Abstract
The error-free repair of double-stranded DNA breaks by homologous recombination requires processing of broken ends. These processed ends are substrates for assembly of DNA strand exchange proteins that mediate DNA strand invasion. Here, we establish that human BLM helicase, a member of the RecQ family, stimulates the nucleolytic activity of human exonuclease 1 (hExo1), a 5′→3′ double-stranded DNA exonuclease. The stimulation is specific because other RecQ homologs fail to stimulate hExo1. Stimulation of DNA resection by hExo1 is independent of BLM helicase activity and is, instead, mediated by an interaction between the 2 proteins. Finally, we show that DNA ends resected by hExo1 and BLM are used by human Rad51, but not its yeast or bacterial counterparts, to promote homologous DNA pairing. This in vitro system recapitulates initial steps of homologous recombination and provides biochemical evidence for a role of BLM and Exo1 in the initiation of recombinational DNA repair.
Keywords: Bloom syndrome, Rad51, recombination, RecQ, DNA pairing
Homologous recombination contributes toward the maintenance of genomic integrity by accurately repairing double-stranded DNA (dsDNA) breaks. The recombinational repair of dsDNA breaks (DSBs) proceeds by successive reactions that start with processing of the broken DNA ends to reveal single-stranded DNA (ssDNA) (1). Processing requires the action of a helicase and/or nuclease. The resultant 3′-terminated ssDNA is used by a DNA strand exchange protein as the substrate for assembly of the nucleoprotein filament that is the active species in the search for homologous DNA and subsequent DNA pairing. It is unclear which enzymes in eukaryotes resect the DNA break to initiate recombination; however, much is known about the process in bacteria. In Escherichia coli, recombinational DNA repair occurs by either the RecBCD or the RecF pathway (1). RecBCD is a helicase/nuclease that processes dsDNA to produce 3′-tailed ssDNA onto which RecA is loaded. In the RecF pathway, a separate helicase and nuclease are used for resection of DSBs and ssDNA-gaps: RecQ, a 3′→5′ helicase, and RecJ, a 5′→3′ exonuclease (1).
Components of the RecF pathway have counterparts in eukaryotes (1). Human exonuclease 1 (hExo1) is a 5′→3′ exonuclease (2, 3) that exists in Ia and Ib isoforms (2) and shares 27% identity with yeast Exo1 (2). Discovered in yeast (4), Exo1 participates in recombination (4, 5), telomere maintenance (6, 7), mismatch repair (2, 3, 8), and processing of stalled replication forks (9). Consistent with its recombination function, mice devoid of Exo1 demonstrate a reduction in ssDNA formation at DSBs (7). Although RecJ and Exo1 have the same DNA degradation polarity, they differ in substrate specificity: RecJ acts preferentially on ssDNA (10), whereas Exo1 acts on dsDNA (4). Nonetheless, both nucleases are envisioned to function similarly in the initiation of recombinational repair (1, 4, 9, 11). Although the Mre11–Rad50–Nbs1 (MRN) (12) complex and CtIP (13) are needed for DSB processing, it is evident that MRN, which possesses a 3′→5′ exonuclease activity, is not involved in extensive resection of DSBs (14).
Humans have 5 helicases related to RecQ (15). BLM is a member of this RecQ family (16) and has been implicated in branch migration (17), disruption of joint molecules (18–20), and processing of double Holliday junctions by interaction with Rmi1 and topoisomerase IIIα (19, 21, 22), all late functions. However, E. coli RecQ functions both early and late in recombination (1, 11, 23) by stimulating RecJ (N. Handa, K. Morimatsu, S. T. Lovett, and S.C.K., unpublished observations) and cooperating with topoisomerase III (24), respectively. Thus, we examined whether BLM might also have multiple functions and could stimulate hExo1 to mimic RecQ and RecJ behavior functionally. Here, we show that hExo1 and BLM cooperate to resect broken DNA and to enable DNA pairing by the human DNA strand exchange protein, hRad51 (25).
Results
BLM Stimulates Resection of dsDNA by hExo1.
To determine whether BLM modulates hExo1 activity, we monitored hExo1-mediated degradation of 3′-end-labeled dsDNA (2.7 kbp). Because hExo1 is a 5′→3′ exonuclease, the use of 3′ end-labeled dsDNA permits visualization of resection intermediates. By itself, hExo1 is not a processive nuclease (5), and it showed only limited resection at this concentration (Fig. 1A, lanes 7–9 and 16–18). The BLM was free of contaminating nucleases (Fig. 1A, lanes 1–3 and lanes 10–12) and was unable to unwind the long 2.7-kbp dsDNA, consistent with its limited helicase activity (26, 27). However, BLM stimulated the hExo1-mediated degradation of DNA to generate resection products ranging from ≈2 kbp to 125 bp (Fig. 1A, lanes 6 and 15). BLM stimulated both isoforms of hExo1 to a similar extent (Fig. 1A, lanes 4–6 and lanes 13–15), which, at this concentration of BLM (40 nM), was ≈3-fold (Fig. 1B). No difference was seen between the 2 isoforms in this and all subsequent assays reported here (data not shown); consequently, only the results with hExo1b are shown hereafter. Similar results were obtained when the reaction products were analyzed by alkaline electrophoresis supporting information (SI) Fig. S1A]. The stimulation of hExo1 by BLM was also evident with 5′ end-labeled dsDNA (Fig. S1B), wherein resection led to simple loss of end label from the substrate. Because resection of 5′ end-labeled substrate precluded radioisotopic detection of processing intermediates or DNA-pairing products (see below), the 3′ end-labeled DNA was used for all ensuing reactions.
