Summary
Yeast peroxisomes multiply by fission. Fission requires two dynamin-related proteins, Dnm1p and Vps1p. We show that Dnm1p-dependent peroxisome fission requires Fis1p, Caf4p and Mdv1p using an in vivo fission assay. Fluorescence microscopy of cells expressing GFP-tagged Caf4p and Mdv1p revealed that their association with peroxisomes relies on Fis1p.
Vps1p-dependent peroxisome fission occurs independently of these factors. Vps1p contributes most to fission of peroxisomes when cells are grown on glucose. Overexpression of Dnm1p suppresses the fission defect as long as Fis1p and either Mdv1p or Caf4p are present. Conversely, overexpression of Dnm1p does not restore the vacuolar fusion defect of vps1 cells, and Vps1p overexpression does not restore the mitochondrial fission defect of dnm1 cells. These data show that Vps1p and Dnm1p are part of independent fission machineries. Since the contribution of Dnm1p to peroxisome fission appears to be more pronounced in cells that proliferate peroxisomes in response to mitochondrial dysfunction, Dnm1p might be part of the mechanism that coordinates mitochondrial and peroxisomal biogenesis.
Keywords: Caf4, Mdv1, Dnm1, peroxisome
Introduction
Eukaryotic cells contain a set of functionally distinct membrane-bound compartments called organelles. The maintenance of a full set of organelles is fundamental to eukaryotic life. With every cell division, organelles are duplicated and segregated between daughter cells. Organelles multiply either by fission of existing organelles or by de novo formation.
Peroxisomes are organelles that are found in almost all eukaryotic cells. The number, size and shape of peroxisomes varies between different cell types and different environmental conditions, from small spherical organelles to large elaborate reticular structures. For instance, in Saccharomyces cerevisiae, peroxisomes proliferate in response to growth on the long-chain fatty acid, oleic acid. Recently, we have shown that in wild type cells peroxisomes multiply by fission of preexisting peroxisomes, but they can also form de novo in cells temporarily devoid of these organelles (Motley and Hettema, 2007).
The family of dynamin-related proteins (DRPs) has been implicated in multiple membrane remodelling events in eukaryotic cells. Whereas some DRPs are specific to a particular organelle, others seem to be more promiscuous. In yeast, Vps1p was identified as being required for transport of vacuolar hydrolases from Golgi to endosomes (Rothman et al., 1990; Vater et al., 1992). Later it was also shown to be involved in vacuole fusion (Luo and Chang, 2000; Peters et al., 2004), secretion of a subset of secretory proteins (Harsay and Schekman, 2002) and to be required for normal peroxisome abundance (Hoepfner et al., 2001). Vps1p requires Pex19 for its recruitment to peroxisomes, although the exact function of Pex19p is unclear (Vizeacoumar et al., 2006). A second yeast DRP, Dnm1p, was shown to be required for mitochondrial fission (Bleazard et al., 1999) and to regulate peroxisome abundance to a minor extent (Kuravi et al, 2006).
Genetic and biochemical approaches have identified three additional proteins (Fis1p, Mdv1p, Caf4p) that are required for mitochondrial fission and that cooperate with Dnm1p (for a review see (Hoppins et al., 2007)). Fis1p recruits Dnm1p to mitochondrial membranes in concert with Mdv1p and Caf4p (Cerveny and Jensen, 2003; Griffin et al., 2005; Mozdy et al., 2000; Tieu and Nunnari, 2000; Tieu et al., 2002). Both Caf4p and Mdv1p bind Dnm1p, and in an mdv1Δ /caf4Δ double mutant much of the Dnm1p is dissociated from mitochondria (Schauss et al., 2006). Mdv1p seems to play a more important role in fission than Caf4p, as mdv1Δ cells show a clear fission defect whereas caf4Δ cells do not (Griffin et al., 2005). Num1p also has a role in recruiting Dnm1p to mitochondria: cells lacking Num1p display a decrease in mitochondrial fission (Cerveny et al., 2007; Schauss and McBride, 2007).
In mammalian cells, a single DRP, (dynamin-like protein 1, DLP1) is thought to be required for peroxisome fission (Koch et al., 2003). DLP1 resembles Dnm1p and is also required for mitochondrial fission (Smirnova et al., 2001). A patient with a mutation in DLP1 has recently been described, and this mutation results in a lethal disorder whereby fission of both peroxisomes and mitochondria is impaired (Waterham et al., 2007).
Human Fis1p is required for a normal mitochondrial and peroxisomal morphology. Both DLP1 and hFis1 partially localise to peroxisomes (Koch et al., 2005; Li and Gould, 2003). Recently, it was shown also in yeast cells that Fis1p and Dnm1p partially localise to peroxisomes (Kuravi et al., 2006). Mutants lacking Dnm1p or Fis1p are not affected in peroxisome abundance when grown on a fermentable carbon source. However, when shifted to oleic acid as carbon source, the number of peroxisomes failed to increase as much as was observed in wild type cells (Kuravi et al., 2006). Additionally, a vps1Δ/dnm1Δ double mutant exhibited a more severe decrease in peroxisome number than the vps1Δ mutant on its own. These results suggest that Vps1p and Dnm1p are partially redundant in the regulation of peroxisome abundance. Using an assay designed to study peroxisome fission in vivo, we showed that the decreased abundance in a vps1Δ/dnm1Δ mutant results from a strong reduction in peroxisome fission (Motley and Hettema, 2007).
Here we describe our analysis of the role of Dnm1p in controlling peroxisome abundance in cells grown on a fermentable carbon source. Under these conditions, a constant peroxisomal number is maintained with no requirement for peroxisome proliferation. We show that Dnm1p-dependent fission of peroxisomes depends on Fis1p, and that similarly to mitochondrial fission, Caf4p and Mdv1p are required for Dnm1p-dependent peroxisome fission, although unlike for mitochondria, their roles in peroxisome fission are redundant. Furthermore, we show that Vps1p-dependent fission is not dependent on the presence of Fis1p, Caf4p or Mdv1p. We are able to suppress the fission defect in vps1Δ cells by 1) overexpression of Dnm1p and 2) redirecting Fis1p from mitochondria to peroxisomes. Dnm1p contributes little to peroxisome fission during fermentative growth, but its contribution appears far greater under conditions of mitochondrial dysfunction. We show that although Dnm1p and Vps1p are partially redundant for peroxisome fission, they also exhibit specific and nonoverlapping functions: overexpression of Vps1p does not rescue the mitochondrial fission defect in a dnm1Δ mutant, and overexpression of Dnm1p does not rescue the vacuolar fusion defect in vps1Δ cells.
