Abstract
During protein synthesis, tRNAs and mRNA move through the ribosome between aminoacyl (A), peptidyl (P), and exit (E) sites of the ribosome in a process called translocation. Translocation is accompanied by the displacement of the tRNAs on the large ribosomal subunit toward the hybrid A/P and P/E states and by a rotational movement (ratchet) of the ribosomal subunits relative to one another. So far, the structure of the ratcheted state has been observed only when translation factors were bound to the ribosome. Using cryo-electron microscopy and classification, we show here that ribosomes can spontaneously adopt a ratcheted conformation with tRNAs in their hybrid states. The peptidyl-tRNA molecule in the A/P state, which is visualized here, is not distorted compared with the A/A state except for slight adjustments of its acceptor end, suggesting that the displacement of the A-site tRNA on the 50S subunit is passive and is induced by the 30S subunit rotation. Simultaneous subunit ratchet and formation of the tRNA hybrid states precede and may promote the subsequent rapid and coordinated tRNA translocation on the 30S subunit catalyzed by elongation factor G.
Keywords: translocation, elongation factor G, cryo-electron microscopy
During the elongation cycle, the tRNAs·mRNA complex is translocated to allow reading of the following codon on mRNA. Translocation is promoted by elongation factor G (EF-G) and accompanied by GTP hydrolysis. During translocation, the ribosome changes from the pretranslocation state with deacylated tRNA in the peptidyl site (P site) and peptidyl-tRNA in the aminoacyl site (A site) to the posttranslocation state where A- and P-site tRNAs have moved to P and exit (E) sites, respectively. The universal design of ribosomes from two subunits inspired early suggestions that translocation may involve a movement of the subunits relative to each other (1, 2). Such models imply the existence of intermediate states for the tRNAs that differ from the classic A, P, and E states. Chemical probing experiments with pretranslocation ribosomes indicated that after peptidyl transfer the acceptor ends of the tRNAs spontaneously moved on the large 50S subunit toward their posttranslocation positions (3), indicating that peptidyl-tRNA and deacylated tRNA entered hybrid A/P and P/E states, respectively. The transition was observed in the absence of EF-G, and the driving force for the movement was attributed to different affinities of the A, P, and E sites on the 50S subunit (4) for the chemically different acceptor ends of the tRNAs (5, 6). Nevertheless, cryo-electron microscopy (cryoEM) of ribosomes in the pretranslocation state showed the tRNAs in their classic A, P, and E states (7). Although this result was difficult to reconcile with the hybrid-state model, it is important to note that in that complex the occupancy of the E site by deacylated tRNA may have prevented the P-site tRNA to enter the P/E-site hybrid state.
CryoEM of ribosomes in complex with EF-G revealed a relative rotation between subunits referred to as ratchet motion (8). Ratcheting included a rotation of the 30S subunit relative to the 50S subunit and large conformational change of the L1 stalk from the 50S subunit toward the intersubunit cavity (7). Using ribosomes with deacylated tRNA in the P site and vacant A site, it was shown that ribosome binding of EF-G with a nonhydrolyzable GTP analog promoted a ratchet-like motion of the subunits and a movement of the tRNA into the P/E hybrid position (7). However, bulk FRET studies suggested that hybrid states coupled to the ratchet-like motion may form spontaneously, i.e., without binding EF-G (9, 10). Further evidence for spontaneous ratcheting was provided by single-molecule FRET studies, which showed individual pretranslocation ribosomes oscillating between the nonrotated and the rotated states (4). Also the tRNAs were observed to fluctuate between classic and hybrid states in dynamic equilibrium (11), possibly sampling through additional intermediate states. The relation between ratcheting and hybrid-state formation was, however, not demonstrated directly.
Results and Discussion
Classification of Pretranslocation Ribosomes.
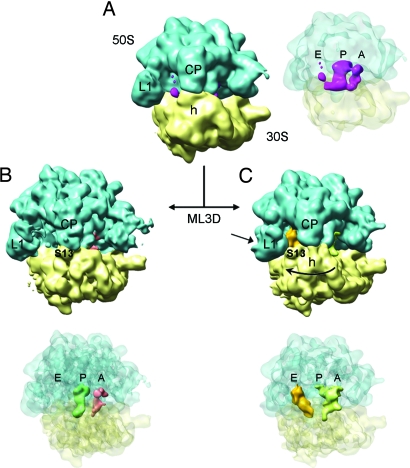
Here, we revisit the structure of pretranslocation ribosomes by cryoEM. We characterized 70S ribosomes from Escherichia coli programmed with mRNA and containing deacylated tRNAfMet in the P site and fMetLeu-tRNALeu in the A site (see Methods). An initial set of 63,539 individual particles resulted in a cryoEM density map that showed no clear indication of ratcheting and with strong density for tRNA in the P site (Fig. 1A). However, weak and scattered density in the A and E sites suggested the presence of a mixture of different structural states in the sample. Subsequently, maximum-likelihood-based classification (ML3D) (12) was applied to sort the data and yielded two groups, corresponding to distinct structural states of the ribosome (Fig. 1 B and C). We note that this classification method does not require prior information about the nature of the structural variability in the sample, thereby avoiding potential bias associated with supervised classification methods (13).