Fig. 1.
BLM stimulates resection of dsDNA by hExo1. Nuclease reactions were performed by using 3′ end-labeled EcoRI-linearized pUC19 as described in Materials and Methods. (A) Gel showing time courses. Lanes: 1–3, BLM; 4–6, hExo1b and BLM; 7–9, hExo1b; 10–12, BLM; 13–15, hExo1a and BLM; and 16–18, hExo1a. (B) Quantification of hExo1 activity. The percentage of intact dsDNA remaining, from experiments as shown in A, was determined relative to the 0-min time points (100%). Error bars indicate variation between multiple preparations and independent experiments and were determined by using GraphPad Prism version 4. (C) Gel showing stimulation of Exo1 (20 nM) as a function of BLM concentration (0, 20, 40, and 80 nM) after 5 min; lanes: 1, absence of proteins; and 2–5, increasing concentration of BLM. The positions of the intact substrate (2.7 kbp), resection products, and molecular size standards (kbp) are indicated.
Stimulation of hExo1-mediated resection of DNA depends on BLM concentration: at the highest BLM concentration tested (80 nM), most of the DNA was resected within the 5-min incubation time, and products were broadly distributed in size (Fig. 1C, lane 5). Because human RPA (hRPA) interacts with and stimulates BLM unwinding activity (27), we tested its effect on BLM-dependent stimulation of hExo1. RPA did not affect the nuclease activity of hExo1 alone (data not shown), nor did it appreciably change substrate utilization by hExo1 in the presence of BLM (Fig. S2); however, RPA did decrease the extent of resection (i.e., increase the size of the DNA intermediates produced) by BLM and hExo1 (Fig. S2). A similar effect of RPA was observed for hMutSα-dependent stimulation of hExo1, and it was determined that RPA reduced the nucleolytic processivity of hExo1 (28).
The Exonuclease Activity of hExo1 Is Essential for BLM-Mediated Stimulation of DNA Resection.
To confirm that the nuclease activity intrinsic to hExo1 protein is essential for BLM-stimulated resection, we initially performed a hExo1 titration in the absence and presence of BLM; resection was indeed a function of hExo1 concentration, and BLM stimulated this activity (Fig. 2A). To verify that BLM stimulated the intrinsic nuclease activity of hExo1 and not a contaminant present in the protein preparation, we also examined a mutated hExo1 that lacks nuclease activity, hExo1(D173A) protein (29). The mutant protein was purified in the same manner as the wild-type protein and was characterized (29). With either hExo1(D173A) alone or both hExo1(D173A) and BLM, nucleolytic degradation was undetectable (Fig. 2B, lanes 3 and 4), showing that resection required the nucleolytic activity that is intrinsic to hExo1. Furthermore, hExo1(D173A) competitively inhibited hExo1–BLM-mediated resection (data not shown), confirming the original observations that the mutant hExo1 was active with regard to DNA-binding function (29).
Fig. 2.
Exonuclease activity of hExo1 is essential for BLM-mediated stimulation of resection. (A) Nuclease reactions as a function of Exo1 concentration. Images show reactions of products with increasing amounts of hExo1 (0, 5, 10, and 20 nM) in the absence (lanes 1–4) or presence of BLM (20 nM, lanes 5–8). Incubation time was 30 min. (B) The mutant hExo1(D173A), devoid of nuclease activity, shows no nucleolytic activity in the presence or absence of BLM. Incubations were for 30 min. Lanes 1 and 2, hExo1 (WT) with and without BLM, respectively; 3 and 4, hExo1(D173A) with and without BLM, respectively. hExo1, preparation B, was used in B; because of the 3-fold higher specific activity of preparation B, more resection is evident. The positions of the intact substrate (2.7 kbp) and resection products are indicated.
Stimulation of hExo1 by BLM Is Specific.