We conclude that fission of peroxisomes in yeast is controlled by two different dynamin-related proteins that are recruited to peroxisomes independently of each other by distinct proteins. Although these DRPs display redundancy for their role in peroxisome fission, at least some of their other roles within the cell are nonoverlapping.
Results
Peroxisome abundance is affected in cells lacking components of the mitochondrial fission machinery
With every cell division peroxisomes duplicate by fission. We have shown previously that the dynamin-related protein Vps1p is required for this process (Motley and Hettema, 2007). Recently, it was shown that peroxisome abundance in a vps1Δ/dnm1Δ double mutant is even lower than in a vps1Δ mutant, suggesting that Dnm1p also plays a role in peroxisome fission. This was further supported by the observation that some colocalisation of Dnm1p and Fis1p was found with peroxisomes (Kuravi et al., 2006).
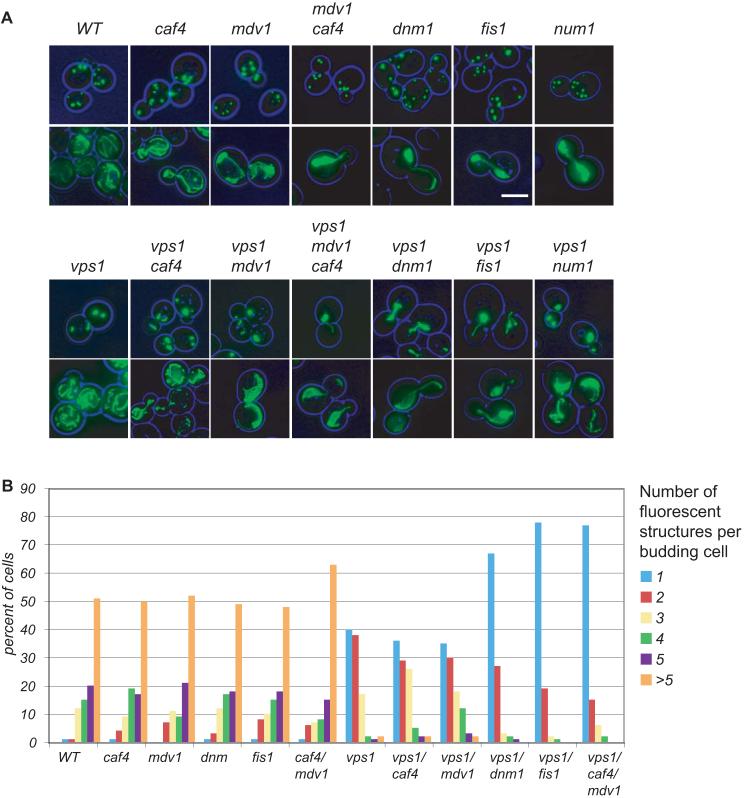
We analysed yeast strains lacking one of the genes encoding mitochondrial fission components. We transformed these strains with a well established fluorescent marker appended with a type I peroxisomal targeting signal (GFP-PTS1), grew them to log phase on glucose medium as described in Materials and Methods, and counted peroxisomes in dnm1Δ, fis1Δ, caf4Δ, mdv1Δ and num1Δ cells (Fig. 1). No clear differences in peroxisome number were observed between these mutants and wt cells, in agreement with previous findings (Kuravi et al., 2006). Since the contribution of Dnm1p to peroxisomal fission only becomes clear when Vps1-dependent fission is blocked, we introduced a vps1Δ mutation into the mitochondrial fission mutants: a vps1Δ/fis1Δ double mutant contains a single peroxisomal structure in the majority of dividing cells (Fig. 1). Neither vps1Δ/caf4Δ nor vps1Δ/mdv1Δ cells displayed a peroxisome abundance phenotype more severe than that of vps1Δ alone. However, the triple mutant vps1Δ/caf4Δ/mdv1Δ showed a phenotype indistinguishable from that of vps1Δ/dnm1Δ or vps1Δ/fis1Δ cells.
Fig. 1.
Peroxisome abundance is affected in cells lacking components of the mitochondrial fission machinery. (A) cells expressing GFP-PTS1 (top row) or mito-GFP (bottom row) were grown to log phase on 2% glucose-containing medium and representative images were captured. (B) Peroxisome numbers were counted from images of >100 budding cells for each strain. Images are flattened z-stacks. Bar represents 5 μm.
We conclude that the machinery used for mitochondrial fission is used also for peroxisomal fission. Our results suggest that Dnm1p is recruited to peroxisomes via Fis1p, Mdv1p and Caf4p, whereby Caf4p and Mdv1p are redundant. Furthermore, our results show that Vps1p operates independently of Fis1p, Caf4p, Mdv1p and Dnm1p. vps1Δ/num1Δ cells did not show a reduction of peroxisome abundance compared to vps1Δ cells, suggesting that Num1p is not required for Dnm1p-dependent peroxisome function. Num1p was not further studied.
Overexpression of Dnm1p restores peroxisome abundance in vps1Δ cells
Our results suggest that two independent systems operate in peroxisome fission, one relying on Dnm1p, and a second relying on Vps1p. Vps1p plays the major role in peroxisome fission as vps1Δ cells have a clear phenotype whereas dnm1Δ cells do not. However, when Dnm1p is overexpressed in a vps1Δ mutant, peroxisome number normalises (Fig. 2). This shows that 1) Dnm1p can substitute for Vps1p in peroxisome fission, and 2) that Dnm1p is limiting in Dnm1p-dependent (peroxisomal) membrane fission. In contrast, Dnm1p overexpression does not restore peroxisome abundance to normal levels in vps1Δ/fis1Δ or vps1Δ/caf4Δ/mdv1Δ mutants. We conclude that peroxisome abundance depends on two redundant machineries of which the Vps1p-containing machinery is the major contributor.
Fig. 2.
