Abstract
Unlike most DNA viruses, poxviruses replicate in the cytoplasm of host cells. They encode enzymes needed for genome replication and transcription, including their own thymidine and thymidylate kinases. Some herpes viruses encode only 1 enzyme catalyzing both reactions, a peculiarity used for prodrug activation to obtain maximum specificity. We have solved the crystal structures of vaccinia virus thymidylate kinase bound to TDP or brivudin monophosphate. Although the viral and human enzymes have similar sequences (42% identity), they differ in their homodimeric association and active-site geometry. The vaccinia TMP kinase dimer arrangement is orthogonal and not antiparallel as in human enzyme. This different monomer orientation is related to the presence of a canal connecting the edge of the dimer interface to the TMP base binding pocket. Consequently, the pox enzyme accommodates nucleotides with bulkier bases, like brivudin monophosphate and dGMP; these are efficiently phosphorylated and stabilize the enzyme. The brivudin monophosphate-bound structure explains the structural basis for this specificity, opening the way to the rational development of specific antipox agents that may also be suitable for poxvirus TMP kinase gene-based chemotherapy of cancer.
Keywords: BVdU, dihalovinyl-dU, dimerization mode, TMP kinase, vaccinia virus
There has been growing concern about poxvirus because of the recent emergence of human monkeypox infections in Africa and the U.S. (1) and possible bioterrorism threats. The human population is now susceptible to smallpox since the vaccination program was stopped in 1979 after eradication of the disease. Poxviruses comprise a large family of double-stranded DNA viruses that replicate in the cytoplasm of the host's cells (2). Vaccinia virus (Vacc), the Orthopoxvirus family prototype (3), has a genome with ≈200 ORFs, and some of the proteins they encode are potential therapeutic targets (4). Recent studies have identified several active compounds, including inhibitors of vaccinia protease (5), envelope protein (6) and DNA polymerase such as thymidine analogs (7), and ester lipid analogs of cidofovir (8, 9).
Orthopoxviruses are the only viruses that encode their own thymidine kinase (TK) and thymidylate kinase (TMPK) although herpes simplex virus type 1 (HSV1) and varicella-zoster virus (VZV) encode a TK that has both activities. The potent selective activity of anti-herpes virus nucleoside analogs such as acyclovir (10), ganciclovir (11), and brivudin (BVdU) (12) relies on a specific phosphorylation by the virus thymidine kinase. After conversion of the nucleoside into the required triphosphate form (13), the nucleotide analog can interact with the virus DNA polymerase as a competitive inhibitor with respect to TTP and as an alternative substrate affecting DNA stability and processing during viral growth. The drugs approved for treating herpes virus infections opened the way to antiviral therapies. The same approach may be used to treat orthopoxviruses.
In contrast to human cytosolic TK1, Vacc-TK phosphorylates efficiently araT (14), BVdU (15), N-methanocarbathymidine (16, 17), and novel bulky 5-substituted deoxyuridine analogs that are active against vaccinia virus and other viruses (18). We have focused on the thymidylate kinase encoded by vaccinia virus (Vacc-TMPK) that is 42% identical to human TMP kinase (hTMPK). A clearer understanding of the differences between Vacc-TMPK and human TMPK should help to improve the design of analogs that are specifically phosphorylated during virus infection. The Vacc-TMPK gene, A48R, is an immediate-early gene of the virus. These genes tend to encode virulence factors (19). Vacc-TMPK phosphorylates TMP, dUMP, and 5-halogenated-dUMP as does the human enzyme, but it also phosphorylates dGMP (20). The 5-I-dU efficiently suppresses vaccinia virus infections in mice (21). The final target of 5-I-dU is vaccinia virus DNA polymerase, also the target of cidofovir, an acyclic cytosine nucleoside phosphonate analog that blocks poxvirus infections (9).
Vacc-TMPK belongs to the NMP kinase family, whose members have a highly conserved fold but less well-conserved sequences (20–40%). The structure of hTMPK (22) has features in common with bacterial TMPKs, such as Mycobacterium tuberculosis (Mt) (23) and Staphylococcus aureus (Sa)v (24) and herpes viral TK (25, 26). They fold with a central 5-stranded β-sheet surrounded by helices that are made up of 3 domains: LID, NMP binding, and CORE. The CORE domain involves the P-loop and the ATP-binding site. The LID domain is rather flexible, closing down on the substrate cleft and allowing phosphate transfer. All known TMP kinases are dimeric and have similar CORE domains.
This report describes the structures of Vacc-TMPK bound to TDP and to BVdU-MP. The poxvirus enzyme is a dimer, like hTMP kinase. The TDP-bound structure carries at the active site TDP plus PPi plus Mg2+. These provide valuable insights into its catalytic mechanism. Its mode of dimerization is surprising: The two subunits interact in an orthogonal fashion, in contrast to the antiparallel interactions of human, herpes, and several bacterial TMPKs and to the parallel interaction of the Mt TMPK. This unusual monomer tilting is closely correlated with enzyme stability and with the presence of a cavity allowing broader substrate specificity, as shown by the structure of the BVdU-MP-bound enzyme.