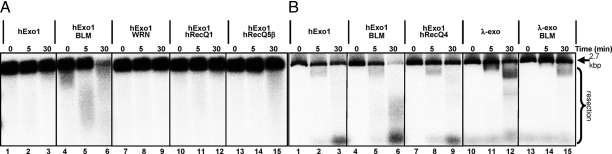
To investigate the specificity of BLM-mediated stimulation of hExo1, we tested other RecQ helicases: human WRN, RecQ1 (hRecQ1), RecQ4 (hRecQ4) and RecQ5β (hRecQ5β), and E. coli RecQ. Fig. 3 shows that none of the other human RecQ homologs stimulated hExo1. Whereas the DNA was almost completely degraded in 30 min in the BLM-stimulated hExo1 reaction (Fig. 3 A and B, lanes 6), there was essentially no stimulation by WRN, hRecQ1, hRecQ5β (Fig. 3A, lanes 9, 12, and 15, respectively), and hRecQ4 (Fig. 3B, lane 9). Although WRN was found to stimulate both the flap endonuclease of hExo1 at a ssDNA–dsDNA junction and the incision of a nucleotide at a nick (30), we did not detect extensive stimulation of exonucleolytic resection by WRN. Furthermore, the intrinsic nuclease activity of WRN was also not evident because the DNA used in our assay is a poor substrate for WRN (31). The E. coli RecQ was also an ineffective stimulator of this reaction; in fact, the bacterial helicase inhibited hExo1 (Fig. S3) most likely by blocking hExo1 access to the ends of DNA. All of the helicases tested [except hRecQ4, which lacks helicase activity (32)] were active as determined by their ability to unwind a forked substrate, and they failed to stimulate even at 2- to 3-fold higher concentrations that provided greater helicase activity than BLM (data not shown). Finally, to determine whether the stimulation by BLM is specific to hExo1 and not a reflection of a general capacity of BLM to stimulate nucleases, we also tested a noncognate functional homolog, λ exonuclease (λ-exo), which is also a 5′→3′ dsDNA-specific exonuclease. BLM failed to stimulate λ-exo; in fact, the presence of BLM reduced λ-exo-mediated resection (Fig. 3B, lanes 12 and 15). These data indicate that BLM-mediated stimulation of hExo1 is quite specific.
Fig. 3.
BLM-mediated stimulation of hExo1 nucleolytic activity is specific. Nuclease reactions are as described in Materials and Methods except, as indicated in the figure, either BLM was replaced with equimolar amounts of WRN, hRecQ1, hRecQ5β, or hRecQ4, or hExo1 was replaced with a comparable amount of nuclease activity (1 unit) of λ exonuclease. (A) Gel showing time courses. (B) Gel showing time courses using hExo1 preparation B, which had a 3-fold higher specific activity. The positions of the intact substrate (2.7 kbp) and resection products are indicated.
Stimulation of hExo1 by BLM Does Not Require ATP.
The inability of other RecQ homologs to stimulate hExo1 prompted us to ask whether unwinding of dsDNA is a prerequisite for BLM-dependent stimulation of hExo1. We therefore compared the stimulation of hExo1 by BLM in the presence and absence of ATP because ATP hydrolysis is required for DNA unwinding. Interestingly, in the absence of ATP, BLM stimulated hExo1 to the same degree as in its presence (Fig. 4A, lanes 2–4 and 5–7). This observation showed that stimulation is not a consequence of DNA unwinding associated with, or followed by, nucleolytic degradation, but rather suggested that it relies on physical association between the 2 proteins.
Fig. 4.
Stimulation of hExo1 by BLM does not require ATP, and BLM physically interacts with hExo1. (A) Nuclease reactions incubated for 5 min with hExo1 (20 nM) and BLM (80 nM) in the presence and absence of ATP. Lanes: 2–4, hExo1 and BLM in the absence of ATP; 5–7, hExo1 and BLM in the presence of ATP; 1 and 8, hExo1 in the absence or presence of ATP, respectively. The positions of the intact substrate (2.7 kbp) and resection products are indicated. (B) Pull-down experiments were performed in the absence of DNA in 3 sets: hExo1 alone, hExo1–BLM, and BLM alone. The wash, eluate, and bead fractions were tested for presence of hExo1 by using the standard nuclease assay: Lane 1, substrate; lanes 2–4, wash fractions; lane 5–7, eluate fractions; lanes 8–10, bead fractions. The positions of the intact substrate (2.7 kbp; linear and nicked) and resection products are indicated. The band labeled nicked DNA arises during Klenow fill-in of nicked DNA. (C) Analysis of eluted proteins by gel electrophoresis and silver staining. Pull-down experiments were performed, and the material bound to the beads was analyzed by silver staining. Lanes: 1 and 2, ≈100 ng of Exo1 and BLM, respectively; 3–5, pull-downs with Exo1 alone, Exo1–BLM, and BLM alone, respectively. The positions of size markers (kDa) BLM and Exo1 are indicated.
BLM Physically Interacts with hExo1.
The specificity of nucleolytic stimulation suggested the existence of a specific interaction between BLM and hExo1. To test this possibility, pull-down experiments with the purified proteins were performed. We used Ni–nitrilotriacetic acid magnetic beads, exploiting the presence of a C-terminal His6 tag on BLM; coelution of hExo1 with BLM was initially detected through its nuclease activity. hExo1 was incubated with BLM–beads, and the bound complex was analyzed. As a control for nonspecific binding, hExo1 was incubated with the beads alone. Washing the complex with binding buffer did not release nuclease activity (Fig. 4B, lanes 2–4). However, when BLM was eluted from the beads, hExo1 activity also coeluted (Fig. 4B, lane 6) but not in the control beads lacking BLM (Fig. 4B, lane 5). Although hExo1 alone did not bind to beads (Fig. 4B, lanes 8), some hExo1 remained bound to BLM on the beads, even after imidazole elution (Fig. 4B, lane 9). No nuclease activity eluted or remained bound to the beads when BLM was incubated with the beads alone (Fig. 4B, lanes 7 and 10). The degradation of nicked DNA, which is the preferred substrate for hExo1 (28), confirms the identity of hExo1 as the nuclease being detected in the pull-down assay.