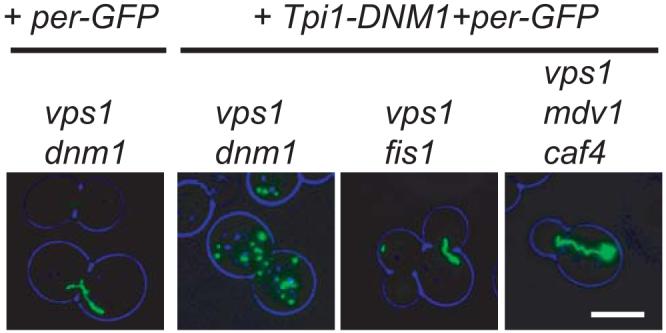
Overexpression of Dnm1p restores peroxisome abundance in vps1Δ/dnm1Δ cells, but not in vps1Δ/fis1Δ or vps1Δ/caf4Δ/mdv1Δ cells. Peroxisomes were visualised using GFP-PTS1. Images are flattened z-stacks. Bar represents 5 μm.
Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p
In the experiments described above, we have analysed peroxisome abundance only. We now tested whether Dnm1p-mediated peroxisome fission is affected in cells lacking Fis1p or Caf4p and Mdv1p using an assay we developed recently (Motley and Hettema, 2007). vps1Δ/dnm1Δ cells were pulse labelled with GFP-PTS1 (see Materials and Methods) and mated with vps1Δ/dnm1Δ cells pulse labelled with HcRed-PTS1 and overexpressing Dnm1p. In the latter strain, the red peroxisomes are small and abundant, whereas in the former strain, the GFP-labelled peroxisome is a single elongated structure. After mating and subsequent cytoplasmic mixing, Dnm1p diffuses from one mating partner to the other, and the prelabeled green peroxisome is divided almost instantly into small peroxisomal structures (Fig. 3AxB). In these mating cells, red peroxisomes coexist with green peroxisomes. However, when vps1Δ/ dnm1Δ cells overexpressing Dnm1p are mated with vps1Δ/fis1Δ or vps1Δ/caf4Δ/mdv1Δ mutants, there is a delay before fission of the prelabelled peroxisome begins to occur: initially the red peroxisomes from the vps1Δ/ dnm1Δ mating partner coexist with green tubulated peroxisome from the vps1Δ/fis1Δ (AxD) or vps1Δ/caf4Δ/mdv1Δ (AxE) mating partner. That this fission of peroxisomes is carried out by Dnm1p is clear from the mating of vps1Δ/dnm1Δ with vps1Δ/dnm1Δ cells in the absence (BxC) or presence (AxB) of overexpressed Dnm1p. We conclude that Fis1p, Caf4p and Mdv1p are required for Dnm1p-dependent peroxisome fission.
Fig. 3.
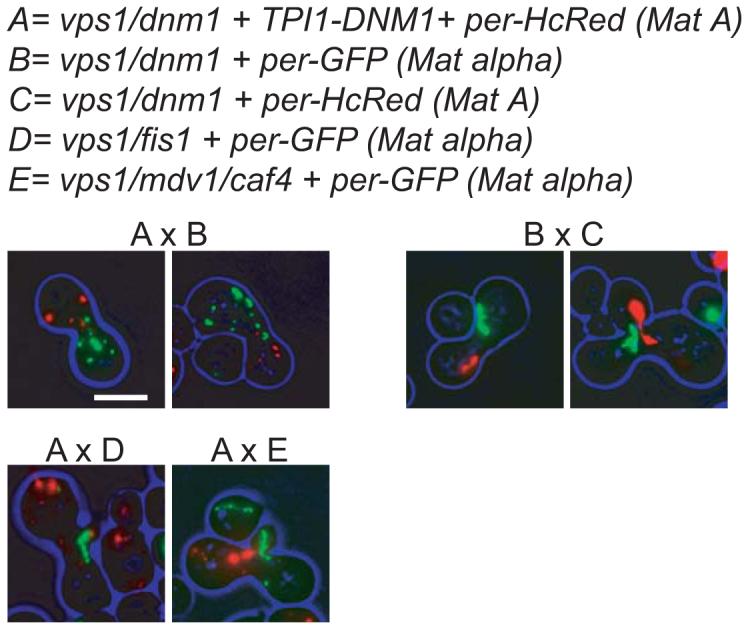
Mating experiment showing Dnm1p-mediated peroxisome fission requires the presence of Fis1p and Mdv1p and Caf4p. Fission of the prelabelled peroxisomal structure occurs when Dnm1p is supplied by cytoplasmic mixing after mating (AxB). Fission occurs soon after cytoplasmic mixing, before zygote formation. When both mating partners are deficient for Vps1p and Dnm1p, the prelabelled tubulated peroxisomes coexist together in the mating cells (BxC). When one of the mating partners is deficient in Fis1p or Mdv1p and Caf4p, the prelabelled tubulated peroxisome (which is lacking Fis1p or Mdv1p/Caf4p) initially fails to divide in spite of Dnm1p being supplied in the cytoplasm of the mating partner (AxD, AxE). Fission starts to occur only when the zygote is being formed. Images are flattened z-stacks. Bar represents 5 μm.
Redirecting Fis1p to peroxisomes rescues a Vps1p-dependent fission defect
A large fraction of Dnm1p is associated with mitochondria in a Fis1p-dependent manner (Cerveny and Jensen, 2003; Mozdy et al., 2000; Tieu et al., 2002). Rescue of the vps1Δ phenotype by overexpression of Dnm1p suggests that Dnm1p is limiting for peroxisome fission. We argued that if the Dnm1p usually associated with mitochondria could be redirected to peroxisomes, then endogenous levels of Dnm1p may be sufficient to overcome the peroxisomal fission defect in vps1Δ cells. To test this hypothesis we made use of the observation that exchanging the C-terminal membrane anchor sequence of Fis1p with that of the peroxisomal membrane protein Pex15p results in an exclusive localisation of the resulting fusion protein to peroxisomes (Halbach et al., 2006). We expressed this fusion protein under control of the FIS1 promoter in vps1Δ /fis1Δ cells and analysed peroxisome and mitochondrial morphology. Whereas expression of Fis1p restored peroxisome abundance to that observed in vps1Δ cells, when expressing the Fis1-Pex15 fusion protein, peroxisome abundance was restored to wild type levels. This indicates an increased level of Dnm1-dependent peroxisome fission. On the other hand, expression of the fusion protein did not restore the mitochondrial fission defect, whereas expression of Fis1p restored mitochondrial fission to wt levels (Fig. 4). These observations show that peroxisomes and mitochondria compete for Dnm1p, and that Fis1p plays a pivotal role in distributing Dnm1p between peroxisomes and mitochondria.
Fig. 4.
