The stability of the native folds of globular proteins is rather remarkable, in that this stability is marginal and restricted to a relatively narrow window of thermodynamic and solution composition conditions (1). The development of a deep understanding of the balance of forces that tip the scales between native and denatured states in terms of the individual roles of electrostatics, hydrophobic interactions, polymer entropy, temperature, and pressure would have a profound impact on our ability to understand native structures and abnormal aggregated states and aid in development of bio-mimetic systems. Determining how unfolding occurs, i.e., the dynamic pathway by which the denatured state is established, is even more demanding but may provide insight into the landscape governing protein folding (2). Earlier simulations combining stress from increased temperature and denaturant cosolvent, urea, have followed this pathway for short times (3).
In this issue of PNAS, Hua et al. (4) present the results of a tour de force simulation comprising several microsecond-long simulations of the dynamics of ambient temperature lysozyme in concentrated urea solution, revealing a mechanistic pathway isolating the impact of urea on protein unfolding for the first time. In particular, the simulations reveal a stepwise process, starting from a state manifesting preferential solvation of the globular state by urea, compared with water, driven at least in part by the greater Van der Waals attraction of the protein for urea. The loss of native structure occurs with an initial intrusion into the tertiary structure predominantly by urea, followed only later by substantial hydration, in contrast to evidence for initial hydration during denaturation of chymotrypsin inhibitor 2 at elevated temperatures (3). Appreciating the lessons provided by these observations requires some reconciliation with related studies on the interactions of urea with simpler solutes in aqueous media.
The principles behind the destabilization of folded protein structures by aqueous urea have been actively discussed for decades, with a literature far too large to summarize here. Although some of the earliest discussions of the mechanism focused on perturbation of water structure per se (5), this so-called “indirect” mechanism has not received much support from experimental (6) or simulation (ref. 7 and references therein) studies of aqueous urea. Such studies imply that urea readily substitutes into the water hydrogen bond network, and that there is no segregation of urea from water (6). The alternative, “direct” mechanism implies a causative interaction between urea and the polypeptide, a characterization clearly evidenced in the simulated pathways (3, 4).
The ability of urea to interact with both nonpolar and polar components of proteins was recognized early on as beneficial to denaturation power (8). Experimental investigations (9) and theoretical studies (10–13) of smaller model systems can provide clues to the molecular-scale elements in the context of proteins. In this context, two seemingly different points of view have been put forward. Because urea acts to enhance the aqueous solubility of all but the smallest hydrocarbons (14), a logical inference was that urea weakened the hydrophobic interaction, by stabilizing the solvation of the unfolded protein state where a greater number of non polar side chains are exposed to solution. The driving force for preferential solvation of peptide by urea arising from Van der Waals interactions, identified clearly in the new simulations (4), enriches the availability of urea for hydrophobic hydration. Solvent entropy gain when water initially constrained by hydrophobic hydration is displaced by the larger urea molecule (10, 11, 13) also appears to enhance this effect (13). Separate studies (12) have provided results that support a predominantly electrostatic basis for urea activity, and destabilization of a polypeptide helix by aqueous urea clearly correlates with preferential association of urea with backbone polar groups and charged side chains. A group of large-scale simulations of each of 22 glycine-capped tri peptides in aqueous urea solution (11) has provided the contact preferences between the atoms of each central residue and urea, relative to water. The finding is that, with the exception of residues with charged side chains, urea is always preferred to water. This preference increases monotonically with increasing side-chain hydrophobicity; the backbone attracts urea, and the hydrophobic side chains enhance this effect.
Returning then to the pathway for denaturation so clearly seen in these new simulations (4), one can ask how to integrate these various careful studies with each other. First, the identification of an important role of Van der Waals attraction in urea's preferential solvation of peptides (4) helps explain the increasing preference of urea for increasingly large nonpolar side chains. More importantly, why does urea dominate water in the earliest stage of structural penetration? In addition to the attractive interactions just mentioned, one suspects the role of confinement. Liquid water forms a space-filling 3D tetrahedral hydrogen bond network (15) that is remarkably adaptable to the presence of both hydrophilic and hydrophobic “intruders.” Nevertheless, confinement in a reduced dimension is incompatible with satisfying the network, with substantial enthalpic and entropic costs, even leading in some cases to ambient temperature freezing (16). Urea can also form a network structure, evident in its neat crystalline form and nonpolar clathrate compounds, but in concentrated solution it appears to readily form chains and clusters (6). The demonstrated ability to interact preferentially with backbone hydrogen bonds (12) adds yet a third driving force for urea's entry into protein fold interfaces. The initial tendencies lead in a fairly short time to an unfolded state (after 100 ns) that clearly reveals a substantial enhancement in the contact between both urea and water with hydrophobic side chains, and the effect is clearly enhanced when solvent or cosolvent hydrogen bonds with the polar backbone (4).