To confirm the interaction between BLM and hExo1 with a nonenzymatic assay, the bound proteins were analyzed by PAGE. Silver staining showed that hExo1 is pulled down only when BLM was bound to the beads (Fig. 4C, compare lanes 3 and 4), demonstrating directly that the 2 proteins physically interact. The ability of BLM to pull down hExo1 was specific for this nuclease because it did not retain λ-exo (data not shown), which was consistent with the failure of BLM to stimulate λ-exo (Fig. 3B). Collectively, these observations demonstrate that the interaction between BLM and hExo1 is specific.
Homologous DNA Pairing Promoted by hRad51, hRPA, hExo1, and BLM.
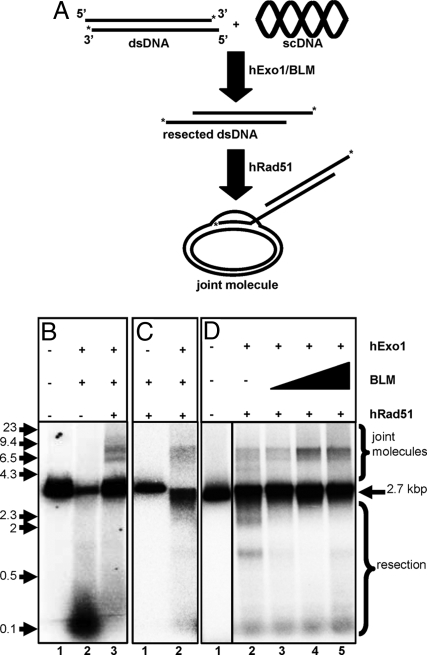
Resection of the dsDNA by BLM and hExo1 generates ssDNA with a 3′ overhang, which should be a substrate for human Rad51 nucleoprotein formation. To determine whether this was the case, the ability of hRad51 to promote pairing between the resected DNA and homologous duplex DNA was tested. In the joint molecule formation assay (Fig. 5A), the processed linear DNA is the invading DNA whereas complementary supercoiled DNA (scDNA) serves as the target for homologous pairing. Our results (Fig. 5B) show that the processed dsDNA is indeed a substrate for Rad51-mediated homologous pairing: whereas no pairing products were seen in the absence of hRad51 (Fig. 5B, lane 2), 10 ± 3% of the linear dsDNA participated in joint molecule formation in the presence of hRad51 (Fig. 5B, lane 3). Control experiments showed that joint molecule formation depended on the presence of homologous scDNA and ATP (data not shown). Because of the varying extents of resection, the joint molecules typically run as a diffuse species with 2 or 3 prominent bands that migrate between the 9,400- and 4,300-bp markers (Fig. 5 B and C). In control reactions with RecA, invasion of scDNA by a resected linear dsDNA (resected by λ-exo) produced diffuse joint molecules with 2 major species that migrated between the 4,300- and 6,500-bp, and the 6,500- and 9,400-bp size markers; invasion by full-length ssDNA produced joint molecules migrating approximately with the 6,500-bp marker (data not shown). Furthermore, the joint molecule products dissociated upon restriction of the scDNA, confirming their identity as simple joint molecules (data not shown). Finally, efficient DNA pairing required higher than stoichiometric amounts of hRad51 with respect to donor DNA, because some hRad51 binds the duplex DNA and, hence, is not productive with regard to DNA pairing (Fig. S4) (33). This binding of hRad51 to the linear duplex is responsible for the reduction of resection evident in the presence of hRad51 (Fig. 5B).
Fig. 5.
Rad51 can use dsDNA processed by hExo1 and BLM for homologous DNA pairing. Joint molecule reactions were performed as described in Materials and Methods. (A) Schematic representation of the reaction. The asterisks indicate 32P label on the 3′ ends. (B) Joint molecule formation requires hRad51. Gel shows reaction products in the absence of hRPA. Lanes: 1, absence of proteins; 2, BLM and hExo1; 3, BLM, hExo1, and hRad51. (C) Joint molecule formation requires hExo1. Lanes 1 and 2, joint molecule reactions in the absence and presence of hExo1, respectively. (D) Joint molecule formation as a function of BLM concentration. Lane 1, absence of proteins; 2–5, increasing amounts of BLM (0, 20, 40, 80 nM). The positions of intact DNA (2.7 kbp), resection products, joint molecules, and molecular size standards (kbp) are indicated.
hExo1 was absolutely required for pairing (Fig. 5C, lane 2), thereby establishing its requirement in resection. BLM stimulated joint molecule formation, but it was not essential: DNA processed by hExo1 alone was sufficient for hRad51-mediated DNA pairing, but the yield was reduced (6 ± 1%) compared with reactions with both BLM and hExo1 (Fig. 5D, compare lane 2 and lanes 3–5). The rate and extent of joint molecule formation depended on both the BLM and hRad51 concentration because of increased processing and pairing, respectively (Fig. 5D and Fig. S4). The joint molecules formed by the simultaneous action of hExo1, BLM, and hRad51 were not disrupted by BLM concentrations as high as 80 nM (Fig. 5D, lane 5). Human RPA had a nearly negligible effect on DNA pairing (from ≈10% to ≈12%; Fig. S5). Alone, hRPA failed to form joint molecules and also to protect the linear dsDNA because it does not bind dsDNA (34) (Fig. S5).