Fig. 1.
3D density maps of pretranslocation ribosomes determined by cryoEM and particle classification. Rendering of density maps for the 70S·mRNA·tRNAfMet·fMetLeu-tRNALeu complex shows the ribosomal subunits solid and in semitransparent display to allow visualization of densities for tRNAs within the intersubunit space. (A) CryoEM map for the total data set containing 63,539 individual images (resolution ≈13 Å). (B and C) The initial set of images was classified (ML3D) into two different groups, and the corresponding maps are displayed. (B) The larger subset contained 47,520 images (resolution ≈13 Å) and resulted in a map with tRNAs in classic state. (C) A subset with 12,590 images (resolution ≈16 Å) revealed tRNAs in hybrid-state positions and a relative rotation between ribosomal subunits. Landmarks: CP, central protuberance; L1, L1 stalk of the 50S subunit; h, head of the 30S subunit; S13, protein S13 on the 30S subunit.
The first group represented ≈80% of the total number of particles and the corresponding density map showed no indication of ratcheting (Fig. 1B). Improved density for the tRNAs was observed at the classic A (solid red in Fig. 1B) and P (dark green in Fig. 1B) sites, although the density at the A site was weaker than that at the P site (corresponding to an occupancy of 70%). Importantly, there was no tRNA density associated with the E site. This observation contrasted with the earlier cryoEM work on pretranslocation ribosomes (7) and was consistent with the fact that HPLC-purified Leu-tRNALeu, devoid of contaminating deacylated tRNA prone to enter the E site, was used to fill the A site, and pretranslocation complexes were additionally purified by ultracentrifugation. The map in Fig. 1B depicts a pretranslocation ribosome in nonrotated configuration with the tRNAs in classic A- and P-site positions.
The second group accounted for the remaining 20% of the particles. The corresponding map in Fig. 1C clearly showed the ribosome in a ratcheted state and with the L1 stalk moved toward the E site. Moreover, the deacylated tRNA was found in the P/E hybrid state bridging the 30S P site with the 50S E site (depicted in solid orange in Fig. 1C), whereas the peptidyl-tRNA (light green in Fig. 1C) was located in the vicinity of the A and P sites. Comparison of this structure with the classic A, P, and E arrangements revealed that the peptidyl-tRNA was present in an A/P hybrid state (Fig. 2). Apparently the two tRNAs have moved, following the rotation of the 30S subunit (small arrows in Fig. 2A) over a distance similar to the 8-Å displacement of the ribosomal small subunit during the ratchet motion measured at the level of the mRNA (7). Thus, both tRNAs still reside in A and P sites on the small subunit, but are shifted toward P and E sites on the 50S subunit. At the current resolution (≈16 Å) the structure of the ratcheted ribosome resembled that of the ribosome interacting with EF-G (7). The relative rotation and the change in connecting bridges between subunits, the large movement of the L1 stalk, and the displacement of the head of the 30S following the path of the mRNA are features displayed in the present cryoEM map. We, therefore, assume that the structure of this subset of ribosomes in the pretranslocation state includes the conformational changes previously described at higher resolution (7).
Fig. 2.