Results
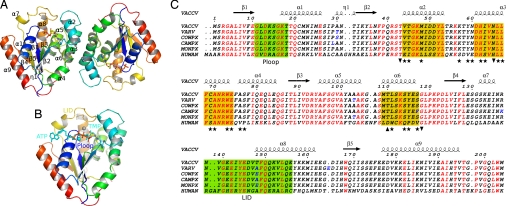
Vacc-TMPK Overall Structure and Oligomerization.
The structure of Vacc-TMPK bound to TDP was determined by molecular replacement and refined to a resolution of 2.4 Å. Data and refinement statistics are summarized in supporting information (SI) Table S1. Arg-93, which is part of the DRX motif of thymidylate kinases, is the only residue lying outside the allowed regions of the Ramachandran plot, in agreement with the structures of other enzymes in this family. The overall Vacc-TMPK structure has an α/β fold with a 5-stranded parallel β-sheet surrounded by 9 α-helices, similar to that of the human enzyme (Fig. 1 A and B) (22). Superimposing the structure of the vaccinia enzyme on that of the human enzyme provided an RMSD of 1.16 Å over 199 Cα atoms. The main differences were in the loops connecting helix α1 and strand β2, helices α5–α6 and helices α7–α8 (the LID region) because of the deletion of residues in Vacc-TMPK (Fig. 1C and Fig. S1). Pox enzymes are well-conserved, Vacc-TMPK differing in only 2 to 3 residues from other poxvirus TMPKs, most of them located within loops (residues 30, 103, and 148 in Fig. 1C). The sequences of TMPKs in the known vaccinia virus strains are identical (Fig. 1C).
Fig. 1.
Quaternary and primary structures of vaccinia virus TMPK. (A) Overall structure of the TMPK dimer. The cartoon of the Vacc-TMPK dimer as seen in the asymmetric unit, rainbow colored with ligands shown as sticks and secondary structure numbering on subunit A. (B) Superimposition of the nucleotides from the human enzyme (PDB ID 1e2q) at the active site of Vacc-TMK chain B. ATP, TMP and magnesium from hTMPK are shown in cyan, whereas TDP and PPi and Mg2+ from Vacc-TMPK are colored according to atom type. (C) Sequence alignment of human, vaccinia (VACCV), variola (VARV), cowpox (COWPX), camelpox (CAMPX), and monkeypox (MONPX) viruses TMP kinases. Identical residues are in red. The TMP kinases of the pox family viruses are identical except for a few conservative substitutions (residues in blue), mostly in loops. Secondary structure elements from the Vacc-TMPK crystal structure are shown above the alignment. The P-loop and LID are boxed in green. Helices involved in dimer contacts are boxed in yellow. The positions of residues involved in the dimer interface of both enzymes are marked with stars, and those unique to human or vaccinia enzyme are marked as up and down triangles, respectively. This figure was created by using ESPript (51).
The vaccinia virus enzyme crystallized as a dimer in the asymmetric unit. The dimer subunits A and B had a noncrystallographic twofold symmetry resulting in the orthogonal stacking of helices α2, α3, and α6 as well as the loop connecting α2 and α3 with their symmetry-related equivalents (symmetry C2). This packing buries ≈880 Å2 of each subunit, i.e., 11.5% of the surface of the monomer. The interface is formed of a closely packed hydrophobic core (74% of nonpolar atoms) with 7 hydrogen bonds. These values are equivalent to those obtained for the buried surfaces of other TMPK dimers (human 780 Å2, Mycobacterium 840 Å2, TK HSV1 900 Å2). They are typical of dimeric protein interfaces that, on average, bury ≈14% of the subunit surface and contain 62% of hydrophobic atoms and ≈1 hydrogen bond per 100 Å2 (27).
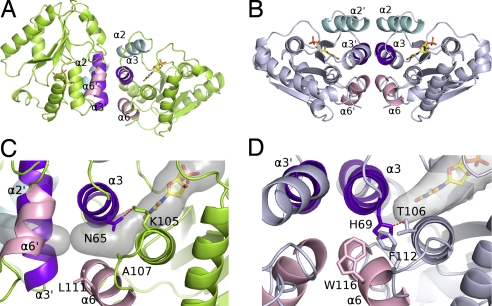
The angle between two equivalent α3 helices is ≈80°, resulting in almost orthogonal dimer packing. This association is quite different from the structures of other TMPKs, where the helix stacking is antiparallel (human, Escherichia coli, Sa) (Fig. 2) or parallel (Mt). The sequences of human and vaccinia helix α3 are very similar (3 differences among 13 residues). There are 14 residue substitutions between the human and vaccinia dimer interfaces contributing to the dimer reorganization (Fig. 1C). Among them, the Pro-120 to Tyr-115 substitution in helix α6 would introduce a steric clash precluding the antiparallel packing of the Vacc-TMPK dimer.
Fig. 2.
Subunit associations of vaccinia virus and human TMP kinases. (A and B) Ribbon representation of the vaccinia virus enzyme in lime green (A) and the human enzyme in pale blue (B) (PDB ID code 1e2d). The interface helices of dimers are in cyan (α2), purple (α3), and pink (α6). (C and D) Close-up of the packing between the helices in vaccinia virus (C) and human (D) enzymes at the dimer interfaces. The α3 helices are packed perpendicularly in the vaccinia enzyme (C) and antiparallel in human TMP kinase (D). His-69 closes the rear of the base binding pocket in the human enzyme and is maintained in this orientation by a hydrogen bond with Thr-106. A secondary shell made of Trp-116 and Phe-112 fills the space between helices α3 and α6 in hTMPK. Asn-65 is rotated through 120° in Vacc-TMPK compared to the human His-69 and is bound to Lys-105. Together with the lower steric hindrance of the secondary shell residues Ala-107 and Leu-111, it creates a cavity [semitransparent gray volume calculated by using CAVER (52)] between helices α3 and α6 connecting the base binding site to the protein surface.