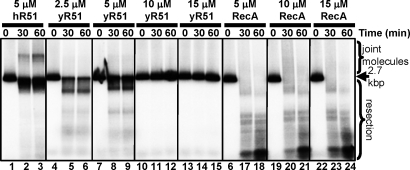
To determine whether joint molecule formation displayed species specificity, hRad51 was replaced with either Saccharomyces cerevisiae Rad51 (yRad51) or E. coli RecA. At an equimolar concentration, DNA pairing demonstrated species specificity: whereas hRad51 mediated efficient (≈10%) pairing, yRad51 and RecA produced only trace amounts (Fig. 6). It is also evident that at equivalent concentrations of the DNA strand exchange proteins, hRad51 limited nucleolytic resection by hExo1–BLM most effectively (Fig. 6). yRad51 failed to produce DNA-pairing products over a wide range of protein concentrations perhaps because of its inability to interact with BLM (Fig. 6, lanes 4–15). For RecA, resection was greatest, likely because RecA binds dsDNA weakly at pH 7.5; the product smear above dsDNA results from annealing of dsDNA resected beyond the midpoint (Fig. 6, lanes 16–24). In contrast to the marked preference for hRad51 in DNA pairing, substitution of yRPA for hRPA resulted in no change (Fig. S6). Hence, only the cognate hRad51 protein can mediate efficient DNA pairing in concert with BLM and hExo1. Accordingly, by using an in vitro reaction with purified proteins, we have successfully reconstituted initial steps of homologous recombination in humans.
Fig. 6.
Joint molecule formation by BLM, hExo1, hRPA, and hRad51 shows species specificity. Joint molecule reactions were performed as described in Materials and Methods. The gel shows the following time courses: Lanes: 1–3, hRad51 (5 μM); 4–6, 7–9, 10–12, 13–15: 2.5, 5, 10, and 15 μM yRad51 respectively; lanes 16–18, 19–21, 22–24: 5, 10, and 15 μM RecA, respectively. The positions of intact DNA (2.7 kbp), resection products, and joint molecules are indicated.
Discussion
Here, we have provided biochemical evidence for a role of BLM and hExo1 in the initiation of recombinational DNA repair. We showed that BLM stimulates dsDNA resection by hExo1 through a physical association between BLM and hExo1. The comparable extents of stimulation for the 2 isoforms of hExo1 indicate that the C-terminal 44 aa, which are missing from isoform 1a because of alternate splicing (2), are not essential for the interaction with BLM. The salient feature of this interaction is the lack of an ATP requirement for BLM-mediated stimulation of hExo1. This characteristic eliminates the possibility that the observed enhancement of hExo1 activity requires BLM-catalyzed DNA unwinding. Interestingly, the ability of a human RecQ family helicase to stimulate the activity of another protein without requiring ATP is not without precedent: BLM stimulates FEN-1 (35), and WRN stimulates hExo1, FEN-1, and translesion DNA polymerases via direct interactions that do not require ATP-driven unwinding (30, 36, 37). WRN was shown to stimulate both the flap endonuclease activity of hExo1 and removal of the 5′-terminal nucleotide at a nick (30). Stimulation of the flap endonuclease activity of both Exo1 and FEN-1 required the C-terminal domain (residues 949-1432) of WRN (30, 36). Interestingly, the C-terminal domains of WRN and BLM share 19% similarity and 12% identity (36). This fact and the observations that BLM also stimulates both hExo1 and FEN-1 make it likely that BLM stimulates both of these nucleases through interactions involving its C-terminal region. Our findings show that BLM increases substrate utilization by hExo1; hence, they suggest that BLM increases the effectiveness of hExo1 by physically associating with the nuclease and enhancing its binding to DNA. Therefore, we propose that this mutual association between BLM and hExo1 serves the function of recruiting either protein to a DNA end, gap, or nick, resulting in increased exonucleolytic processing.
