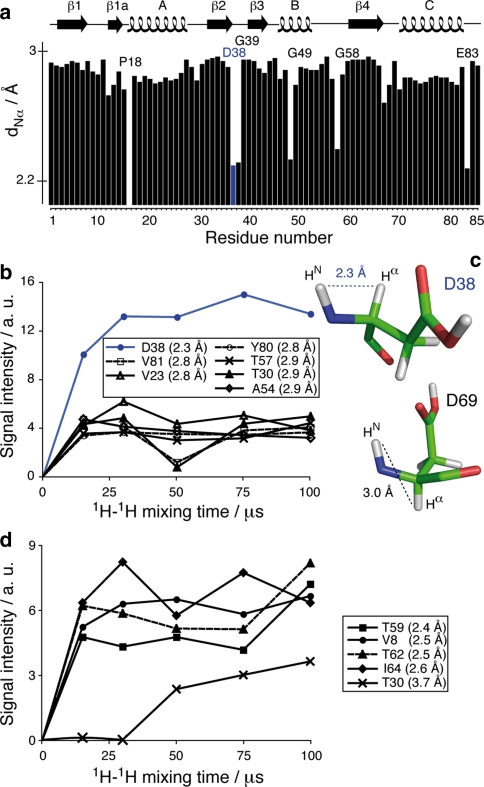
The intra-residue 1H–1H distance dNα between the amide and α proton is directly related to the torsion angle ϕ. However, no statistically significant variations are observed between α-helix and β-sheet secondary structures, from around 2.6 to 2.8 Å. This is illustrated in Fig. 1a, where dNα is plotted against the amino acid sequence for the Crh protein. In addition to glycines, which have a shorter dNα due to their two Hα protons, residues Asp38 and Glu83 show unusual dNα values, around 2.2 Å. Figure 1b displays magnetization build-up curves for isolated N–Cα cross signals in NHHC spectra. Stronger signals are observed for Asp38 at all mixing times, allowing distinction between the cis conformation of the HN, Hα protons of this residue from residues in trans conformation showing lower signal intensities. Due to spectral overlap, no build-up curve could be obtained for Glu83.
Fig. 1.
(a) Intra-residue HN–Hα distances dNα as a function of the residue number, according to the crystal structure. (b) Build-up curves for resolved intra-residue N–Cα cross signals in NHHC spectra. (c) Residues Asp38 and Asp69 as examples for typical amino acid showing HN–Hα in cis and trans conformations. (d) Build-up curves for resolved N–Cβ cross signals in NHHC spectra. For the sake of clarity, connecting lines are shown in the graphs. All indicated distances were measured on the crystal structure
The same type of analysis can be conducted on intra-residue HN–Hβ correlations; the corresponding distance dNβ is distributed between 2 and 4 Å for the here considered Ile, Thr and Val residues showing a unique β-proton. We could identify isolated cross signals for Val8, Ile64, Thr59 and 62, where dNβ is around 2.5 Å, and for Thr30, with dNβ of 3.6 Å. The magnetization build-up curves are shown in Fig. 1d. The magnetization build-up for short distances is accordingly faster, enabling to distinguish short distances dNβ of 2–3 Å from longer distances (3–4 Å).
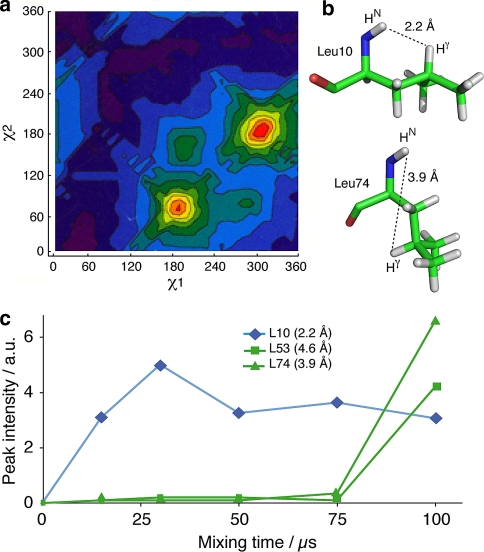
For intra-residue contacts involving Hγ protons, the proton-proton distance dNγ depends on the torsion angle ϕ, as well as on the values of χ1 and χ2 (Markley et al. 1998). As ϕ values are constant for the large majority of residues, dNγ mainly depends on the two side chain angles χ1 and χ2. The conformations of side chain atoms have been studied in detail (Ramachandran et al. 1963), and are described in rotamer libraries which are compiled in reference databases (Lovell et al. 2000; Dunbrack et al. 1997). We here concentrate on leucine residues, since this is the only amino-acid which possesses a unique Hγ. For these residues, two rotamers dominate (Lovell et al. 2000) with a probability of 59 % for (χ1 = −65°, χ2 = 174°) and 29% for (χ1 = −177°, χ2 = 65°), as shown in Fig. 2a. The distances dNγ corresponding to these two conformers are around 2.2 Å and 3.9 Å (Fig. 2b). Among the ten leucine residues of Crh, three have isolated N–Cγ cross peaks, and their magnetization build-up curves are shown in Fig. 2c. The build-up for Leu10, corresponding to the shorter distance of 2.2 Å, is faster than those from Leu53 and Leu74, whose cross signals only appear at mixing times of 100 μs. This shows that the magnetization build-up correlates well with interatomic distances, and allows the direct distinction between the two major rotamers in leucine residues.
Fig. 2.