Active Site.
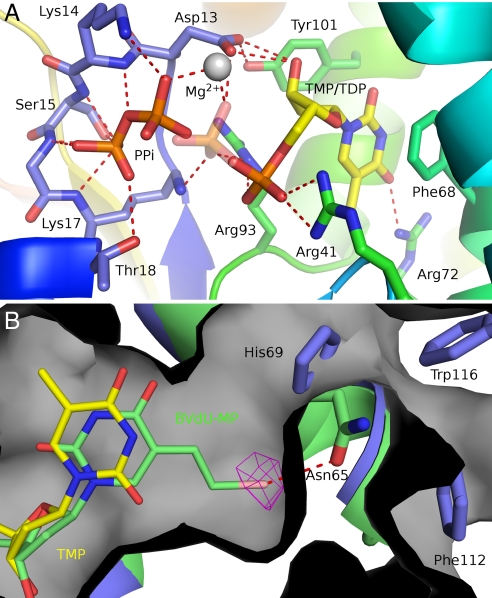
Vacc-TMPK crystals were grown in the presence of TDP, the product of TMP phosphorylation. TDP was present in the NMP site of subunits A and B of the asymmetric unit. A strong extra density was seen in subunit B at the position expected for the β and γ phosphates of the phosphoryl donor ATP, as inferred by superimposition on the human enzyme in complex with TDP and ATP (Fig. 1 A and B). This density was modeled as a pyrophosphate. Excess TDP was presumably bound to the ATP site, but nucleoside moiety and α phosphate were not visible in the electron density. Because these bound species were reactive, we saw an average density, representing either TMP plus PPi or TDP plus Pi (Fig. S2). Among the 4 phosphates at the B subunit active sites, the two central ones had a lower electron density, reflecting half-occupation because of the phosphotransfer. This feature was refined by using alternate conformations with the transferred phosphoryl group on either the acceptor site (TDP) or the donor site (PPi). The difference between the 2 active sites in the A and B subunits could be due to the crystal packing that stabilizes the lid in B and allows lower B-factors.
The binding of TDP at the NMP binding (acceptor) site was similar to the binding of TMP to hTMPK. The base was stacked to Phe-68 with its oxygen O4 bound to Arg-72, both residues belonging to the α3 helix. The sugar sat between Tyr-101 and Leu-53, and the P-loop appeared to have a closed conformation (22) with the carboxylate of Asp-13 hydrogen bonded to the 3′ hydroxyl of TDP (Fig. 3A). The P-loop was held in this closed conformation by the H-bond between Tyr-101 and Asp-13. The α-phosphate was bound to Arg-93 (from the DRX motif) and Arg-41, whereas the β-phosphate was bound to Arg-41, Lys-17, and the magnesium ion.
Fig. 3.
The Vacc-TMP kinase active site. (A) View of the active site with the two alternated conformations of the TDP-PPi ligands. The enzyme is rainbow colored, and TMP and PPi are shown as sticks and colored by atom type. Polar interactions are indicated by red dashes. The transferred phosphoryl group here shown at the donor site (PPi) has an alternate conformation (semitransparent phosphate) bound to the thymidine nucleotide. (B) The superimposed crystallographic structures of BVdU-MP (green sticks) and TDP (yellow sticks) at the active site of Vacc-TMPK (green). The bromovinyl group fits into a cavity (gray surface) at the rear of the base binding pocket and is specifically recognized by a halogen bond to Asn-65 (red dashes). The bromine atom is shown in its anomalous difference electron density map contoured at 5 σ. His-69, Trp-116, and Phe-112 close this cavity in the human enzyme (blue).
At the donor site, the pyrophosphate was bound to the P-loop. Its first phosphate occupied the position of the β-phosphate of ATP and was bound to all main-chain nitrogens from Lys-14 to Thr-18, and to Thr-18 side chain. Its second phosphate occupied the position of the γ-phosphate of ATP and was bound to the Lys-14 side-chain and main-chain nitrogens and to Mg2+. The magnesium ion in the B subunit had octahedral coordination geometry but was pentacoordinated with an apical ligand missing (Fig. S3). A magnesium ion was also found near the TMP binding site of Mt-TMPK (23).
There was a major difference between the acceptor sites of the vaccinia and human TMPKs at the rear of the base binding pocket. In hTMPK, the pocket is closed by the H-bonded side chains of His-69 (in α3 helix) and Thr-106 (in α6 helix). The equivalent residues in Vacc-TMPK are Asn-65 and Ala-102, which do not interact. Instead, Asn-65 made polar contact with Lys-105, so that the Asn-65 side chain was rotated by 120° compared with that of His-69. The residues in the secondary shell around the active site (and near the dimer interface) were Leu-111 and Ala-107 (α6 helix) in Vacc-TMPK and Trp-116 and Phe-112 in hTMPK. Consequently, the cavity of the base binding pocket was much larger in Vacc-TMPK and was connected to the solvent through a channel that ended in the vicinity of the dimer interface (Fig. 3B). This cleft at the back of Vacc-TMPK extended between α3 and α6 and greatly reduced contact between these helices, creating a packing defect within the monomer. The packing between α3 and α6 involved only half as many residues in the Vacc-protein compared with the human enzyme and buried at least 40% less surface (420 versus 750 Å2).