Whereas the need for Exo1 during the initial stages of recombination has been documented (5, 7), there had been no clear precedent for BLM acting in the resection of DNA ends. However, recent evidence shows that BLM localizes at sites of DSBs almost immediately on breakage (38). Also, BLM is a component of the BASC complex and is phosphorylated by ATM kinase upon DNA damage (39). These experiments suggest an early function for BLM in addition to its better-established late functions (17–21). In agreement, in S. cerevisiae, mutation of Sgs1 protein blocks DNA-pairing interactions between a DSB and its target (40). However, most genetic analyses have failed to place Sgs1 at the initiation step of recombinational repair. We believe that this negative result can be explained by the existence of overlapping functions involving several functionally redundant helicases that at least partially complement Sgs1 (and BLM) function in initiation, but not in its later functions. This explanation is exemplified in E. coli, wherein multiple partially complementing helicases (RecQ, UvrD, and helicase IV) can act to initiate recombination in the RecF pathway (41).
On the basis of our observations, we propose the model for initiation depicted in Fig. 7. Although hExo1 can degrade dsDNA, by itself, it does not efficiently resect dsDNA (5) (step I). However, BLM also binds DNA, and our data suggest that the association between BLM and hExo1 tethers the nuclease to the dsDNA end and increases the efficiency of processing (step II); this mechanism was initially invoked to explain stimulation by hMutSα, which forms a clamp-like structure that increases both loading and processivity of hExo1 (28). The requirement for BLM is not absolute because hExo1 can process the ends in its absence, albeit with lower efficiency. This is similar to the RecQ–RecJ interaction, wherein the helicase stimulates, but is not required for, RecJ activity (N. Handa, K. Morimatsu, S. T. Lovett, and S.C.K., unpublished observations). However, because hExo1 also acts on nicked DNA as part of DNA mismatch repair, we suggest that BLM might activate Exo1-mediated resection of DNA nicks, ssDNA gaps, and dsDNA breaks to funnel the broken DNA into an appropriate repair pathway. The ssDNA generated by the action of hExo1 and BLM is most efficiently used by hRad51 to form a nucleoprotein filament (step III). The interaction of BLM and hRad51 (42) can explain the species specificity that we observed for hRad51 in DNA pairing and offers the testable hypothesis that BLM also recruits hRad51 to the resected DNA through its shared interaction with hExo1. Given that the N- and C-terminal domains of BLM can independently interact with hRad51 (42) and that the C-terminal domain of BLM likely interacts with hExo1 (30, 36), such a mutual recruitment scheme has some molecular support. The nucleoprotein filament then searches for DNA sequence homology and mediates DNA strand invasion and exchange (step IV).
Fig. 7.
Model for the role of BLM and hExo1 in the initiation of recombinational DNA repair. Step I, inefficient processing of dsDNA by hExo1 (red oval) generates limited amounts of ssDNA. Step II, the efficiency of hExo1-mediated degradation is enhanced by its association with BLM (gray hexameric ring). Step III, the BLM–hExo1 association efficiently generates ssDNA, onto which Rad51 assembles (green ovals). Step IV, the Rad51–ssDNA filament then seeks a homologous intact DNA molecule (blue) and mediates joint molecule formation. RPA (tricolored ovals) stabilizes the joint molecules by binding to the displaced DNA strand.
One of the proposed late functions of BLM is disruption of replicated joint molecules as a prelude to the annealing of their ssDNA tails, resulting in DSB repair by a pathway called synthesis-dependent strand annealing. Indeed, BLM can disrupt joint molecules that are devoid of proteins (18). Our observation here, that BLM does not disrupt D-loops when Rad51 is present and active, complements the report that BLM can disrupt joint molecules when Rad51 is inactivated (20). BLM-mediated disruption of joint molecules occurred after free Rad51 was removed and the nucleoprotein filament was allowed to hydrolyze ATP and become inactive because of ADP formation (20). These findings suggest to us that Rad51 needs to be removed from the joint molecule before BLM can unwind the invading strand from the joint molecule. Rad51 can be removed by either Rad54 (43) or BLM (20). These biochemical observations support a role for BLM in preventing chromosome crossing-over by disrupting joint molecules before they mature into Holliday junctions. Our results and model do not deny the participation of BLM in these additional late functions; rather, the pleiotropic role of BLM in recombination is comparable with that of E. coli RecQ, which can act both early to initiate recombination and late to disrupt joint molecules and decatenate junctions (23, 24).
Based on the physical analysis of DSB formation and processing in meiosis (see ref. 44), resection of DNA breaks is seen to occur in 2 distinct biochemical phases. First, the MRE11 complex, which possesses 3′→5′ exonuclease and 5′-specific endonuclease activities (12), acts in a currently unspecified manner to produce a partially processed DNA intermediate that is a precursor to the resection step; a recently identified factor, CtIP (13), may stimulate this reaction. In the second phase, this intermediate is processed by other nucleases and helicases to generate a longer stretch of ssDNA with a 3′ overhang. We propose that hExo1 and BLM are 2 of the proteins involved in this resection. Recent physical analysis shows that the exo1 sgs1 mutant of S. cerevisiae generates partially resected intermediates that are inefficient substrates for Rad51-dependent DNA strand invasion (45). A specific 2-step model for DSB processing was proposed wherein the Mre11 complex and Sae2 (CtIP/Ctp1) are involved in the initial processing of a DSB to generate an intermediate that is then rapidly and extensively processed by Sgs1 and Exo1 in a second step. Sgs1 and Exo1 were found to function in separate pathways rather than the same pathway but, as in the case of E. coli, it remains plausible that Exo1 also functions with redundant nucleases requiring Sgs1 in one pathway or that Sgs1 functions with redundant helicases in the pathway requiring Exo1; hence, their interaction would have escaped genetic detection. Alternatively, given that yeast has only 1 RecQ homolog, whereas humans have 5 homologs, it remains possible that the interaction with hExo1 is a specialized property of human BLM. Although more analysis is needed, these data lend strong support for an early role for BLM and Exo1 in DNA break repair.