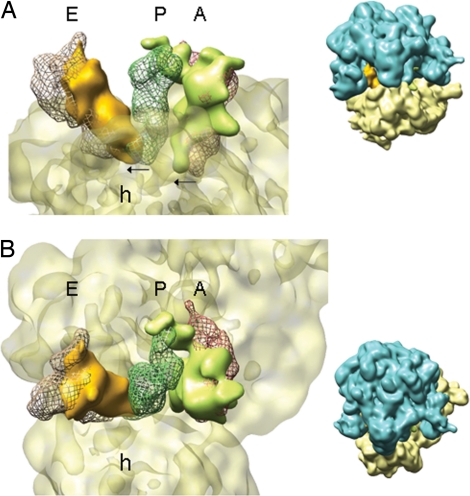
Hybrid states of tRNAs. The 30S subunit and the tRNAs from the map depicted in Fig. 1C (same color code) are rendered in a close-up view. For reference, tRNAs in classic states in A, P, and E sites (taken from the cryoEM map of 70S·mRNA·tRNAfMet·fMetPhe-tRNAPhe (7) after alignment of both maps by their 50S subunit) are depicted in mesh representation (A, red; P, dark green; E, brown). Thumbnails at the right show the orientation of the ribosome. (A) A view along the anticodon arms reveals the movement of tRNAs together with the 30S subunit. The arrows indicate the direction and magnitude of the movement. (B) A view from the 50S subunit along the acceptor arms of the tRNAs clearly shows that the 3′ ends of the tRNAs reach into P and E sites. Landmark: h, head of the 30S subunit.
Hybrid tRNA in A/P Position.
In the map displayed in Fig. 2B, the P/E hybrid state of the deacylated tRNA is obvious, because the distance between the 3′ ends in the P and E sites is large, and the acceptor arm of the present P/E hybrid (solid and orange in Fig. 2B) clearly reaches the classic E site tRNA (brown mesh in Fig. 2B). Although the difference between the A and P sites is smaller, the acceptor arm of the peptidyl-tRNA (solid and light green in Fig. 2B) deviates from the trajectory of a classic A-site tRNA (red mesh in Fig. 2B), being directed toward the P site on the 50S subunit.
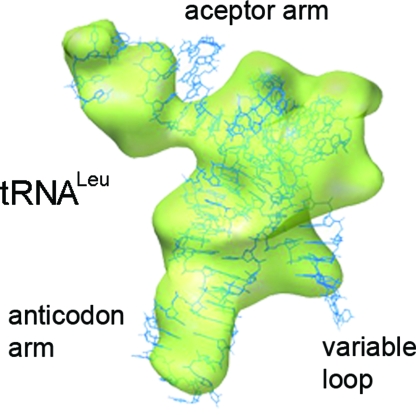
The 20% particle subset of the present cryoEM map represents a pretranslocation ribosome with simultaneous ratcheting of subunits and hybrid-state formation. The conformational homogeneity of the subset and, with that, the quality of the classification, can be judged by the similarity of the cryoEM density with known atomic coordinates. In Fig. 3, the atomic structure of tRNALeu (14) is shown fitted within the density for fMetLeu-tRNALeu in the A/P site. The quality of the fit is obvious, as the cryoEM density accounts for the entire tRNA, including the characteristic long variable loop of tRNALeu. Small differences are restricted to the acceptor arm. These characteristics may indicate that the acceptor end of the peptidyl-tRNA is passively moved into the 50S P site by the 30S subunit rotation, involving slight changes in its acceptor arm configuration. This contention is consistent with the modest effect on translocation of the mutations that impair the formation of the A/P hybrid state by disrupting interactions between the acceptor end of tRNA and the 23S rRNA residues (15).
Fig. 3.
Peptidyl-tRNALeu in the A/P position. Semitransparent rendering of the density for tRNALeu in the A/P hybrid state (green) is extracted from the cryoEM map presented in Fig. 1C. The atomic structure of tRNALeu (blue) from Pyrococcus horikoshii [Protein Data Bank ID code 1WZ2 (14)] is fitted to the cryoEM density.
Translocation Intermediate.
Our results indicate that the pretranslocation ribosome can spontaneously assume either the nonratcheted conformation with the tRNAs in classic states or the ratcheted conformation with the tRNAs in hybrid states. The observed ratio between the classic and hybrid tRNA states is exactly the same as predicted by single-molecule methods based on the average lifetimes of the respective states at elevated Mg2+; at low Mg2+, the distribution of tRNAs between the classic and hybrid states (16) and the ribosomes between the nonratcheted and ratcheted conformations (4) is close to 50%. Subunit ratcheting and the ability of the P-site tRNA to interact with the E site on the 50S subunit are essential for EF-G-promoted translocation (5, 17), suggesting that the combined hybrid-ratcheted state is an authentic early intermediate of translocation. On the other hand, the spontaneous formation of this state as such is not sufficient to move the acceptor end of peptidyl-tRNA into the proper posttranslocation position, as indicated by low reactivity with puromycin (15, 18). Therefore, we postulate that the hybrid-ratcheted configuration is the natural substrate for EF-G, and EF-G may either bind to that state preferentially or shift the ribosomes to that state before the tRNA movement on the 30S subunit. Translocation is then completed by coupling conformational changes of the factor with further ribosome rearrangements, resulting in tRNA movement and, presumably, the reverse rotation of the 30S subunit (19–21).