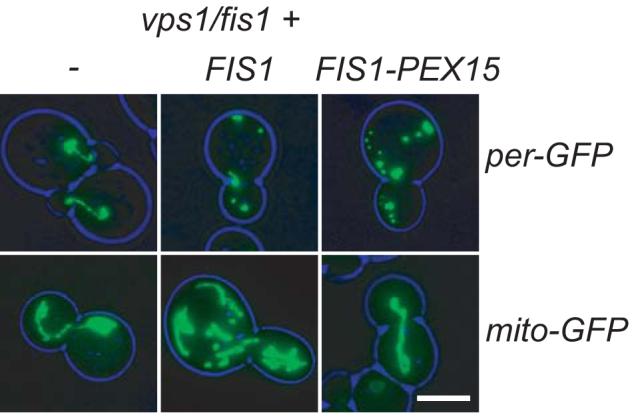
Redirecting Fis1p to peroxisomes by expression of a Fis1-Pex15p fusion protein rescues the peroxisomal fission defect in vps1Δ /fis1Δ cells. Expression of Fis1p rescues the mitochondrial phenotype to that of wt cells, but restores the peroxisome phenotype to that of vps1Δ cells. Expression of Fis1-Pex15p does not rescue the mitochondrial phenotype, but restores the peroxisome phenotype to that of wt cells. Images are flattened z-stacks. Bar represents 5 μm.
Fis1p recruits Caf4p and Mdv1p to peroxisomes
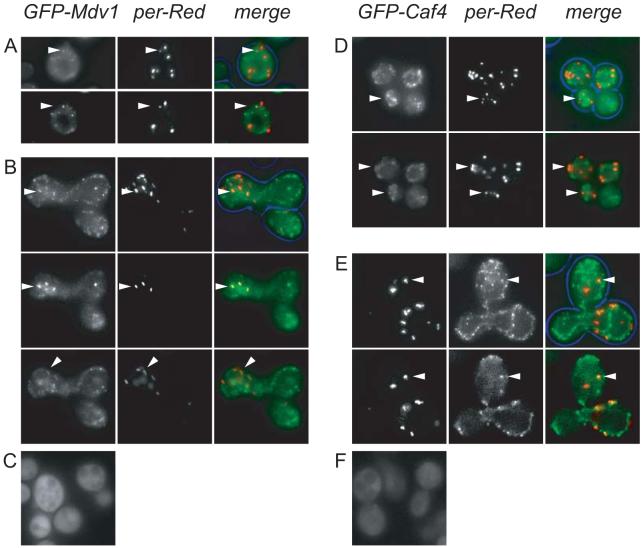
Our results suggest that similarly to mitochondria, Fis1p recruits Mdv1p and Caf4p to peroxisomal membranes. To test this, we analysed GFP fusions of both Caf4p and Mdv1p for their localisation in vps1Δ/caf4Δ /mdv1Δ cells (Fig. 5). Peroxisome number was restored indicating that the GFP fusions are functional. These cells gave clearer localisations of GFP-Mdv1p and GFP-Caf4p than wt cells, presumably due to lack of endogenous protein competing for localisation. Double labelling of GFP-Mdv1p or GFP-Caf4p with HcRed-PTS1 in vps1Δ/mdv1Δ /caf4Δ cells showed a limited amount of colocalisation. As expected, most GFP punctae were distinct from peroxisomes and most likely represent the mitochondrion-associated pool of these proteins. Peroxisomal labelling was hard to detect in flattened Z-stacks but was easier to detect in single focal planes (Fig 5A and D). GFP-Mdv1p or GFP-Caf4p was not detected on all peroxisomes.
Fig. 5.
GFP-Mdv1p and -Caf4p partially localise to peroxisomes in a Fis1p-dependent manner. (A and D) caf4Δ /mdv1Δ/vps1Δ cells coexpressing GFP-Mdv1p (A) or GFP-Caf4p (D) and HcRed-PTS1 (per-Red). Top row: flattened z-stack showing faint colocalisation of GFP fusion with peroxisome. Bottom row: colocalisation is easier to detect in individual slices of the z-stack. (B and E) Mating experiment showing enhanced levels of Mdv1p and Caf4p on peroxisomes as a result of Fis1-Pex15 expression. fis1Δ /vps1Δ cells expressing both Pex15-Fis1p and either GFP-Mdv1p (B) or GFP-Caf4p (E) were mated with pex3Δ cells expressing HcRed-PTS1. Top row: flattened z-stack showing colocalisation with peroxisomes. Bottom row, colocalisation is easier to detect in individual slices of z-stack. Peroxisome labelling with GFP is much stronger compared to (A and D). (C and F) Both GFP fusions were mainly cytosolic when expressed in fis1Δ cells. Bar represents 5 μm.
We have shown above that expression of Fis1-Pex15p increases Dnm1p-dependent peroxisome fission in an Mdv1p- and Caf4p-dependent manner. Therefore, we expected GFP-Caf4p and -Mdv1p levels on peroxisomes to be increased in cells expressing Fis1-Pex15p. To test this we performed a mating assay: one mating partner comprised vps1Δ/fis1Δ cells expressing Fis1-Pex15p in combination with either GFP-Caf4p or GFP-Mdv1p; the other mating partner is a mutant lacking peroxisomes (pex3Δ) and expressing HcRed-PTS1. Upon cell fusion and cytoplasmic mixing, HcRed-PTS1 is imported into the Fis1-Pex15-containing peroxisomes of the mating partner (Fig 5B and E). Now a more pronounced colocalisation of GFP-Mdv1p and GFP-Caf4p with peroxisomes is observed (compare Figs. 5B with A and E with D). However, both GFP fusion proteins were for the most part cytosolic when expressed in fis1Δ cells, showing their dependence on Fis1p for membrane association (Fig 5C and F). We conclude that Mdv1p and Caf4p associate with peroxisomes and this association depends on Fis1p.