Vacc-TMPK Enzyme Activity and Acceptor Site Specificity.
The large cavity of the base binding pocket should allow broader substrate specificity. We therefore examined the capacity of Vacc-TMPK to accommodate bulky chains in the 5 position of the pyrimidine ring. (E)-5-(2 bromovinyl)-dU (BVdU) has already been reported to be a substrate for Vacc-TK (15). The monophosphate of BVdU is indeed phosphorylated by Vacc-TMP kinase with a KM of 190 ± 20 μM (10 times higher than KMTMP) and a turnover number (0.86 ± 0.10 s−1) approximately one third the kcat for TMP. Both substrates are processed without any apparent cooperativity. The catalytic efficiency of BVdU-MP (CE = 4.5 × 103 M−1s−1) and dGMP (20) are quite similar, at ≈5% of the TMP efficiency (CE = 105 M−1s−1) (Fig. S4).
We investigated the acceptor site by comparing the inhibition of Vacc- and hTMPK by several nucleosides. Vacc-TMPK was more sensitive to dT than was the human enzyme (Table 1). This agrees with the previous determination of dT affinity by a competition assay using fluorescent TDP analog as a probe: KDdT was ≈2 μM for Vacc-TMPK and 40 μM for the human enzyme (20, 28). The better affinity of dT for the vaccinia enzyme is probably due to the P-loop Asp-13 maintained in closed conformation by H-bonding to Tyr-101, a residue replaced by Phe in hTMPK. The absence of a 5-methyl resulted in a much poorer inhibition of Vacc-TMPK activity (KIdU = 1 mM compared with KIdT = 25 μM) (Table 1) but inserting a halogen atom in the 5 position restores inhibition. 5-Br and 5-I-dU were among the most efficient inhibitors (Table 1). These data correlate with the catalytic efficiencies of their monophosphate form, kcat/KM = 5 × 104 and 3.8 × 104 M−1s−1 for 5-Br-dUMP and 5-I-dUMP, respectively (20). Substrate inhibition was observed at high concentration for 5-I-dUMP (Fig. S4), an inhibition devoid of physiological significance and generally attributed to unproductive binding of the ligand at the same site or at a secondary site. The 5-bromovinyl moiety was accommodated by the Vacc-TMPK (KIBVdU = 0.5 mM) better than by the human enzyme (KIBVdU = 1.1 mM). However, the presence of 2 halogens on the vinyl moiety reduced its specificity for the viral TMP kinase (Table 1). Further measurements of the phosphorylation of these derivatives as monophosphates are needed to confirm these results. Our data agree with the observed plasticity of the Vacc-TMPK acceptor site.
Table 1.
Inhibition potencies of thymidine analogs for vaccinia virus and human TMP kinases
| Inhibitor | Ki for Vacc TMP kinase | Ki for human TMP kinase, mM |
|---|---|---|
| dT | 25 ± 5 μM | 0.25 |
| dU | 1.0 ± 0.1 mM | 2.5 |
| 5F-dU | 0.7 ± 0.1 mM | >5 |
| 5Br-dU | 7 ± 1 μM | 0.2 |
| 5Cl-dU | 15 ± 5 μM | 0.37 |
| 5I-dU | 23 ± 2 μM | 0.35 |
| BVdU | 0.5 ± 0.1 mM | 1.1 ± 0.1 |
| 5I-BrV-dU | 0.5 ± 0.05 mM | 0.3 ± 0.05 |
| 5diIV-dU | 0.6 ± 0.1 mM | 0.35 ± 0.15 |
Activities were measured in standard conditions with [ATP] = 1 mM, [TMP] from 10 μM to 1 mM, and varying inhibitor concentrations (0-5-50 μM for the more efficient). KI values were obtained from double reciprocal plots. Shown in italics are data from ref. 41.
We solved the structure of Vacc-TMPK bound to BVdU-MP at a resolution of 2.9 Å (Fig. S5) to obtain further insight into this phenomenon. We measured the anomalous signal originating from the bromine atom of the nucleotide analog so as to unambiguously position it in the active site, Fig. 3B shows BVdU-MP in the TMP site channel. The presence of a bromovinyl group in position 5 caused the glycosyl bond to rotate from anti to syn to accommodate the bulky group in the extra cavity of the vaccinia enzyme active site. The replacement of His-69 and Thr-106 in the human enzyme by Asn-65 and Ala-102, respectively in the pox enzyme, and the resulting side chain flipping, clearly opens the active site to bulky groups. The BVdU-MP is specifically recognized by a bromine halogen bond with the Asn-65 side chain (29).
Conformational Stability of Vacc-TMPK.