As mentioned, our results and interpretations do not exclude additional nonredundant late functions for BLM in recombinational DNA repair. In fact, a unique molecular aspect of RecQ, Sgs1, and BLM is their singular ability to interact with a cognate topoisomerase III to promote a novel type of DNA strand passage (19, 24, 46, 47). Their combined activities can result in the dissolution of Holliday junctions without the formation of cross-overs (19). Therefore, one expected consequence of BLM/Sgs1 mutation is an increase in cross-overs; in fact, this phenotype is seen for both human cells and S. cerevisiae (19, 48). In BLM-deficient cells, this defect is manifested as an increase in sister chromatid exchanges and quadriradial chromosomes (49), but not in the complete loss of recombination as might be expected if BLM was solely required for the DNA resection step. The likely explanation for this behavior is that the loss of initiation function can be compensated by alternative helicases and nucleases, which are yet to be fully defined in eukaryotes, whereas the resolution without cross-over function cannot be replaced.
Although we have depicted BLM and hExo1 functioning in DSB repair, the same activities can repair ssDNA gaps generated by stalled replication forks as proposed for E. coli RecQ and RecJ (11). These processed ssDNA gaps can be used by hRad51 to mediate DNA pairing that would subsequently restart stalled replication (9). Thus, these studies have revealed a function for BLM and Exo1 in dsDNA break and ssDNA gap repair, processes that are conserved from bacteria to humans.
Materials and Methods
Additional procedures are detailed in SI Materials and Methods.
Nuclease Assays.
Unless otherwise noted, hExo1 (isoform b; 20 nM) and BLM (40 nM) were incubated with 32P-labeled (3′ end) EcoRI-linearized pUC19 [1.4 nM ends; 4 μM nucleotides (nt)] in standard buffer [20 mM Na-Hepes (pH 7.5), 5 mM MgCl2, 0.1 mM DTT, 100 μg/mL BSA, 0.05% Triton X-100, and 1 mM ATP] at 37 °C. Reactions were stopped with 7 μg/μL proteinase K, 50 mM EDTA, and 2% SDS (final concentrations) by incubation for 30 min. Products were analyzed by native agarose gel (1%) electrophoresis with TAE [40 mM Tris-acetate (pH 8.00), 1 mM EDTA] at 9 V/cm for 80 min. Gels were dried on DE81 paper (Whatman), visualized, and quantified with a Molecular Dynamics Storm 860 using ImageQuant version 5.2.
Pull-Down Assays.
hExo1 and BLM (20 nM each) were incubated in standard buffer lacking both ATP and BSA and containing 20 mM imidazole at 4 °C for 1 h. Ni–nitrilotriacetic acid magnetic beads (Qiagen) were added to a concentration of 1% and incubated at 4 °C for 1 h. The beads were isolated by using a magnet, and washed (once with 50 μL and twice with 100 μL) with standard buffer containing 50 mM imidazole. Bound proteins were eluted with standard buffer containing 250 mM imidazole, and beads were resuspended in standard buffer. Fractions were assayed for hExo1 nuclease activity or analyzed by 10% SDS/PAGE and silver staining (Bio-Rad).
Joint Molecule Formation Assays.
hExo1 (20 nM), BLM (40 nM), hRPA (0.5 μM), and hRad51 (5 μM) were incubated with 32P-labeled (3′ end) EcoRI-linearized pUC19 (3.5 nM ends; 10 μM nt) and supercoiled pUC19 DNA (17.5 nM molecules; 100 μM nt) in 20 mM Na-Hepes (pH 7.5), 10 mM MgCl2, 1 mM ATP, 0.1 mM DTT, 100 μg/mL BSA, and 0.05% Triton X-100. Reactions were incubated at 37 °C for 1 h, stopped, and analyzed as described for the nuclease assays with the exception that electrophoresis was for 120 min.
Supplementary Material
Acknowledgments.
We are grateful to Dr. Ian Hickson (Oxford University, U.K.) for hRecQ1, hRecQ5, and the overexpression plasmid for BLM (pJK1); Dr. David Chen (University of Texas Southwestern, Dallas, TX) for WRN; Dr. Patrick Sung (Yale University, New Haven, CT) for RecQ4; Behzad Rad (University of California, Davis) for RecQ; and Aura Carreira (University of California, Davis) for hRad51 and hRPA. This work was supported by National Institutes of Health Grants GM-41347 and GM-62653 (to S.C.K.) and Tobacco-Related Disease Research Program Postdoctoral Fellowship 16FT-0069 (to A.V.N.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809380105/DCSupplemental.