(a) Major conformers observed for leucine residues (Lovell et al. 2000). (b) Conformations and distances corresponding to the major conformers. (c) Magnetization build-up curves for resolved Ni–Cγi cross signals in NHHC spectra. For the sake of clarity, connecting lines are shown in the graph
Intra-residue cross signals containing conformational information are also present in CHHC spectra. Indeed, contacts involving magnetization transfer between Hα and Hβ are readily observed in the CHHC spectrum shown in Fig. 3a. Cα–Cβ cross signals are indicated with circles, in blue for residues with dαβ from 2.2 to 2.7 Å, and in red for those between 2.7 and 3.1 Å. Signals corresponding to alanine residues are shown in gray. One can observe cross signals for all residues showing short 1H–1H distances, with the exception of Arg17, Val55 and Asp69. Val55 and Asp69 are located in loops, and Arg17 already shows weak cross signal intensities in PDSD spectra, indicating increased dynamics. As given from bond geometry, distances larger than 2.7 Å are exclusively found for residues with only one Hβ proton. Except for Val33 and Thr57, these residues show very weak signals or are absent in the spectrum taken with 100 μs mixing time. Figure 3b shows magnetization build-up curves corresponding to magnetization transfers between Hα and the unique Hβ of Thr, Val and Ile residues with resolved cross signals. Magnetization build-up is faster for residues Val2, Val42, Val61 (which Cα, Cβ resonate at the same frequency) and Thr12, whose Hα–Hβ are in gauche conformation, and which reach maximum intensity at 100 μs mixing time. Hα–Hβ of residues Thr57, Val8, Val23 and Ile64, which are in trans conformation, show significantly slower magnetization build-up. In the case of a two-spin system, the 1H–1H distances corresponding to the experimental magnetization build-up curves shown in Fig. 3b are in good agreement with theoretical curves shown in green (Lange et al. 2003). In Fig. 3c, we compare the magnetization build-up curves for residues with two protons attached to the β-carbon. Globally, the magnetization build-up still correlates well to the expected behavior, as most of the experimental curves fall into the 2.6–2.8 Å distance range predicted theoretically.
Fig. 3.
(a) Extract of the Cα–Cβ region of the CHHC spectrum recorded at 700 MHz with a mixing time of 100 μs and short CP steps of 125 μs. Indicated are all intra-residue Cα–Cβ cross signals; blue circles corresponding to dαβ < 2.7 Å, and red circles to dαβ > 2.7 Å. In light grey are shown alanine cross signals which are not further analyzed since their Hα–Hβ distance does not show a conformational dependence. Assignments are given for all residues with long Hα–Hβ distances, as well as for the three pairs having short dαβ, but for which no cross signal is observed. (b) Magnetization build-up curves for resolved cross signals from residues with only one β-proton. In green are shown theoretical results obtained by calculation of an exponential signal build-up using a 1H–1H distance of 2.2, 2.6, 3.0, 3.4 and 3.8 Å respectively (Lange et al. 2003). (c) Magnetization build-up curves for resolved Cα–Cβ signals with two protons attached, as well as the same theoretical magnetization build-up curves as in (b). Only the shortest Hα–Hβ distance is indicated in the case of carbons with two β-protons attached. For the sake of clarity, connecting lines are shown in the graphs
Considering intra-residue Hα–Hγ contacts of the leucine residues, with 1H–1H distances ranging from 2.9 to 3.3 Å, the isolated intra-residue Cα–Cγ signals of Leu10, Leu14 and Leu53 are absent from the spectrum shown in Fig. 3a, which is consistent with the observation of Hα–Hβ contacts mainly up to a distance of 2.7 Å in this spectrum, as discussed above.
Sequential contacts
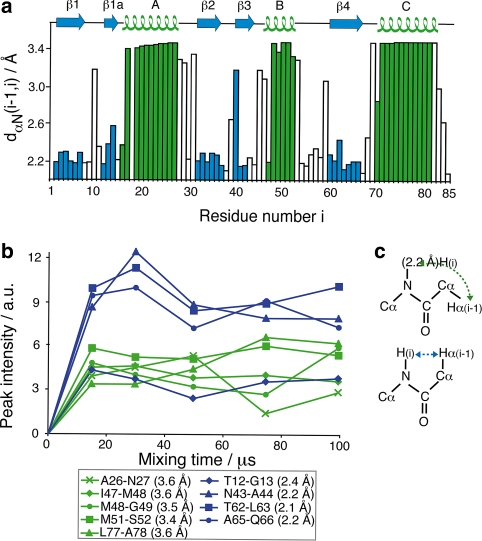
According to the previously described protocol, 46 resolved sequential contacts could be identified in NHHC spectra between the backbone amide hydrogen of residue i and hydrogens bonded to carbons of residue i−1. Among these contacts, the distance dαN between the amide hydrogen i and the α-proton of carbon i−1 directly correlates to the value of the torsion angle ψ (Billeter et al. 1982) and enables the identification of α-helical or β-sheet secondary structure. Figure 4a shows the values of dαN as observed in the Crh X-ray structure (Juy et al. 2003), together with the corresponding secondary structure. Residues involved in β-sheet conformation show short dαN, those in α-helical conformation long dαN. For loops and turns, there is no direct correlation with the inter-nuclear distance. Build-up curves for resolved peaks encoding sequential HN(i)–Hα(i−1) contacts are shown in Fig. 4b. At very short mixing times, direct nitrogen to alpha-proton magnetization transfer seems to dominate the signals, resulting from the poor selectivity of the 200 μs N–H CP transfer step. With longer proton mixing times, the gain of magnetization via direct transfers over the short 1H–1H distances becomes clear-cut. Signal intensities at mixing times around 30 μs allow the distinction between short and long distances. All sequential HN(i)–Hα(i−1) pairs with long distances show weak peak intensities; and peaks corresponding to short distances show stronger signals. The only exception is the Thr12-Gly13 pair, which shows a weak intensity despite the short HN–Hα distance. This could be due to increased dynamics of this residue pair, which is located in the hinge region of the domain-swapped dimer.