We have considered the relationship between the unusual active-site geometry, the dimerization mode and the stability of TMP kinases by studying the thermal unfolding of Vacc-TMPK and hTMPK. Unfolding was analyzed by microcalorimetry between 10 °C and 80 °C, at pH 7.5 at a protein concentration of 1.5 mg/ml. The thermal unfolding of both proteins was irreversible, as for HSV-1-TK and Mt-TMP kinase (30, 31). The midtemperature (Tm) of thermal denaturation was 42.5 °C for the viral enzyme and 52 °C for the human enzyme. Mt-TMPK is reported to be much more stable with a Tm = 68 °C without ligand, and 75 °C with 1 mM TMP (31). Adding 1 mM TMP increased the Tm of Vacc-TMPK by 4.9 °C and that of hTMPK by 7.7 °C, in agreement with findings for another viral kinase, HSV-1-TK, that is also not very temperature stable (Tm = 43.5 °C) and was stabilized by its substrate T (Tm = 52.6 °C) (32). The BVdU-MP stabilized the Vacc-TMPK by 2.8 °C and marginally modified the human TMPK Tm, whereas dGMP increased the Vacc-TMPK Tm by 2.3 °C but by only 1 °C for hTMPK (Table S2). These results demonstrate the preferential binding of BVdUMP and dGMP to vaccinia enzyme.
Discussion
Although the vaccinia and human TMPKs have similar sequences and monomer folds, their quaternary structures are remarkably different. Like the human enzyme, Vacc-TMPK is a homodimer. Because close homologs (≥30–40% identity) should interact in almost the same way (33), we originally proposed an identical dimeric association (20). The dimer interfaces of both enzymes involve the same helices (α2, α3, and α6), but their arrangements are different. The packing of the α3 helices is antiparallel in the human (22), yeast (34), and Sa (24) enzymes but almost orthogonal in Vacc-TMPK (Fig. 2). This unusual structural property of the Vacc-TMPK appears to be related to a broader substrate specificity, and this may now be exploited for rational drug development.
The Mt enzyme (22% identity with hTMPK) also differs from the human enzyme in that the dimer is parallel. The buried areas are similar (≈1,700 Å2) in all 3 types of packing, but the stabilities of the enzymes differ greatly. The Mt dimer is resistant to temperatures >65 °C because of a salt bridge between monomers (23). Although the dimer interfaces of the human and vaccinia enzymes have very similar physicochemical properties, their thermal stabilities differ by >10 °C: hTMPK has a Tm of 52 °C, whereas that of Vacc-TMPK is 42.5 °C. A similar marginal stability has also been reported for HSV1-TK (32).
An important finding was the smaller contact area between helices α3 and α6 within the Vacc-TMPK monomer. The residues involved in the interaction of helices α3 and α6 in hTMPK constitute a second shell of bulky residues at the rear of the active site, and their absence creates a large canal between the thymine-binding pocket of Vacc-TMPK and the surface of the protein, emerging at the edge of the dimer interface (Fig. 2B). The stability and flexibility of a protein are directly correlated with its packing density (35, 36), and this cavity should make a major contribution to the decreased stability of the enzyme. The orthogonal dimerization probably counterbalances this effect. In the context of a packing defect between helices α3 and α6 in the vaccinia monomers, the orthogonal dimerization, which is unprecedented among the NMP kinases, brings these helices perpendicular to their equivalents (α3′ and α6′) of the other subunit. This geometry avoids propagating the packing defect between monomers and thus helps stabilizing the interaction between these helices. Together, these properties make the active site more plastic and able to accommodate bulky molecules like pyrimidines with large groups at the C-5 position (BVdU-MP) or even purine nucleotides (dGMP). The binding of these bulky substrates to the TMP site is demonstrated by their ability to be phosphorylated and protect the enzyme from thermal denaturation. The shape of the TMP-binding pockets of these related enzymes vary remarkably, as described for protein pockets binding the same ligand (37). This diversity is most important for designing specific drugs. The structure of the BVdU-MP bound to the vaccinia enzyme shows the key role of the halogen bond in its recognition and explains the molecular basis for such specificity.
The differences in the specificities of vaccinia TK and TMPK and their human counterparts must be studied to understand analog phosphorylation by vaccinia virus-infected cells. Vacc-TK accepts major changes in the substrate 5 position (14, 18). BVdU-MP prodrugs like cyclosal-BVdU monophosphate are active in vitro against vaccinia virus infection of Vero cells (38), in agreement with our structural and kinetic findings. Moreover, BVdU is a better inhibitor (EC50 ≈1 μM) of the infection of human embryonic lung (HEL) cells in culture by vaccinia virus than is cidofovir (result not shown).
Our findings also provide a rational basis for the use of vaccinia virus TMPK genes in combined suicide gene/chemotherapy in conjunction with specific substrates such as BVdU derivatives. Vaccinia viruses are already considered to be efficient vectors for gene therapy and as oncolytic viruses that specifically target tumors (39). A combination of both approaches could result in synergistic inhibitory effects.
In conclusion, our studies have shown that the unusual dimeric packing of Vacc-TMPK is correlated with an enlarged substrate-binding site, which should allow the rational design and development of novel Vacc-TMPK substrates. We have already demonstrated that nucleotides like brivudin monophosphate are a promising lead and provided the structural basis for further specific drug development.
Materials and Methods
Chemicals.