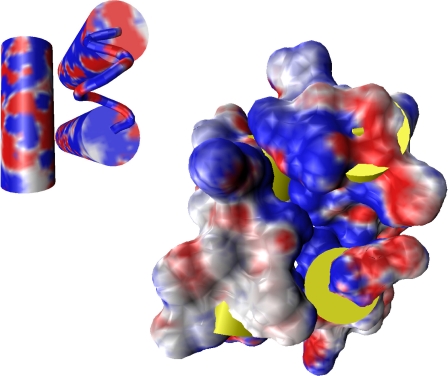
The new protein simulations, combined with the earlier studies of model systems, provide a clear picture of the activity of urea both on the thermodynamics of native and denatured states and along the pathway between them. It appears clear that the ability of aqueous urea to act as a solvent for both nonpolar and polar groups plays a vital role. It is the critical quality determining the urea-first stepwise pathway. The penetration of a molecule such as urea, having an affinity for both polar and nonpolar solvation, into protein interior interfaces is readily understood from visualization of a prototype. In Fig. 1, an interface between the helices in a particular folded protein core is shown with a map of surface electrostatic potentials. As is evident in this map, there is considerable polarity present at such interfaces, even though they are normally assigned a primarily hydrophobic origin. The propensity for a significant positive electrostatic potential in protein interiors has been identified as typical of a variety of fold motifs (18). It is worth emphasizing that the collective evidence shows that all of the key components of the intermolecular forces (Van der Waals attraction, electrostatics, hydrogen bonding, and hydrophobic interactions) play a significant part in creating these relative affinities. Correspondingly, one must remain somewhat skeptical of quantitative calculations based on model potentials when operating in uncharted thermodynamic and composition domains.
Fig. 1.
Electrostatic potentials mapped onto the surface of a simplified secondary structure cylinder representation (Left) and onto the solvent-accessible surface area of each helix (Right; cylindrical helices yellow) in the core formed by the A, G, and H helices in apomyoglobin (17). Electrostatic potentials from red to blue correspond to the range −5 kT/e to +5 kT/e, where k is Boltzmann's constant, e is the magnitude of the electron charge, and T is taken as 298 K. (Figure courtesy of Dr. Carlos F. Lopez, Department of Systems Biology, Harvard Medical School, Boston.)
It is of great interest to ask about the ability to generalize the urea denaturation mechanism. For example, important approaches to the study of protein folding dynamics are initiated from states obtained by various denaturing conditions (19). In what way are these initial states potentially different? Does the guanidinium ion denature by substantially the same mechanism and pathway as urea? Simulation results show that this ionic cosolvent has a more dramatic effect on the interaction between charged solutes than does urea (12). Experimental structural studies show that the guanidinium ion is extremely weakly hydrated (9), so that this bulky ion could also associate with hydrophobic surfaces (9, 12). A more subtle question is the relationship of urea denaturation to the “indirect” effects of pressure and temperature. Is there a rational molecular-scale or thermodynamic comparison to be made between the solvation of nonpolar and polar groups in cold or high-pressure water and in denaturant solutions, and is there a close structural analogy in the denaturation pathways? It has been argued that cold- and pressure-induced denaturation should be viewed as the penetration of water into hydrophobic domains, rather than in terms of the solubility of hydrophobes in liquid water (20), so it is not a stretch of imagination to believe that there are connections to be made. When two or more perturbations are combined, such as urea-induced denaturation with modest heating, how similar are the pathways and end states that are accessed? Comparison of results so far (3, 4) suggest that the pathways are different. With the evident strides being made in the processes that can be simulated and measured on the same length and time scales, we should not have to wait too long to learn the answers to these questions.
Footnotes
The author declares no conflict of interest.
See companion article on page 16928.