Fig. 4.
(a) Hα(i−1)–HN(i) distance dαN as a function of the residue number, according to the crystal structure. (b) Magnetization build-up curves corresponding to sequential contacts involving residues in α-helices (green) or in β-sheets (blue). (c) Sketches of α-helical (top) and β-sheet (bottom) conformation. For the sake of clarity, connecting lines are shown in the graph. All indicated HN–Hα distances were measured on the crystal structure
In CHHC spectra, numerous sequential contacts between α-carbons can be identified. The corresponding sequential Hα–Hα distances dαα(i,i + 1) in the Crh protein range from 4.26 to 4.88 Å. As a consequence, the difference between the distance dαα(i,i + 1) for the different secondary elements is small. Moreover, although the sequential Hα–Hα contacts involve carbon atoms with only one proton attached, they correspond to 1H–1H distances longer than 4 Å. The observed 1H–1H contacts could therefore result from relayed transfer mechanisms involving other protons, as will be discussed further below. It is thus difficult to extract detailed information on the secondary structure from the presence or absence of sequential Hα–Hα cross signals at different (1H,1H) mixing times. As the longer distances correspond to proton pairs in α-helical secondary structure, only few sequential contacts in helices A and C can be detected even at relatively short (1H,1H) mixing times, whereas for helix B, no signals at all are observed even at longer mixing times (see Fig. 11 in the supporting information). The absence of cross signals for helix B is probably due to increased dynamics observed for this helix (see also below) (Favier et al. 2002).
Medium and long-range contacts
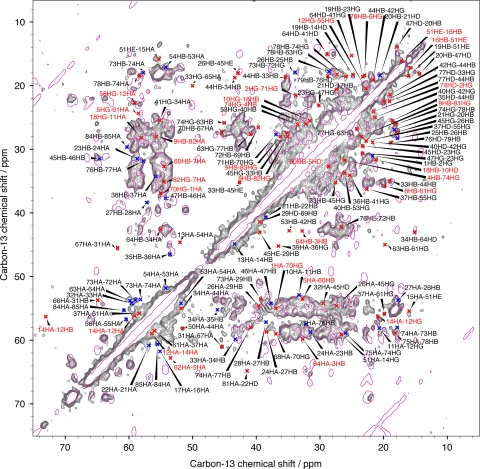
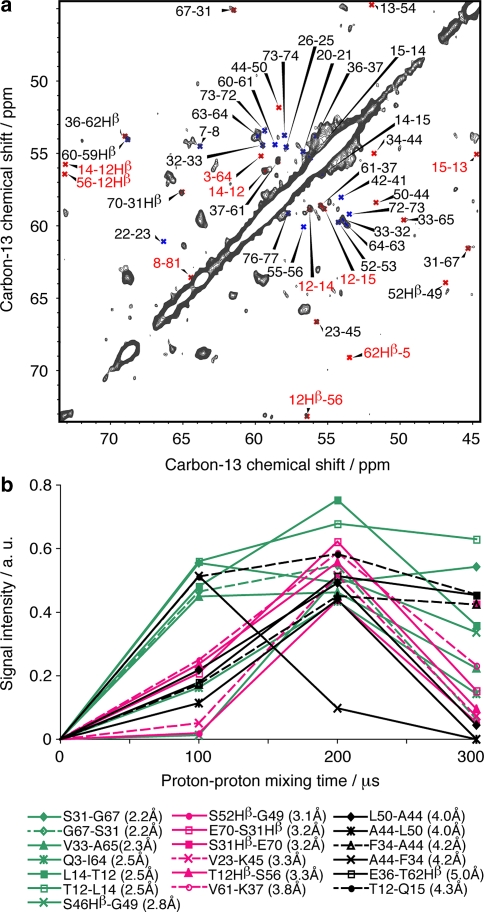
Tertiary structure determination by NMR spectroscopy relies to a large degree on the observation of medium and long-range contacts, which serve a critical role in structure calculations for the determination of the 3D fold. Figure 5 shows a CHHC spectrum with the assignment of the resolved cross signals. The spectrum was recorded at 500 MHz with 150 μs mixing time. It is superimposed on a proton-driven spin diffusion (PDSD) spectrum recorded with 20 ms mixing time showing only intra-residue cross signals. In this CHHC spectrum, a total of 125 CHHC signals connecting protons up to a distance of 5 Å could be identified, representing 34 sequential (27%), 17 medium-range (14%) and 74 long-range constraints (59%), amongst which were identified 30 inter-monomer constraints. Correlations overlapping with intra residue cross signals were not considered here.
Fig. 5.