Nucleoside analogs were from Sigma. BVdU monophosphate was synthesized by L. Kerremans (Rega Institute for Medical Research, Leuven, Belgium). Cidofovir was from Gilead. 5-(1,2-dihalo)vinyl-dU was synthesized as reported (40).
Protein Purification and TMPKs Assays.
His-tagged Vacc-TMPK and human TMPK were synthesized and purified to homogeneity as previously described (20, 41). The purified proteins were equilibrated by dialysis against 50 mM Tris·HCl (pH 7.5) buffer containing 20 mM NaCl, 1 mM DTT, and 50% glycerol. The forward reaction of TMP kinase at 37 °C was followed at 340 nm by measuring ADP formation (20).
Crystallization and Data Collection.
TDP/MgCl2 or BVdU-MP/MgCl2 were added to Vacc-TMPK before initial screening with NeXtal kits. A Cartesian Technology pipetting robot was used to set up 200-nl sitting drops in 96-well microplates at 20 °C (Greiner). Final TDP cocrystals were grown by mixing 1 μl of 5 mg/ml Vacc-TMPK, 5 mM TDP, 10 mM MgCl2, and 1 μl of the reservoir solution containing 23% (wt/vol) polyethylene glycol 400 (PEG400), 100 mM sodium acetate, and 100 mM Mes buffer (pH 6.5). Crystals were soaked in the reservoir solution supplemented with PEG400 (final concentration: 25% wt/vol) and then frozen at 100 K. Data were collected on beamline ID14–1 at the European Synchrotron Radiation Facility (ESRF) in Grenoble, France, by using a wavelength of 0.934 Å. The crystal system is orthorhombic, space group P212121, with 2 molecules per asymmetric unit. Final BVdU-MP cocrystals were grown by mixing 100 nl of 10 mg/ml Vacc-TMPK, 28 mM BVdU-MP, 5 mM MgCl2 and 100 nl of 24% PEG 2000 MME, 0.1M Tris·HCl (pH 8.5). Crystals were soaked in the reservoir solution supplemented with 20% (wt/vol) glycerol, 5 mM MgCl2 and 10 mM BVdU-MP and frozen at 100 K. Microcrystals were screened on the ID23-2 microfocus beamline, and anomalous data were collected near the Br K-edge absorption wavelength (0.9198 Å) on beamline ID23-1 (ESRF). The crystal system is orthorhombic, space group P212121, with 2 molecules per asymmetric unit. Data were processed and scaled with the XDS package (42) (Table S1).
Structure Determination.
The structure of the TDP/Vacc-TMPK complex was solved by molecular replacement with PHASER (43), by using human TMPK (PDB ID 1e2g) as search model. This structure was subsequently used to solve the BVdU-MP/Vacc-TMPK complex by using the same method. The nucleotides in the 2 molecules of the asymmetric unit immediately gave clear electronic density in both cases. The initial model was rebuilt by using COOT (44). CNS (45) and Phenix (46) were used for refinement, which was monitored by using the free R factor (47). Simulated annealing plus a light noncrystallographic constraint were initially used. In the final steps, we placed water molecules in residual densities >2.5 standard deviations. The final TDP-TMPK model contained 2 polypeptide chains, 2 TDP molecules, and a pyrophosphate molecule, and the final BVdUMP-TMPK model contained 2 polypeptides and 2 analog molecules. The models were evaluated by using COOT validation tools and PROCHECK (48). Models were superimposed with lsqkab from the CCP4 suite (49). The surface buried by protein–protein interaction was calculated by using CNS with a molecular probe of 1.4-Å diameter. A summary of the refinement and data statistics is given in Table S1. The structure representations were realized by using PyMOL (50).
Differential Scanning Microcalorimetry.
Differential scanning microcalorimetry was carried out on a Model 6300 Nano-DSC III (Calorimetry Sciences). Samples were dialyzed against 50 mM Tris·HCl (pH 7.5) and degassed for 5 min under vacuum before loading. The scan rate was 1.0 °C/min. After the first heating scan, the samples were scanned for a second time to estimate the reversibility of the unfolding transition. Buffer baselines were measured under identical conditions and were removed from corresponding sample data.
Supplementary Material
Acknowledgments.
We thank the staff on beamlines ID14-1, ID23-1 and ID23-2 of European Synchrotron Radiation Facility (Grenoble, France), particularly Didier Nurizzo and David Flot, for their assistance; Pierre Nicolas Université Marie et Pierre Curie-Paris 6 (UMPC-Paris 6) and Francis Schaeffer (Institut Pasteur, Paris, France) for helpful discussions; Michele Reboud (UPMC-Paris 6) for laboratory facilities, Aurélien Montaigu and Vincent Roy (Institut de Chimie Organique et Analytique, Orléans) for synthesizing dihalovinyl-dU. This work was supported by UMPC-Paris 6 and the Centre National de la Recherche Scientifique (Unité Propre de Recherche 3082 and Formation de Recherche en Evolution 2852), Agence Nationale de Recherche Grant ANR-05-BLAN-0368-02 (to L.A.A. and D.D.-B.), the Association pour la Recherche Contre le Cancer (P.M.), the Fondation pour la Recherche Médicale (C.C.), and a Geconcerteerde Onderzoeksacties grant (Krediet no. 05/19) of the Katholieke Universiteit Leuven (to J.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2v54 and 2w0s).