Mitochondrial dysfunction results in dynamin-related protein-dependent peroxisome proliferation
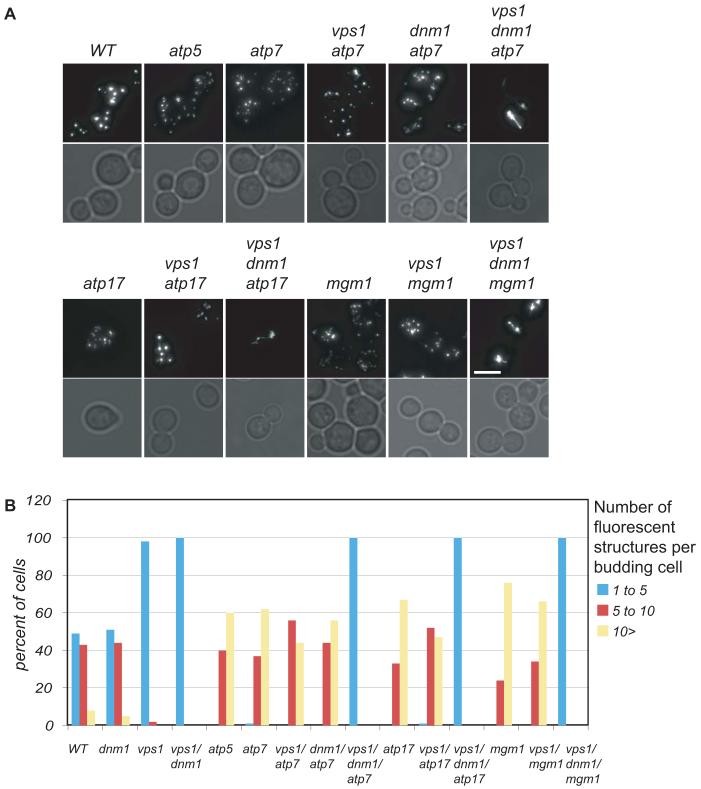
Peroxisomes have been shown to multiply in response to mitochondrial dysfunction (Butow and Avadhani, 2004). In a genome-wide screen for peroxisome morphology mutants (G.P.Ward and E.H.Hettema, unpublished data), we observed many mutants in mitochondrial proteins to display increased peroxisome abundance, and these mutants included mitochondrial ribosomal proteins (for instance MRP5, MRPL8, MRPL49 and the nuclear encoded F1F0-ATP synthase subunits, fig.6). We tested whether the increased peroxisome abundance in the atp7Δ and atp17Δ cells is dependent on DRPs. First we disrupted the ATP7 and ATP17 gene in vps1Δ cells that normally have a reduced number of peroxisomes. Strikingly, vps1Δ/atp7Δ cells and vps1Δ/atp17Δ cells display an increased peroxisome abundance compared to both vps1Δ cells and wt cells implying that Vps1p is not necessary for this proliferation. On the other hand in vps1Δ/dnm1Δ/atp7Δ cells and vps1Δ/dnm1Δ/atp17Δ cells a single peroxisomal structure is observed. These results indicate that DRPs are required for peroxisome proliferation during mitochondrial dysfunction, with Dnm1p having a major contribution. To test whether Dnm1p is solely responsible for this proliferation or whether in its absence, Vps1p can take over we constructed a dnm1Δ/atp7Δ strain. This strain also displays an increased peroxisome abundance. We conclude that both Dnm1p and Vps1p are involved in peroxisome proliferation in response to mitochondrial dysfunction. Whereas Dnm1p only contributes to a minor extent under non-proliferative conditions, under peroxisome proliferation conditions the contribution of Dnm1p is much greater.
Fig. 6.
Peroxisome abundance in cells with mitochondrial dysfunction. (A) Cells expressing GFP-PTS1 were grown on 2% glucose medium to log phase and representative images were captured. Images are flattened z-stacks. Bar represents 5 μm. (B) Peroxisome numbers were counted from images of >100 budding cells for each strain.
We analysed a third DRP, Mgm1p, for its function in peroxisome multiplication. Mgm1p has been shown to be required for mitochondrial fusion and proper assembly of F1F0-ATPase and cristae formation (Amutha et al., 2004); (Meeusen et al., 2006). We found that mgm1Δ cells show increased peroxisome abundance (Fig. 6), although the abundance of peroxisomes in these cells was very sensitive to growth conditions (see Materials and Methods). A vps1Δ/mgm1Δ mutant displays an increased peroxisome abundance similar to that seen in mgm1Δ cells and atpΔ cells (Fig. 6). A vps1Δ/dnm1Δ/mgm1Δ triple mutant, however, showed a huge decrease in peroxisome number: most cells contain a single peroxisome. Since mature peroxisomes do not fuse (Motley and Hettema, 2007), the phenotype of vps1Δ/dnm1Δ/mgm1Δ cells implies that the increased peroxisome number in mgm1Δ and mgm1Δ/vps1Δ cells is a result of excessive fission by DRPs.
Vps1p and Dnm1p are only partially redundant
We tested whether Dnm1p and Vps1p are redundant in other functions besides peroxisomal fission. vps1Δ cells have a vacuolar fusion defect which can be visualised by allowing the cells to take up FM 4-64 to steady state levels (Vida and Emr, 1995). In wt cells, FM 4-64 accumulates in the vacuolar membrane, whereas in vps1Δ cells staining is most intense in the smaller, fragmented vacuolar structures (Fig. 7). Overexpression of Dnm1p does not rescue the vacuolar fusion defect observed in these cells (Fig. 7). Similarly, we find that the mitochondrial fission defect in dnm1Δ cells is not rescued by overexpression of Vps1p (Fig. 7). We conclude that Vps1p and Dnm1p are partially redundant for their role in peroxisome fission, with their functions on other organelles being nonoverlapping.
Fig. 7.
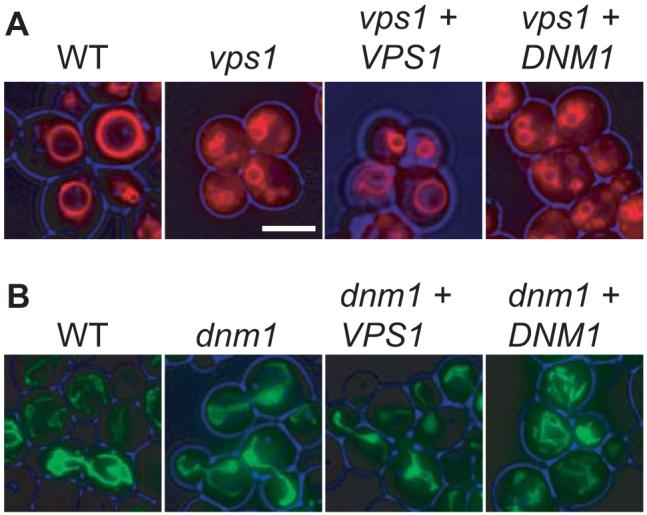
Dnm1p and Vps1p have distinct functions. Dnm1p overexpression does not restore the vacuolar fusion defect in vps1Δ cells, and Vps1p overexpression does not rescue the mitochondrial fission defect in dnm1Δ cells. Top panel: Vacuoles were visualised by allowing cells to take up FM 4-64 to steady state levels. The vacuolar membrane is clearly visible in wt cells and in vps1Δ cells expressing Vps1p, whereas unfused, fragmented vacuoles are visible in vps1Δ cells and vps1Δ cells overexpressing Dnm1p. Bottom panel: Cells expressing mito-GFP were grown to log phase on 2% glucose. dnm1Δ cells overexpressing Vps1p show the mitochondrial fusion defect typical of dnm1Δ cells. Bar represents 5 μm.