CHHC spectrum recorded at 500 MHz with a mixing time of 150 μs. Sequential contacts are indicated in blue, medium and long-range contacts in red. Red labels correspond to inter-monomer contacts. A proton-driven spin diffusion (PDSD) spectrum recorded with 20 ms mixing time is shown in pink with one contour line
The NHHC spectrum shown in Fig. 6 was recorded with 100 μs (1H,1H) mixing time. A total of 138 contacts corresponding to 1H–1H distances up to 5 Å could be identified in this spectrum, amongst them 32 intra-residue (23%) 46 sequential (33%), and 21 medium- and long-range (44%).
Fig. 6.
NHHC spectrum recorded with a mixing time of 100 μs. Intra-residue and sequential contacts are indicated in blue, medium- and long-range contacts in red. For the signal assignments, refer to Fig. 12 in the supporting information
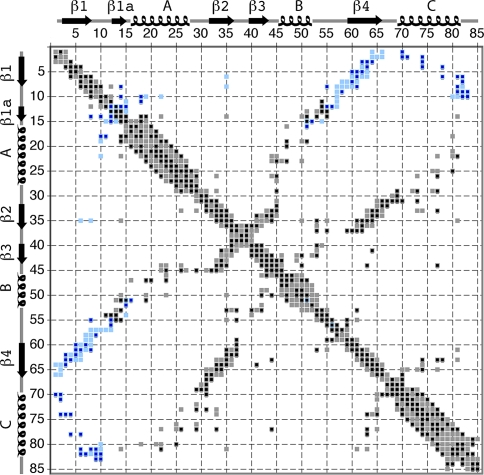
Figure 7 sums up all the contacts that could be identified in the different NHHC and CHHC spectra and shows that contacts are observed for all parts of the protein. A total of 164 intra-residue, 112 sequential, and 66 medium-range contacts could be identified. Among the 186 long-range contacts, we identified 62 inter-monomer contacts, mainly in the CHHC spectra.
Fig. 7.
Contacts identified are shown in black and are superimposed on predicted intra-monomer (light grey) and inter-monomer (light blue) contacts along the sequence of Crh, all corresponding to 1H–1H distances up to 5 Å
Contacts between α-protons give rise to cross signals within a delimited region in the CHHC spectra where no intra-residue correlations are located (except Hα-Hβ from the serines and threonines). With the exception of glycines, Hα–Hα contacts involve two carbon atoms with only one bound proton. To explore whether this allows for a detailed analysis even in the case of medium- and long-range contacts, we measured magnetization build-up curves for isolated signals with unique assignments, which are indicated in Fig. 8a. A large majority (about 80%) of the expected medium- and long-range Hα–Hα contacts corresponding to a 1H–1H distance up to 4 Å can be detected. However, roughly half of the signals are located close to the diagonal, and thus difficult to identify. The magnetization build-up curves, which are shown in Fig. 8b, can be roughly divided into two categories. Most curves indicating fast magnetization build-up correspond to distances shorter than 3 Å. Exceptions are the intra-monomer contact between Phe34 and Ala44 and the inter-monomer contact between Thr12 and Gln15, which give rise to fast magnetization build-up despite their long Hα–Hα distances of 4.17 and 4.33 Å, respectively. This might be due to relayed magnetization transfer mechanisms between the two spins (see also below). On the other hand, the short inter-monomer Hα–Hα distance of 2.45 Å between Gln3 and Ile64 leads to a slow build-up, probably due to increased dynamics near the N-terminal part of the protein. The contact between Ser46 and Gly49, corresponding to a relatively short Hα–Hα distance of 2.82 Å, also shows a slow build-up. Both residues belong to helix B, which shows increased dynamic behavior in the monomeric Crh variant (Favier et al. 2002). These observations indicate that besides by distance, magnetization build-up for medium and long-range contacts is scaled as well by the relative dynamics of the two protons, but allows still for a qualitative distance analysis. Additional magnetization build-up curves corresponding to long-range contacts and/or to carbons with several protons attached, and for which no clearcut correlation between magnetization build-up and distance can be observed are presented in Fig. 13 in the supporting information.
Fig. 8.
(a) Extract of a CHHC spectrum recorded at 700 MHz with 200 μs (1H,1H) mixing time. Except otherwise stated, indicated cross signals correspond to Hα–Hα contacts. Sequential contacts are shown in blue, medium- and long-range contacts in red. Labels for inter-monomer contacts are in red. (b) Magnetization build-up curves corresponding to Hα–Hα contacts are shown. Green curves correspond to 1H–1H distances shorter than 3 Å, pink curves to distances between 3 and 4 Å, black curves to distances longer than 4 Å
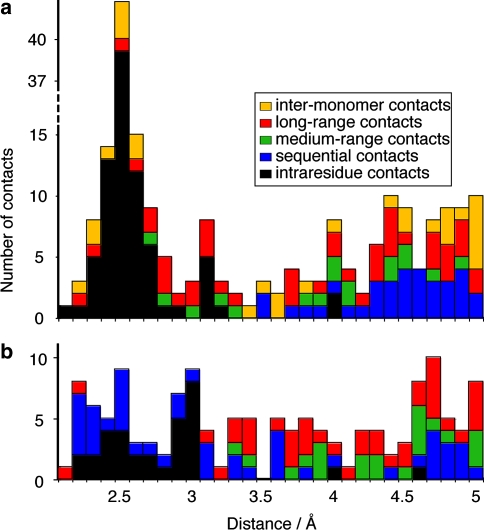
In order to evaluate the distribution of distances between the protons which give rise to the cross signals observed in CHHC and NHHC spectra, we plotted the number of identified correlation peaks as a function of the corresponding proton-proton distance, as shown in Fig. 9. We set the upper limit to 5 Å. We did not consider signals in crowded regions of the spectra nor peaks near the diagonal of the CHHC spectrum. Figure 9a shows all the contacts which could be identified in a CHHC spectrum recorded at 500 MHz with a (1H,1H) mixing time of 150 μs, and Fig. 9b shows the contacts for a NHHC spectrum recorded with a 100 μs mixing time. It can be seen that contacts all the way up to 5 Å can be identified on both spectra, and no clear limit is reached before. A detailed analysis taking into account longer distances is hampered by the associated increase in ambiguities. It is thus difficult to correlate the proton-proton mixing time with an upper distance limit corresponding to the maximal distance reached by 1H–1H spin diffusion. As in carbon correlation spectra, cross signals corresponding to longer 1H–1H distances often reflect relayed magnetization transfer mechanisms, and lead to a decrease in accuracy of the distance measurements.