This article contains supporting information online at www.pnas.org/cgi/content/full/0804525105/DCSupplemental.
References
- 1.Nalca A, Rimoin AW, Bavari S, Whitehouse CA. Reemergence of monkeypox: Prevalence, diagnostics, and countermeasures. Clin Infect Dis. 2005;41:1765–1771. doi: 10.1086/498155. [DOI] [PubMed] [Google Scholar]
- 2.Moss B. In: Virology. 2nd Ed. Fields BN, et al., editors. New York: Raven Press; 1990. pp. 2079–2111. [Google Scholar]
- 3.De Clercq E. Vaccinia virus inhibitors as a paradigm for the chemotherapy of poxvirus infections. Clin Microbiol Rev. 2001;14:382–397. doi: 10.1128/CMR.14.2.382-397.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Resch W, Hixson KK, Moore RJ, Lipton MS, Moss B. Protein composition of the vaccinia virus mature virion. Virology. 2007;358:233–247. doi: 10.1016/j.virol.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 5.Byrd CM, et al. New class of orthopoxvirus antiviral drugs that block viral maturation. J Virol. 2004;78:12147–12156. doi: 10.1128/JVI.78.22.12147-12156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang G, et al. An orally bioavailable antipoxvirus compound (ST-246) inhibits extracellular virus formation and protects mice from lethal orthopoxvirus challenge. J Virol. 2005;79:13139–13149. doi: 10.1128/JVI.79.20.13139-13149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smee DF, et al. Efficacy of N-methanocarbathymidine in treating mice infected intranasally with the IHD and WR strains of vaccinia virus. Antiviral Res. 2007;76:124–129. doi: 10.1016/j.antiviral.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lebeau I, et al. Activities of alkoxyalkyl esters of cidofovir (CDV), cyclic CDV, and (S)-9-(3-hydroxy-2-phosphonylmethoxypropyl)adenine against orthopoxviruses in cell monolayers and in organotypic cultures. Antimicrob Agents Chemother. 2006;50:2525–2529. doi: 10.1128/AAC.01489-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magee WC, Hostetler KY, Evans DH. Mechanism of inhibition of vaccinia virus DNA polymerase by cidofovir diphosphate. Antimicrob Agents Chemother. 2005;49:3152–3162. doi: 10.1128/AAC.49.8.3153-3162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colby BM, Furman PA, Shaw JE, Elion GB, Pagano JS. Phosphorylation of acyclovir [9-(2-hydroxyethoxymethyl)guanine] in Epstein–Barr virus-infected lymphoblastoid cell lines. J Virol. 1981;38:606–611. doi: 10.1128/jvi.38.2.606-611.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith KO, Galloway KS, Kennell WL, Ogilvie KK, Radatus BK. A new nucleoside analog, 9-[[2-hydroxy-1-(hydroxymethyl)ethoxyl]methyl]guanine, highly active in vitro against herpes simplex virus types 1 and 2. Antimicrob Agents Chemother. 1982;22:55–61. doi: 10.1128/aac.22.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Clercq E. Discovery and development of BVDU (brivudin) as a therapeutic for the treatment of herpes zoster. Biochem Pharmacol. 2004;68:2301–2315. doi: 10.1016/j.bcp.2004.07.039. [DOI] [PubMed] [Google Scholar]
- 13.Fyfe JA. Differential phosphorylation of (E)-5-(2-bromovinyl)-2′-deoxyuridine monophosphate by thymidylate kinases from herpes simplex viruses types 1 and 2 and varicella zoster virus. Mol Pharmacol. 1982;21:432–437. [PubMed] [Google Scholar]
- 14.El Omari K, Solaroli N, Karlsson A, Balzarini J, Stammers DK. Structure of vaccinia virus thymidine kinase in complex with dTTP: Insights for drug design. BMC Struct Biol. 2006;6:22. doi: 10.1186/1472-6807-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solaroli N, Johansson M, Balzarini J, Karlsson A. Substrate specificity of three viral thymidine kinases (TK): Vaccinia virus TK, feline herpesvirus TK, and canine herpesvirus TK. Nucleosides Nucleotides Nucleic Acids. 2006;25:1189–1192. doi: 10.1080/15257770600894451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marquez VE, et al. Nucleosides with a twist. Can fixed forms of sugar ring pucker influence biological activity in nucleosides and oligonucleotides? J Med Chem. 1996;39:3739–3747. doi: 10.1021/jm960306+. [DOI] [PubMed] [Google Scholar]
- 17.Smee DF, et al. Cell line dependency for antiviral activity and in vivo efficacy of N-methanocarbathymidine against orthopoxvirus infections in mice. Antiviral Res. 2007;73:69–77. doi: 10.1016/j.antiviral.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Prichard MN, et al. Selective phosphorylation of antiviral drugs by vaccinia virus thymidine kinase. Antimicrob Agents Chemother. 2007;51:1795–1803. doi: 10.1128/AAC.01447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assarsson E, et al. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc Natl Acad Sci USA. 2008;105:2140–2145. doi: 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Topalis D, et al. Substrate specificity of vaccinia virus thymidylate kinase. FEBS J. 2005;272:6254–6265. doi: 10.1111/j.1742-4658.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- 21.Neyts J, Verbeken E, De Clercq E. Effect of 5-iodo-2′-deoxyuridine on vaccinia virus (orthopoxvirus) infections in mice. Antimicrob Agents Chemother. 2002;46:2842–2847. doi: 10.1128/AAC.46.9.2842-2847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostermann N, et al. Insights into the phosphoryltransfer mechanism of human thymidylate kinase gained from crystal structures of enzyme complexes along the reaction coordinate. Structure (London) 2000;8:629–642. doi: 10.1016/s0969-2126(00)00149-0. [DOI] [PubMed] [Google Scholar]