References
- 1.Spies M, Kowalczykowski SC. Homologous recombination by RecBCD and RecF pathways. In: Higgins NP, editor. The Bacterial Chromosome. Washington, DC: Am Soc Microbiol; 2005. pp. 389–403. [Google Scholar]
- 2.Tishkoff DX, Amin NS, Viars CS, Arden KC, Kolodner RD. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res. 1998;58:5027–5031. [PubMed] [Google Scholar]
- 3.Schmutte C, et al. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res. 1998;58:4537–4542. [PubMed] [Google Scholar]
- 4.Szankasi P, Smith GR. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem. 1992;267:3014–3023. [PubMed] [Google Scholar]
- 5.Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maringele L, Lydall D. EXO1 plays a role in generating type I and type II survivors in budding yeast. Genetics. 2004;166:1641–1649. doi: 10.1534/genetics.166.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaetzlein S, et al. Exonuclease 1 deletion impairs DNA damage signaling and prolongs life span of telomere-dysfunctional mice. Cell. 2007;130:863–877. doi: 10.1016/j.cell.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J Biol Chem. 2002;277:13302–13311. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- 9.Cotta-Ramusino C, et al. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell. 2005;17:153–159. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 10.Lovett ST, Kolodner RD. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc Natl Acad Sci USA. 1989;86:2627–2631. doi: 10.1073/pnas.86.8.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courcelle J, Hanawalt PC. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol Gen Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- 12.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 13.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Llorente B, Symington LS. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol. 2004;24:9682–9694. doi: 10.1128/MCB.24.21.9682-9694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hickson ID. RecQ helicases: Caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 16.Ellis NA, et al. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 17.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc Natl Acad Sci USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 20.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plank JL, Wu J, Hsieh TS. Topoisomerase IIIα and Bloom's helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc Natl Acad Sci USA. 2006;103:11118–11123. doi: 10.1073/pnas.0604873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu L, et al. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci USA. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harmon FG, Kowalczykowski SC. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: A conserved mechanism for control of DNA recombination. Mol Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- 25.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 26.Karow JK, Chakraverty RK, Hickson ID. The Bloom's syndrome gene product is a 3′–5′ DNA helicase. J Biol Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 27.Brosh RM, Jr, et al. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J Biol Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 28.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 29.Dzantiev L, et al. A defined human system that supports bidirectional mismatch-provoked excision. Mol Cell. 2004;15:31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 30.Sharma S, et al. The exonucleolytic and endonucleolytic cleavage activities of human exonuclease 1 are stimulated by an interaction with the carboxyl-terminal region of the Werner syndrome protein. J Biol Chem. 2003;278:23487–23496. doi: 10.1074/jbc.M212798200. [DOI] [PubMed] [Google Scholar]
- 31.Shen JC, Loeb LA. Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res. 2000;28:3260–3268. doi: 10.1093/nar/28.17.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund–Thomson syndrome. DNA Repair. 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Sigurdsson S, Trujillo K, Song B, Stratton S, Sung P. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J Biol Chem. 2001;276:8798–8806. doi: 10.1074/jbc.M010011200. [DOI] [PubMed] [Google Scholar]
- 34.Wold MS. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 35.Sharma S, et al. Stimulation of flap endonuclease-1 by the Bloom's syndrome protein. J Biol Chem. 2004;279:9847–9856. doi: 10.1074/jbc.M309898200. [DOI] [PubMed] [Google Scholar]
- 36.Brosh RM, Jr, et al. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamath-Loeb AS, Lan L, Nakajima S, Yasui A, Loeb LA. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc Natl Acad Sci USA. 2007;104:10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karmakar P, et al. BLM is an early responder to DNA double-strand breaks. Biochem Biophys Res Commun. 2006;348:62–69. doi: 10.1016/j.bbrc.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, et al. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 40.Houston PL, Broach JR. The dynamics of homologous pairing during mating type interconversion in budding yeast. PLoS Genet. 2006;2:e98. doi: 10.1371/journal.pgen.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mendonca VM, Klepin HD, Matson SW. DNA helicases in recombination and repair: Construction of a ΔuvrD ΔhelD ΔrecQ mutant deficient in recombination and repair. J Bacteriol. 1995;177:1326–1335. doi: 10.1128/jb.177.5.1326-1335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu L, Davies SL, Levitt NC, Hickson ID. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 43.Solinger JA, Kiianitsa K, Heyer WD. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- 44.Hunter N. Meiotic recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination: Topics in Current Genetics. Vol 17. Berlin: Springer; 2007. pp. 381–442. [Google Scholar]
- 45.Mimitou EP, Symington LS. Sae2/CtIP, Exo1, and Sgs1/BLM collaborate in double-strand break processing. Nature. 2008 doi: 10.1038/nature07312. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu L, Hickson ID. The Bloom's syndrome helicase stimulates the activity of human topoisomerase IIIα. Nucleic Acids Res. 2002;30:4823–4829. doi: 10.1093/nar/gkf611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci USA. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.