Fig. 9.
(a) Number of identified correlation peaks as a function of the corresponding proton–proton distance in the CHHC spectrum recorded at 500 MHz at 150 μs mixing time and in the (b) NHHC spectrum at 100 μs mixing time. In both cases, only isolated signals were considered
We have analyzed possible relay mechanism in a more detailed manner for unique CH···HC contacts identified in the spectrum recorded at 500 MHz with 200 μs (1H,1H) mixing time. Magnetization transfer between protons is mostly direct up to 3–3.5 Å, as previously observed by Baldus and coworkers (Lange et al. 2003). For longer distances, magnetization transfer is often relayed via a third spin. A succession of two short 1H–1H transfer steps, generally an intra-residue contact, followed by an inter-residue contact, may often result in the observation of long inter-residue contacts. Relayed transfers seem however not to introduce significant bias in distances; as can be seen from the buildup curves, the number of attached protons, as well as dynamic behavior (see below) play a preponderant role with respect to the degree of correlation between cross signal intensity and inter-nuclear distance.
Absence of predicted signals
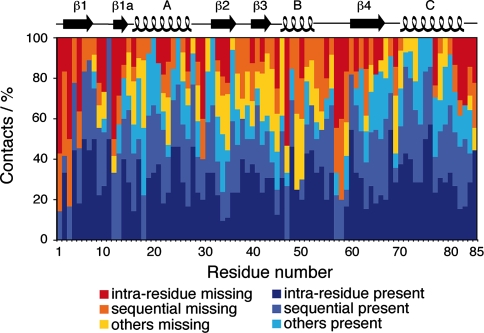
In order to identify conditions that lead to an attenuation in magnetization transfers, we analyzed how well the experimental data match the theoretical predictions from the crystal structure. We analyze the absence of predicted contacts up to 4 Å in the NHHC and CHHC spectra. A contact was considered to be missing when no signal was observed in the spectrum within a chemical shift range of ±0.5 ppm around the predicted peak. Possibly missing peaks in crowded regions were therefore not considered. We first evaluated the proportion of observed and missing NH ··· HC contacts for each residue of the protein (Fig. 10), considering predicted contacts up to a distance of 4 Å involving amide, α- and β-protons. Observed means here that there is signal present at the expected peak position, which might result in some bias for unresolved peaks. This analysis does not take into account 15N–13C cross signals resulting from intermonomer contacts, as a detailed analysis of these contacts is published elsewhere using a uniformly, but heterogeneously [15N:13C] labeled Crh sample (Etzkorn et al. 2004). NHHC experiments potentially yield information on all parts of the protein, as approximately 71% of the expected peaks can be observed, and the majority of residues show more than 50% of the expected contacts. The regions where less than 50% of the predicted contacts are observed include the N-terminal portion, β1a strand, helix B, loop 2 and the C-terminal region. Indeed, all these regions (with exception of the C-terminus) undergo major structural rearrangements on domain swapping (Juy et al. 2003). Increased dynamics have been observed for helix B in the monomeric protein and for loop 2 in the monomeric and dimeric protein (Favier et al. 2002; Giraud et al. 2004, 2005). Besides during proton spin diffusion, motional processes also reduce transfer efficiencies during cross polarization. Thus, methyl protons are difficult to polarize in short CP steps, which explains the absence of 65% methyl protons in the NHHC spectrum at100 μs mixing time.
Fig. 10.
Distribution of the different types of present and missing proton-proton contacts in the spectrum recorded at 100 μs mixing time, namely intra-residue, sequential and medium- and long-range contacts. Only isolated signals involving amide, α- and β-protons and corresponding to 1H–1H distances shorter than 4 Å were considered
We also analyzed predicted but not observed cross signals in the CHHC experiments. Missing cross signals in the distance range up to 4 Å mainly concern methyl groups (42%), which are difficult to polarize with short CP steps, but also labile sidechains of lysines and arginines (11%), or other Hγ (11%). Missing contacts corresponding to 1H–1H distances shorter than 4 Å, with the exception of methyl groups and arginine/lysine sidechains, are represented in Fig. 14 in the supporting information. Missing CH···HC contacts are mostly located in helix B, a result consistent with the identified missing NH···HC contacts. Several other missing signals are located in the hinge region, as well as in β1 and β4 strands and are also probably explained by dynamic features, these parts of the protein being involved in the dimer interface. As in solution NMR, the ultimate proof that dynamic behaviour is at the origin of missing signals can only be brought (else than by backcalculation of expected signals from an existing structure) by dynamic measurements, which can confirm that regions with missing cross peaks indeed show increased dynamics. Several of such measurements have been proposed for solid proteins over the past years (Giraud et al. 2004, 2005; Hologne et al. 2005, 2006; Giraud et al. 2006, 2007; Reif et al. 2006; Chevelkov et al. 2007a, b; Xue et al. 2007). Measurements at low temperatures could also be used to establish if dynamics is at the origin of missing signals; however, line broadening hampers measurements as soon as freezing occurs.