Discussion
In this paper, we establish that the accessory proteins required for Dnm1p-dependent peroxisome fission are the same as those required for Dnm1-dependent mitochondrial fission, whereas Vps1p-mediated peroxisome fission is independent of these proteins. We show that effective regulation of peroxisome fission seems to require both Vps1p and Dnm1p, and that their relative contributions vary with growth conditions.
The role of the dynamin-related protein Dnm1p in mitochondrial fission is well established (for a review see Hoppins et al (2007)). Dnm1p-mediated mitochondrial fission requires the mitochondrial outer membrane protein Fis1p, and the adaptors Mdv1p and Caf4p. The two paralogous DRPs Vps1p and Dnm1p have been shown to be important for a normal abundance of peroxisomes and both have been found associated with peroxisomes (Hoepfner et al., 2001; Vizeacoumar et al., 2006) (Kuravi et al., 2006). We have recently shown that Vps1p and Dnm1p control peroxisome abundance by stimulating fission (Motley and Hettema, 2007). Whereas the association of Vps1p with peroxisomes requires interaction with Pex19p (Vizeacoumar et al., 2006), Kuravi et al (2006) showed that Fis1p is required for Dnm1p recruitment to peroxisomes. In this paper we show that not only Dnm1p but also Caf4p and Mdv1p are recruited to peroxisomes, and that recruitment is dependent on Fis1p. Only a small amount of GFP-Mdv1p and GFP-Caf4p was found to colocalise with some peroxisomes, suggesting that the association is only temporary. Using our recently developed mating assay, we showed a requirement for Fis1p, Mdv1p and Caf4p in Dnm1p-dependent peroxisome fission. When vps1Δ/dnm1Δ cells overexpressing Dnm1p were mated with vps1Δ/dnm1Δ cells, fusion of the peroxisomal structure in the Dnm1p-deficient mating partner occurs soon after cytoplasmic mixing. However, when vps1Δ/dnm1Δ cells overexpressing Dnm1p were mated with either vps1Δ/fis1Δ or vps1Δ/caf4Δ/ mdv1Δ cells, the peroxisomal structure lacking Fis1p or Caf4p and Mdv1p initially failed to divide in spite of the presence of Dnm1p in the (now mixed) cytoplasm of the mating cells. Only after several hours, when the zygote is being formed, did the vps1Δ/ fis1Δ peroxisome begin to divide. The delay in fission suggests that Fis1p, Caf4p and Mdv1p do not associate with the peroxisomal structures directly after cell fusion, and that equilibration of the existing pool is slow or that synthesis of these factors is required. This is not surprising since Fis1p is an integral membrane protein and Caf4p and Mdv1p are peripheral membrane proteins.
We found that Mdv1p and Caf4p can substitute functionally for each other on peroxisomes. This is not the case for mitochondria, as mdv1Δ cells have a mitochondrial fission defect that is much more pronounced than that of caf4Δ cells (Griffin et al., 2005). Indeed, Caf4p has been shown to be required for a more peripheral distribution of Dnm1p. This pool of Dnm1p is not immediately involved in the fission process (Schauss et al., 2006). Another factor that acts in concert with Dnm1p is Num1p. Num1p is required for normal mitochondrial morphology and seems to couple mitochondrial inheritance to fission (Cerveny et al., 2007). Our data do not support a role for this protein in peroxisome fission.
Since Dnm1p- and Vps1p-dependent peroxisome fission appear to be partially redundant, we tested whether Dnm1p could compensate for a Vps1p deficiency. We found that overexpression of Dnm1p can rescue the partial peroxisome fission defect observed in cells lacking Vps1p. This suppression depends on the presence of Fis1p and on Caf4p or Mdv1p. Our results imply that peroxisomes normally have to compete with mitochondria for Dnm1p. Cells lacking either Fis1p or Caf4p display an increase of cytosolic Dnm1p (Schauss et al., 2006). The same is observed in cells lacking Num1p (Cerveny et al., 2007). However, a vps1Δ/num1Δ double mutant does not restore peroxisome number to wild type levels, neither does a caf4Δ/vps1Δ double mutant. This implies that simply increasing the level of Dnm1p in the cytosol is not sufficient to restore peroxisome fission in vps1Δ cells. Since Fis1p is essential for Dnm1-dependent peroxisomal and mitochondrial fission, we redirected Fis1p to an exclusively peroxisomal location. A Fis1p-Pex15p fusion expressed in vps1Δ/fis1Δ cells failed to rescue the mitochondrial fission defect, whereas peroxisome abundance was restored to normal. This shows the importance of Fis1p in localising Dnm1p to mitochondria versus peroxisomes.
We analysed the third DRP present in yeast, Mgm1p. We show that cells lacking Mgm1p have increased peroxisome abundance and that this increase is a result of excessive DRP-dependent fission. Is this reflecting a direct role of Mgm1 on peroxisome dynamics? We favour the interpretation that this increase is not a direct effect of Mgm1p on peroxisomes but rather a response to mitochondrial dysfunction. Firstly, we were unable to colocalise Mgm1p with peroxisomes (not shown). Furthermore, peroxisome proliferation has been reported in response to mitochondrial dysfunction and mgm1Δ cells display a variety of mitochondrial defects, including a failure to assemble their F1F0-ATPase properly and increased loss of mtDNA. Indeed, all ATP synthase mutants we tested show an increase in peroxisome number and this increase is dependent on DRPs. Our data are compatible with the interpretation that the increase in peroxisome abundance in mgm1Δ cells is a result of the mitochondrial dysfunction. However, we can’t exclude a more direct role of Mgm1p in peroxisome dynamics.