References
- 1.Zhang J, et al. NMR-study of the cold, heat, and pressure unfolding of ribonuclease A. Biochemistry. 1995;34:8631–8641. doi: 10.1021/bi00027a012. [DOI] [PubMed] [Google Scholar]
- 2.Torrent J, et al. Distinct unfolding and refolding pathways of ribonuclease A revealed by heating and cooling temperature jumps. Biophys J. 2008;94:4056–4065. doi: 10.1529/biophysj.107.123893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennion BJ, Daggett V. The molecular basis for the chemical denaturation of proteins by urea. Proc Natl Acad Sci USA. 2003;100:5142–5147. doi: 10.1073/pnas.0930122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hua L, Zhou R, Thirumalai D, Berne BJ. Urea denaturation by stronger dispersion interactions with proteins than water implies a 2-stage unfolding. Proc Natl Acad Sci USA. 2008;105:16928–16933. doi: 10.1073/pnas.0808427105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frank H, Franks F. Structural approach to the solvent power of water for hydrocarbons; urea as a structure breaker. J Chem Phys. 1968;48:4746–4757. [Google Scholar]
- 6.Soper AK, Castner EW, Luzar A. Impact of urea on water structure: A clue to its properties as a denaturant. Biophys Chem. 2003;105:649–666. doi: 10.1016/s0301-4622(03)00095-4. [DOI] [PubMed] [Google Scholar]
- 7.Astrand PO, et al. Properties of urea–water solvation calculated from a new ab initio polarizable intermolecular potential. J Chem Phys. 1991;95:8419–8429. [Google Scholar]
- 8.Roseman M, Jencks WP. Interactions of urea and other polar compounds in water. J Am Chem Soc. 1975;97:631–640. [Google Scholar]
- 9.Mason PE, et al. The interaction of guanidinium ions with a model peptide. Biophys J. 2007;93:L4–L6. doi: 10.1529/biophysj.107.108290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuharski RA, Rossky PJ. Solvation of hydrophobic species in aqueous urea solution: A molecular dynamics study. J Am Chem Soc. 1984;106:5794–5800. [Google Scholar]
- 11.Stumpe MC, Grubmueller H. Interaction of urea with amino acids: Implications for urea-induced protein denaturation. J Am Chem Soc. 2007;129:16126–16131. doi: 10.1021/ja076216j. [DOI] [PubMed] [Google Scholar]
- 12.O'Brien EP, et al. Interactions between hydrophobic and ionic solutes in aqueous guanidinium chloride and urea solutions: Lessons for protein denaturation mechanism. J Am Chem Soc. 2007;129:7346–7353. doi: 10.1021/ja069232+. [DOI] [PubMed] [Google Scholar]
- 13.Trzesniak D, Van Der Vegt NFA, Van Gunsteren WF. Analysis of neo-pentane-urea pair potentials of mean force in aqueous urea. Mol Phys. 2007;105:33–39. [Google Scholar]
- 14.Wetlaufer DB, et al. Nonpolar group participation in the denaturation of proteins by urea and guanidinium salts. Model compound studies. J Am Chem Soc. 1964;86:508–514. [Google Scholar]
- 15.Eisenberg D, Kauzmann W. The Structure and Properties of Water. New York: Oxford Univ Press; 1969. [Google Scholar]
- 16.Koga K, Zeng XC, Tanaka H. Freezing of confined water: A bilayer ice phase in hydrophobic nanopores. Phys Rev Lett. 1997;79:5262–5265. doi: 10.1103/physreve.60.5833. [DOI] [PubMed] [Google Scholar]
- 17.Lopez CF, et al. Mechanistic elements of protein cold denaturation. J Phys Chem B. 2008;112:5961–5967. doi: 10.1021/jp075928t. [DOI] [PubMed] [Google Scholar]
- 18.Gunner MR, et al. Backbone dipoles generate positive potentials in all proteins: Origins and implications of the effect. Biophys J. 2000;78:1126–1144. doi: 10.1016/S0006-3495(00)76671-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabelko J, Ervin J, Gruebele M. Cold-denatured ensemble of apomyoglobin: Implications for the early steps of folding. J Phys Chem B. 1998;102:1806–1819. [Google Scholar]
- 20.Hummer G, et al. Role of hydrophobic interactions in pressure denaturation of proteins. Biophys J. 1998;74:A233–A233. [Google Scholar]