Conclusion
We have shown here that proton mediated rare-spin detected experiments yield distance information including intra-residue, short, medium and long-range contacts, and are thus valuable for constraining 3D protein structures. In contrast to carbon correlation spectra, every single cross signal present in the spectra potentially contains distance information, as has already been shown for proton contacts in solution NMR. Cross signals connecting carbon or nitrogen spins with only one proton attached reflect very well the distance encompassed. This is especially true for intra-residue and sequential connections, and, to a slightly lesser extent, also for medium- and long-range contacts. In general, build-up curves for local contacts correlate better to the distances than long-range constraints. The dipolar couplings at the origin of the cross signals depend on the distance as well as the orientation of the spin-pair. If molecular motion occurs during the mixing time, it may provoke a change in orientation leading to partial averaging of the dipolar coupling, resulting in signal attenuation. This is typically the case for cross signals involving lysine and arginine side chains. Larger scale flexibility between secondary structure elements is often involved in function, like molecular recognition, catalysis site function or conformational change in the global folding of the protein, and may lead to the absence of predicted cross signals, as illustrated above. Even if dynamic behavior might attenuate magnetization transfer, we did not observe, in this microcrystalline Crh protein sample, regions completely devoid of corresponding cross signals. This situation seems similar to solution NMR, where less contacts are observed for mobile regions of the protein, without however compromising structural information for well-folded regions of the protein. Another factor, possibly limiting the direct extraction of distance classes from the magnetization build-up curves, is that medium- and long-range contacts may be more subject to relay mechanisms than intra-residue or sequential contacts, as already observed in solution NMR. Consequently, the model of an isolated spin pair is not valid any more, preventing a detailed comparison of the magnitude of the observed cross signals. Even if present, relay mechanisms are still less important for proton connectivities than in carbon correlation experiments, where magnetization transfer between directly bonded carbon spins is complete within a short time compared to the mixing times required for long-range magnetization transfers. Long-range information up to 5 Å can easily be detected in the here presented NHHC and CHHC spectra. It is difficult to evaluate if contacts involving longer distances are present as well, as spectral crowding and assignment ambiguities make an analysis difficult. Our analysis in Fig. 9 shows however that no clear limit is indicated by the cross signals which could be identified, and potentially cross signals corresponding to longer distances might be present. The accuracy of these constraints could however be limited by relayed magnetization transfer mechanisms, and might not add essential information for high-resolution structure determination. Sensitivity remains an issue in proton mediated experiments, but the necessary amount of sample and acquisition time should decrease with the use of higher fields.
We identified a large number of constraints in the CHHC and NHHC spectra for resolved signals, using the information available from the crystal structure and a maximum 1H–1H transfer range of 5 Å. Without structural information, unambiguous signal assignment is impossible for the high number of spins in the Crh protein combined with linewidths of 0.5–1.0 ppm, even if three-dimensional experiments would be used. Structure calculation for this size of proteins from proton-mediated (and also from carbon or nitrogen correlation) experiments will probably mainly be carried out using protocols including automated iterative assignment procedures to resolve ambiguities in the distance restraints, as implemented in software routines such as ARIA (Nilges 1995), SOLARIA (Fossi et al. 2005) or CYANA (Güntert et al. 1991). We will report on this for the Crh protein in a forthcoming paper.
Electronic supplementary material
A graph presenting Hα–Hα distances identified in CHHC spectra, assignments of the NHHC spectrum, additional magnetization build-up curves as well as an analysis of absent signals for the CHHC experiments are given.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work has been supported by the Centre National de la Recherche Scientifique (PICS no. 2424), the ANR (JC05_44957) and the French Ministry (ACI Biologie cellulaire moléculaire et structurale). CG acknowledges a Claudie-Heigneré post-doctoral grant.
Abbreviations
- ssNMR
Solid state NMR
- MAS
Magic angle spinning
- Crh
Catabolite repression HPr-like protein
- PEG
Polyethylene glycol
- CP
Cross polarization
- PDSD
Proton driven spin diffusion
- r.f.
Radio frequency
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s10858-008-9229-3) contains supplementary material, which is available to authorized users.