Whereas fission of peroxisomes in man and Hansenula polymorpha (Nagotu et al., 2007) depends on a single DRP, the presence of two homologous DRPs on peroxisomes in S.cerevisiae is intriguing (Schrader and Yoon, 2007). Despite their extensive amino acid sequence identity, Dnm1p and Vps1p are recruited independently to peroxisomes. Furthermore, they cannot substitute for each other in their other functions (for instance vacuole fusion and mitochondrial fission). This illustrates that these DRPs are functionally distinct and suggests a functional significance for the presence of these two DRPs on peroxisomes in yeast. We can only guess what the significance is.
Since mitochondria and peroxisomes are metabolically linked, a coordinated regulation of their biogenesis is not surprising and has been described in both humans and yeast (for review see (Schrader and Yoon, 2007)). Peroxisomes proliferate in cells with dysfunctional mitochondria (Butow and Avadhani, 2004). We show that this proliferation is dependent on DRPs. Whereas Dnm1p makes only a minor contribution to peroxisome fission under standard growth conditions, in mutants with mitochondrial dysfunction, Dnm1p’s contribution appears to be much greater (compare vps1Δ with an vps1Δ /atp7Δ). This suggests that yeast is able to coordinate the division of peroxisomes with the functional state of mitochondria via Dnm1p. We are currently investigating the molecular basis for this.
Materials and methods
Strains and plasmids
Yeast strains were derivatives of BY4741 (MATA his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0) or BY4742 (MATa his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0) obtained from the EUROSCARF consortium. Double or triple gene deletions were made by replacing the entire coding sequence of the mutated genes with a marker (Schizosaccharomyces pombe HIS5, or the Klebsiella pneumoniae hygromycin B phosphotransferase gene cassette that confers resistance to Hygromycin B (Goldstein and McCusker, 1999). dnm1Δ /vps1Δ, mdv1Δ /vps1Δ, caf4Δ /vps1Δ, mdv1Δ /caf4Δ/vps1Δ, num1Δ/vps1Δ and Δfis1Δ /vps1 were generated by replacing the VPS1 reading frame with the HIS5 cassette in the dnm1Δ, mdv1Δ, caf4Δ, mdv1Δ /caf4Δ, num1Δ and fis1Δ, respectively. The MGM1, ATP7 and ATP17 open reading frames were replaced by that of the Hygromycin cassette to generate dnm1Δ /vps1Δ /mgm1Δ, dnm1Δ /vps1Δ /atp7Δ, dnm1Δ /vps1Δ /atp17Δ, /vps1Δ /atp7Δ, /vps1Δ /atp17Δ and dnm1Δ /atp7Δ,. The mdv1Δ /caf4Δ double mutant was constructed by replacing the CAF4 open reading frame with that of the Hygromycin cassette in the mdv1Δ mutant.
URA3 and LEU2 centromere plasmids were derived from Ycplac33 and Ycplac111 (Gietz and Sugino, 1988). GFP-PTS1 is a peroxisomal luminal GFP marker protein appended with the well-characterised peroxisomal targeting signal type 1 (PTS1) (Gould et al., 1988). A far-red peroxisomal luminal marker was made by appending a variant of the Heteractis crispa Chromoprotein (HcRed) with the PTS1. As source of HcRed we used HcRed-Tandem with optimised yeast codon usage (Evrogen, Moscow, Russia).
Constitutive expression of GFP-PTS1 and HcRed-PTS1 was under control of the TPI1 promoter and the HIS3 promoter, respectively. Dnm1p overexpression was achieved using the TPI1 promoter. All constitutive expression constructs contained the PGK1 terminator. The Fis1-Pex15p fusion protein was expressed from a construct containing the FIS1 promoter, and the fusion protein contained the cytoplasmic domain of Fis1p and the C-terminal tail anchor of Pex15. Caf4p and Mdv1p were N-terminally tagged with GFP. Conditional expression constructs contained the GAL1 promoter. In order to reduce the half-life of the transcript we replaced the PGK1 terminator with the MFA2 terminator (Duttagupta et al., 2003; LaGrandeur and Parker, 1999)
Growth conditions and mating assay
For all experiments, cells were grown overnight in selective glucose medium. For analysis of phenotypes by microscopy, cells were subsequently diluted to OD 0.1 in 2% glucose medium + casamino acids and grown for 2 - 3 cell divisions (4 - 6 h), so that phenotypes were analysed under conditions whereby cells are actively maintaining their peroxisome number. In the case of the mgm1Δ and ATP synthase mutants, peroxisome number varied significantly depending on growth media but reproducible results were obtained using 2% glucose + casamino acid medium. Cells were fixed (see below) for 5 minutes before imaging. For the experiment described in Fig. 3, an overnight culture was used to inoculate selective galactose medium at an OD600 of 0.1 to allow induction of reporter proteins for 3 h. Cells were then switched to selective glucose medium for 2 hours, to shut down expression of the gal-inducible reporter protein, before mating. For mating, cells were collected by filtration onto a 0.22 micron Millipore nitrocellulose filter (type GS, 25 mm diameter) and this filter was incubated, cells side up, on a pre-warmed YPD plate at 30°C. 1×107 cells of each strain were collected per 25 mm filter.
After 2 hours, cells were harvested by vortexing the filter in selective glucose medium, and fixed for 5 min by adding formaldehyde to 3.6%. Free formaldehyde groups were quenched in 0.1 M ammonium chloride/1xPBS. Cells were imaged within 1 h of fixing as loss of fluorescence intensity and increase of autofluorescence was seen in fixed cells left for extended periods. For each experiment, >100 cells were examined and images are representative of findings. For Fig, 1, budding cells were counted as single cells.
FM4-64 vacuole staining
Cells were grown in 2% glucose selective medium to log phase (OD 0.5). 1 ml of log phase cells were pelleted, resuspended in YPD containing 20 μM FM 4-64 and incubated at 30°C for 15 min. They were then pelleted, and resuspended in 1 ml of YPD and incubated at 30°C for 30 min. Subsequently the cells were washed in 1 ml of water before being resuspended in 2% glucose medium, ready for imaging.
Image acquision
Live and fixed cells were analysed at with an Axiovert 200M (Zeiss) equipped with Exfo X-cite 120 excitation light source, band pass filters (Zeiss and Chroma) and alpha Plan-Fluar 100 x/1.45 NA or A-Plan 40 X/0.65 NA Ph2 objective lens (Zeiss) and Hamamatsu Orca ER digital camera. Image acquisition was performed using Openlab software (Improvision) at 21°C. Fluorescence images were collected as 0.2 μm z-stacks and merged into one plain after contrast enhancing in Openlab, and processed further in Photoshop except when stated differently in text or figure legends. Bright field images were collected in one plain. In bright field image was added into the blue channel in Adobe Photoshop. The level of the bright field images was modified and the image was blurred, sharpened and blurred again before one more round of level adjustment so that only the circumference of the cell was visible.