- 23.Li de la Sierra I, Munier-Lehmann H, Gilles AM, Barzu O, Delarue M. X-ray structure of TMP kinase from Mycobacterium tuberculosis complexed with TMP at 1.95 A resolution. J Mol Biol. 2001;311:87–100. doi: 10.1006/jmbi.2001.4843. [DOI] [PubMed] [Google Scholar]
- 24.Kotaka M, et al. Structures of S. aureus thymidylate kinase reveal an atypical active site configuration and an intermediate conformational state upon substrate binding. Protein Sci. 2006;15:774–784. doi: 10.1110/ps.052002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wild K, Bohner T, Folkers G, Schulz GE. The structures of thymidine kinase from herpes simplex virus type 1 in complex with substrates and a substrate analog. Protein Sci. 1997;6:2097–2106. doi: 10.1002/pro.5560061005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardberg A, Shuvalova L, Monnerjahn C, Konrad M, Lavie A. Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases. Structure (London) 2003;11:1265–1277. doi: 10.1016/j.str.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Ponstingl H, Kabir T, Gorse D, Thornton JM. Morphological aspects of oligomeric protein structures. Prog Biophys Mol Biol. 2005;89:9–35. doi: 10.1016/j.pbiomolbio.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Alexandre JA, et al. Enantioselectivity of human AMP, dTMP and UMP-CMP kinases. Nucleic Acids Res. 2007;35:4895–4904. doi: 10.1093/nar/gkm479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voth AR, Hays FA, Ho PS. Directing macromolecular conformation through halogen bonds. Proc Natl Acad Sci USA. 2007;104:6188–6193. doi: 10.1073/pnas.0610531104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wurth C, Thomas RM, Folkers G, Scapozza L. Folding and self-assembly of herpes simplex virus type 1 thymidine kinase. J Mol Biol. 2001;313:657–670. doi: 10.1006/jmbi.2001.5060. [DOI] [PubMed] [Google Scholar]
- 31.Munier-Lehmann H, Chaffotte A, Pochet S, Labesse G. Thymidylate kinase of Mycobacterium tuberculosis: A chimera sharing properties common to eukaryotic and bacterial enzymes. Protein Sci. 2001;10:1195–1205. doi: 10.1110/ps.45701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wurth C, et al. The effect of substrate binding on the conformation and structural stability of Herpes simplex virus type 1 thymidine kinase. Protein Sci. 2001;10:63–73. doi: 10.1110/ps.27401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J Mol Biol. 2003;332:989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Lavie A, et al. Crystal structure of yeast thymidylate kinase complexed with the bisubstrate inhibitor P1-(5′-adenosyl) P5-(5′-thymidyl) pentaphosphate (TP5A) at 2.0 A resolution: Implications for catalysis and AZT activation. Biochemistry. 1998;37:3677–3686. doi: 10.1021/bi9720787. [DOI] [PubMed] [Google Scholar]
- 35.Eriksson AE, et al. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255:178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- 36.Halle B. Flexibility and packing in proteins. Proc Natl Acad Sci USA. 2002;99:1274–1279. doi: 10.1073/pnas.032522499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kahraman A, Morris RJ, Laskowski RA, Thornton JM. Shape variation in protein binding pockets and their ligands. J Mol Biol. 2007;368:283–301. doi: 10.1016/j.jmb.2007.01.086. [DOI] [PubMed] [Google Scholar]
- 38.Sauerbrei A, et al. In vitro activity of cycloSal-nucleoside monophosphates and polyhydroxycarboxylates against orthopoxviruses. Antiviral Res. 2005;67:147–154. doi: 10.1016/j.antiviral.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Thorne SH, et al. Rational strain selection and engineering creates a broad-spectrum, systemically effective oncolytic poxvirus, JX-963. J Clin Invest. 2007;117:3350–3358. doi: 10.1172/JCI32727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Escuret V, et al. Synthesis of 5-haloethynyl- and 5-(1,2-dihalo)vinyluracil nucleosides: Antiviral activity and cellular toxicity. Bioorg Med Chem. 2005;13:6015–6024. doi: 10.1016/j.bmc.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 41.Pochet S, Dugué L, Labesse G, Delepierre M, Munier-Lehmann H. Comparative study of purine and pyrimidine nucleoside analogs acting on the thymidylate kinases of Mycobacterium tuberculosis and of humans. ChemBioChem. 2003;4:742–747. doi: 10.1002/cbic.200300608. [DOI] [PubMed] [Google Scholar]
- 42.Kabsch B. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 43.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 47.Brünger A. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:372–375. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 48.Laskowski RA, et al. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 49.CCP4. The CCP4 suite: programs for protein crystallography Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 50.DeLano W. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 51.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 52.Petrek M, et al. CAVER: A new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinformatics. 2006;7:316. doi: 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.