References
- Billeter M, Braun W, Wuthrich K (1982) Sequential resonance assignments in protein H-1 nuclear magnetic-resonance spectra – computation of sterically allowed proton proton distances and statistical-analysis of proton proton distances in single-crystal protein conformations. J Mol Biol 155:321–346 [DOI] [PubMed]
- Böckmann A (2006) Structural and dynamic studies of proteins by high resolution solid state NMR. C R Chim 9:381–392
- Böckmann A, McDermott AE (2002) Assignments and prospects for structure determination of solid proteins using MAS and isotopic enrichment. In: Grant DM, Harris RK (eds)The encyclopedia of NMR. Wiley, London
- Böckmann A, Lange A, Galinier A, Luca S, Giraud N, Juy M, Heise H, Montserret R, Penin F, Baldus M (2003) Solid-state NMR sequential resonance assignments and conformational analysis of the 2 × 10.4 kDa dimeric form of the Bacillus subtilis protein Crh. J Biomol NMR 27:323–339 [DOI] [PubMed]
- Böckmann A, Juy M, Bettler E, Emsley L, Galinier A, Penin F, Lesage A (2005) Water-protein hydrogen exchange in the micro-crystalline protein Crh as observed by solid state NMR spectroscopy. J Biomol NMR 32:195–207 [DOI] [PubMed]
- Carravetta M, Zhao X, Böckmann A, Levitt MH (2003) Coherence transfer selectivity in two-dimensional solid-state NMR. Chem Phys Lett 376:515–523 [DOI]
- Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H (2002) Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature 420:98–102 [DOI] [PubMed]
- Chevelkov V, Faelber K, Schrey A, Rehbein K, Diehl A, Reif B (2007a) Differential line broadening in MAS solid-state NMR due to dynamic interference. J Am Chem Soc 129:10195–200 [DOI] [PubMed]
- Chevelkov V, Zhuravleva AV, Xue Y, Reif B, Skrynnikov NR (2007b) Combined analysis of (15)N relaxation data from solid- and solution-state NMR spectroscopy. J Am Chem Soc 129:12594–12595 [DOI] [PubMed]
- DePaëpe G, Giraud N, Lesage A, Hodgkinson P, Böckmann A, Emsley L (2003) Transverse dephasing optimized solid-state NMR spectroscopy. J Am Chem Soc 125:13938–13939 [DOI] [PubMed]
- Duma L, Hediger S, Brutscher B, Böckmann A, Emsley L (2003) Resolution enhancement in multidimensional solid-state NMR of 13C-labeled proteins using spin-state selection. J Am Chem Soc 125:11816–11817 [DOI] [PubMed]
- Dunbrack RL, Cohen FE (1997) Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci 6:1661–1681 [DOI] [PMC free article] [PubMed]
- Ernst M, Detken A, Böckmann A, Meier BH (2003) NMR spectra of a micro-crystalline protein at 30 kHz MAS. J Am Chem Soc 125:15807–15810 [DOI] [PubMed]
- Etzkorn M, Böckmann A, Lange A, Baldus M (2004) Probing molecular interfaces using 2D magic-angle-spinning NMR on protein mixtures with different uniform labeling. J Am Chem Soc 126:14746–14751 [DOI] [PubMed]
- Favier A, Brutscher B, Blackledge M, Galinier A, Deutscher J, Penin F, Marion D (2002) Solution structure and dynamics of Crh, the Bacillus subtilis catabolite repression HPr. J Mol Biol 317:131–144 [DOI] [PubMed]
- Fossi M, Castellani F, Nilges M, Oschkinat H, van Rossum BJ (2005) SOLARIA: a protocol for automated cross-peak assignment and structure calculation for solid-state magic-angle spinning NMR spectroscopy. Angew Chem Int Ed Engl 44:6151–6154 [DOI] [PubMed]
- Fung BM, Khitrin AK, Ermolaev K (2000) An improved broadband decoupling sequence for liquid crystals and solids. J Magn Reson 142:97–101 [DOI] [PubMed]
- Galinier A, Haiech J, Kilhoffer MC, Jaquinod M, Stulke J, Deutscher J, Martin-Verstraete I (1997) The Bacillus subtilis crh gene encodes a Hpr-like protein involved in carbon catabolite repression. Proc Natl Acad Sci USA 94:8439–8444 [DOI] [PMC free article] [PubMed]
- Giraud N, Böckmann A, Lesage A, Penin F, Blackledge M, Emsley L (2004) Site-specific backbone dynamics from a crystalline protein by solid-state NMR spectroscopy. J Am Chem Soc 126:11422–11423 [DOI] [PubMed]
- Giraud N, Blackledge M, Goldman M, Böckmann A, Lesage A, Penin F, Emsley L (2005) Quantitative analysis of backbone dynamics in a crystalline protein from nitrogen-15 spin-lattice relaxation. J Am Chem Soc 127:18190–18201 [DOI] [PubMed]
- Giraud N, Sein J, Pintacuda G, Böckmann A, Lesage A, Blackledge M, Emsley L (2006) Observation of heteronuclear overhauser effects confirms the 15N-1H dipolar relaxation mechanism in a crystalline protein. J Am Chem Soc 128:12398–12399 [DOI] [PubMed]
- Giraud N, Blackledge M, Böckmann A, Emsley L (2007) The influence of nitrogen-15 proton driven spin diffusion on the measurement of nitrogen-15 longitudinal relaxation times. J Magn Reson 184:51–61 [DOI] [PubMed]
- Grommek A, Meier BH, Ernst M (2006) Distance information from proton-driven spin diffusion under MAS. Chem Phys Lett 427:404–409 [DOI]
- Güntert P, Qian YQ, Otting G, Müller M, Gehring W, Wüthrich K (1991) Structure determination of the Antp (C39----S) homeodomain from nuclear magnetic resonance data in solution using a novel strategy for the structure calculation with the programs DIANA, CALIBA, HABAS and GLOMSA. J Mol Biol 217:531–540 [DOI] [PubMed]