Acknowledgements
The authors want to thank Peter Piper and Stephan Mills for help with the robotics, and Nina Rajala and Naomi Saggers for help with strain construction. This research was supported by a Wellcome Trust Career Development Fellowship in Basic Biomedical Sciences awarded to EH and a Royal Society Research Grant.
Abbreviations
- DRP
dynamin-related protein
References
- Amutha B, Gordon DM, Gu Y, Pain D. A novel role of Mgm1p, a dynamin-related GTPase, in ATP synthase assembly and cristae formation/maintenance. Biochem J. 2004;381:19–23. doi: 10.1042/BJ20040566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- Cerveny KL, Jensen RE. The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol Biol Cell. 2003;14:4126–39. doi: 10.1091/mbc.E03-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny KL, Studer SL, Jensen RE, Sesaki H. Yeast mitochondrial division and distribution require the cortical num1 protein. Dev Cell. 2007;12:363–75. doi: 10.1016/j.devcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Duttagupta R, Vasudevan S, Wilusz CJ, Peltz SW. A yeast homologue of Hsp70, Ssa1p, regulates turnover of the MFA2 transcript through its AU-rich 3′ untranslated region. Mol Cell Biol. 2003;23:2623–32. doi: 10.1128/MCB.23.8.2623-2632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–34. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–53. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Gould SJ, Keller GA, Subramani S. Identification of peroxisomal targeting signals located at the carboxy terminus of four peroxisomal proteins. J Cell Biol. 1988;107:897–905. doi: 10.1083/jcb.107.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EE, Graumann J, Chan DC. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J Cell Biol. 2005;170:237–48. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach A, Landgraf C, Lorenzen S, Rosenkranz K, Volkmer-Engert R, Erdmann R, Rottensteiner H. Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J Cell Sci. 2006;119:2508–17. doi: 10.1242/jcs.02979. [DOI] [PubMed] [Google Scholar]
- Harsay E, Schekman R. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J Cell Biol. 2002;156:271–85. doi: 10.1083/jcb.200109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D, van den Berg M, Philippsen P, Tabak HF, Hettema EH. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J Cell Biol. 2001;155:979–90. doi: 10.1083/jcb.200107028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu Rev Biochem. 2007;76:751–80. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- Koch A, Thiemann M, Grabenbauer M, Yoon Y, McNiven MA, Schrader M. Dynamin-like protein 1 is involved in peroxisomal fission. J Biol Chem. 2003;278:8597–605. doi: 10.1074/jbc.M211761200. [DOI] [PubMed] [Google Scholar]
- Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2005;16:5077–86. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuravi K, Nagotu S, Krikken AM, Sjollema K, Deckers M, Erdmann R, Veenhuis M, van der Klei IJ. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J Cell Sci. 2006;119:3994–4001. doi: 10.1242/jcs.03166. [DOI] [PubMed] [Google Scholar]
- LaGrandeur T, Parker R. The cis acting sequences responsible for the differential decay of the unstable MFA2 and stable PGK1 transcripts in yeast include the context of the translational start codon. Rna. 1999;5:420–33. doi: 10.1017/s1355838299981748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gould SJ. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J Biol Chem. 2003;278:17012–20. doi: 10.1074/jbc.M212031200. [DOI] [PubMed] [Google Scholar]
- Luo W, Chang A. An endosome-to-plasma membrane pathway involved in trafficking of a mutant plasma membrane ATPase in yeast. Mol Biol Cell. 2000;11:579–92. doi: 10.1091/mbc.11.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S, DeVay R, Block J, Cassidy-Stone A, Wayson S, McCaffery JM, Nunnari J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–95. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Motley AM, Hettema EH. Yeast peroxisomes multiply by growth and division. J Cell Biol. 2007;178:399–410. doi: 10.1083/jcb.200702167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–80. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagotu S, Saraya R, Otzen M, Veenhuis M, van der Klei IJ. Peroxisome proliferation in Hansenula polymorpha requires Dnm1p which mediates fission but not de novo formation. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbamcr.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Peters C, Baars TL, Buhler S, Mayer A. Mutual control of membrane fission and fusion proteins. Cell. 2004;119:667–78. doi: 10.1016/j.cell.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Rothman JH, Raymond CK, Gilbert T, O’Hara PJ, Stevens TH. A putative GTP binding protein homologous to interferon-inducible Mx proteins performs an essential function in yeast protein sorting. Cell. 1990;61:1063–74. doi: 10.1016/0092-8674(90)90070-u. [DOI] [PubMed] [Google Scholar]
- Schauss AC, Bewersdorf J, Jakobs S. Fis1p and Caf4p, but not Mdv1p, determine the polar localization of Dnm1p clusters on the mitochondrial surface. J Cell Sci. 2006;119:3098–106. doi: 10.1242/jcs.03026. [DOI] [PubMed] [Google Scholar]
- Schauss AC, McBride HM. Mitochondrial fission: a non-nuclear role for Num1p. Curr Biol. 2007;17:R467–70. doi: 10.1016/j.cub.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Schrader M, Yoon Y. Mitochondria and peroxisomes: Are the ‘Big Brother’ and the ‘Little Sister’ closer than assumed? Bioessays. 2007;29:1105–14. doi: 10.1002/bies.20659. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–56. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q, Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol. 2000;151:353–66. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q, Okreglak V, Naylor K, Nunnari J. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J Cell Biol. 2002;158:445–52. doi: 10.1083/jcb.200205031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vater CA, Raymond CK, Ekena K, Howald-Stevenson I, Stevens TH. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J Cell Biol. 1992;119:773–86. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995;128:779–92. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizeacoumar FJ, Vreden WN, Fagarasanu M, Eitzen GA, Aitchison JD, Rachubinski RA. The dynamin-like protein Vps1p of the yeast Saccharomyces cerevisiae associates with peroxisomes in a Pex19p-dependent manner. J Biol Chem. 2006;281:12817–23. doi: 10.1074/jbc.M600365200. [DOI] [PubMed] [Google Scholar]
- Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–41. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]