- Hediger S, Meier BH, Ernst RR (1995) Adiabatic passage Hartmann-Hahn cross polarization in NMR under magic angle sample spinning. Chem Phys Lett 240:449–456 [DOI]
- Heise H, Seidel K, Etzkorn M, Becker S, Baldus M (2005) 3D NMR spectroscopy for resonance assignment and structure elucidation of proteins under MAS: novel pulse schemes and sensitivity consideration. J Magn Reson 173:64–74 [DOI] [PubMed]
- Hologne M, Faelber K, Diehl A, Reif B (2005) Characterization of dynamics of perdeuterated proteins by MAS solid-state NMR. J Am Chem Soc 127:11208–11209 [DOI] [PubMed]
- Hologne M, Chen Z, Reif B (2006) Characterization of dynamic processes using deuterium in uniformly 2H,13C,15N enriched peptides by MAS solid-state NMR. J Magn Reson 179:20–28 [DOI] [PubMed]
- Hong M, Jakes K (1999) Selective and extensive 13C labeling of a membrane protein for solid-state NMR investigations. J Biomol NMR 14:71–74 [DOI] [PubMed]
- Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG (2001) Frequency selective heteronuclear dipolar recoupling in rotating solids: accurate (13)C-(15)N distance measurements in uniformly (13)C,(15)N-labeled peptides. J Am Chem Soc 123:3507–3519 [DOI] [PubMed]
- Juy M, Penin F, Favier A, Galinier A, Montserret R, Haser R, Deutscher J, Böckmann A (2003) Dimerization of Crh by reversible 3D domain swapping induces structural adjustments to its monomeric homologue HPr. J Mol Biol 332:767–776 [DOI] [PubMed]
- Ladizhansky V, Griffin RG (2004) Band selective carbonyl to aliphatic side chain distance measurements. J Am Chem Soc 126:948–958 [DOI] [PubMed]
- Lange A, Luca S, Baldus M (2002) Structural constraints from proton-mediated rare-spin correlation spectroscopy in rotating solids. J Am Chem Soc 124:9704–9705 [DOI] [PubMed]
- Lange A, Seidel K, Verdier L, Luca S, Baldus M (2003) Analysis of proton-proton transfer dynamics in rotating solids and their use for 3D structure determination. J Am Chem Soc 125:12640–12648 [DOI] [PubMed]
- Lange A, Becker S, Seidel K, Giller K, Pongs O, Baldus M (2005) A concept for rapid protein-structure determination by solid-state NMR spectroscopy. Angew Chem Int Ed Engl 44:2–5 [DOI] [PubMed]
- Lesage A, Böckmann A (2003) Water-protein interactions in microcrystalline Crh measured by 1H-13C solid-state NMR spectroscopy. J Am Chem Soc 125:13336–13337 [DOI] [PubMed]
- Lesage A, Emsley L, Penin F, Böckmann A (2006) Investigation of dipolar-mediated water-protein interactions in microcrystalline Crh by solid-state NMR spectroscopy. J Am Chem Soc 128:8246–8255 [DOI] [PubMed]
- Lovell SC, Word JM, Richardson JS, Richardson DC (2000) The penultimate rotamer library. Proteins 40:389–408 [DOI] [PubMed]
- Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. Eur J Biochem 256:1–15 [DOI] [PubMed]
- Marulanda D, Tasayco ML, Cataldi M, Arriaran V, Polenova T (2005) Resonance assignments and secondary structure analysis of E. coli Thioredoxin by magic angle spinning solid-state NMR spectroscopy. J Phys Chem B 18135–18145 [DOI] [PubMed]
- Metz G, Wu XL, Smith SO (1994) Ramped-amplitude cross-polarization in magic-angle-spinning Nmr. J Magn Reson Ser A 110:219–227 [DOI]
- Nilges M (1995) Calculation of protein structures with ambiguous distance restraints. Automated assignment of ambiguous NOE crosspeaks and disulphide connectivities. J Mol Biol 245:645–660 [DOI] [PubMed]
- Penin F, Favier A, Montserret R, Brutscher B, Deutscher J, Marion D, Galinier A (2001) Evidence for a dimerisation state of the Bacillus subtilis catabolite repression HPr-like protein, Crh. J Mol Microbiol Biotechnol 3:429–432 [PubMed]
- Ramachandran GN, Ramakrishnan C, Sasisekharan V (1963) Stereochemistry of polypeptide chain configurations. J Mol Biol 7:95 [DOI] [PubMed]
- Reif B, Xue Y, Agarwal V, Pavlova MS, Hologne M, Diehl A, Ryabov YE, Skrynnikov NR (2006) Protein side-chain dynamics observed by solution- and solid-state NMR: comparative analysis of methyl <sup>2</sup>H relaxation data. J Am Chem Soc 128:12354–12355 [DOI] [PubMed]
- Seidel K, Etzkorn M, Heise H, Becker S, Baldus M (2005) High-resolution solid-state NMR studies on uniformly [(13)C,(15)N]-labeled ubiquitin. Chembiochem 6(9):1638–1647 [DOI] [PubMed]
- Sonnenberg L, Luca S, Baldus M (2004) Multiple-spin analysis of chemical-shift-selective (13C, 13C) transfer in uniformly labeled biomolecules. J Magn Reson 166:100–110 [DOI] [PubMed]
- Xue Y, Pavlova MS, Ryabov YE, Reif B, Skrynnikov NR (2007) Methyl rotation barriers in proteins from 2H relaxation data. Implications for protein structure. J Am Chem Soc 129:6827–6838 [DOI] [PubMed]
- Zech SG, Wand AJ, McDermott AE (2005) Protein structure determination by high-resolution solid-state NMR spectroscopy: application to microcrystalline ubiquitin. J Am Chem Soc 127:8618–8626 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.