Methods
Pretranslocation Complex.
Ribosomes, EF-Tu, and initiation factors from E. coli were prepared as described (22); f[3H]Met-tRNAfMet and [14C]Leu-tRNALeu5 were purified by HPLC (23). The mRNA was a 122-nt-long derivative of m022 mRNA with the coding sequence AUGUUG. Initiation complex (70S·mRNA·f[3H]Met-tRNAfMet) and ternary complex (EF-Tu·GTP·[14C]Leu-tRNALeu5) were prepared as described (24). The pretranslocation complex with deacylated tRNAfMet in the P site and fMetLeu-tRNALeu in the A site was prepared by mixing initiation and ternary complexes in buffer A [50 mM Tris·HCl (pH 7.5), 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2] at 37 °C (24). The complex was stabilized by increasing the concentration of MgCl2 to 21 mM and purified by centrifugation through a sucrose cushion (25). Ribosomes were dissolved in buffer A (21 mM MgCl2), shock-frozen in liquid nitrogen, and stored at −80 °C. The extent of tRNA binding (>90%) was determined by nitrocellulose filtration and radioactivity counting; the extent of dipeptide formation (>90%) was verified by HPLC.
CryoEM and Image Processing.
Pretranslocation complexes were diluted to 32 nM final concentration in buffer A (21 mM MgCl2 plus 2 mM spermine). CryoEM grids were prepared following standard procedures, and micrographs were taken in low-dose conditions on a JEM-2200FS electron microscope (JEOL). Images were recorded on a 4kx4k CCD camera at a magnification of 67,368×, resulting in a 2.2-Å final pixel size. The 3D reconstruction followed reference-based projection matching in Spire-Spider package (26, 27), and amplitude correction was applied to the maps (28). Resolution was estimated by using a cutoff of 0.15 in the Fourier shell correlation (29). Nonsupervised maximum-likelihood classification (12) of the images for ribosomes was used from the Xmipp package (30). Based on the maxima of the probability functions upon convergence of the likelihood optimization, the data set was separated into four groups; three of these were similar and were rejoined to form one group. The rigid-body fitting of coordinates for tRNALeu, Protein Data Bank ID code 1WZ2 (14), within the cryoEM density was performed in Chimera (31), which was also used to produce our figures.
Acknowledgments.
We thank Simone Möbitz, Astrid Böhm, Carmen Schillings, and Petra Striebeck for expert technical assistance and the Barcelona Supercomputing Center (Centro Nacional de Supercomputación) for providing computing resources. This work was supported by the Etortek Research Programs 2007/2009 (Department of Industry, Tourism and Trade of the Government of the Autonomous Community of the Basque Country) (M.V.), the Innovation Technology Department of Bizkaia County (M.V.), the German Research Council (M.V.R. and W.W.), and the Federal Ministry of Education and Research (M.V.R.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The cryoEM maps have been deposited in the Electron Microscopy Data Bank, www.ebi.ac.uk/msd-srv/docs/emdb (accession codes EMD-1551, EMD-1553, and EMD-1554).
References
- 1.Bretscher MS. Translocation in protein synthesis: A hybrid structure model. Nature. 1968;218:675–677. doi: 10.1038/218675a0. [DOI] [PubMed] [Google Scholar]
- 2.Spirin AS. A model of the functioning ribosome: Locking and unlocking of the ribosome subparticles. Cold Spring Harbor Symp Quant Biol. 1969;34:197–207. doi: 10.1101/sqb.1969.034.01.026. [DOI] [PubMed] [Google Scholar]
- 3.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 4.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lill R, Robertson JM, Wintermeyer W. Binding of the 3′ terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO J. 1989;8:3933–3938. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noller HF, Yusupov MM, Yusupova GZ, Baucom A, Cate JH. Translocation of tRNA during protein synthesis. FEBS Lett. 2002;514:11–16. doi: 10.1016/s0014-5793(02)02327-x. [DOI] [PubMed] [Google Scholar]
- 7.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 8.Frank J, Agrawal RK. A ratchet-like intersubunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 9.Ermolenko DN, et al. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 10.Ermolenko DN, et al. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 11.Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheres SH, et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 13.Gao H, Valle M, Ehrenberg M, Frank J. Dynamics of EF-G interaction with the ribosome explored by classification of a heterogeneous cryo-EM dataset. J Struct Biol. 2004;147:283–290. doi: 10.1016/j.jsb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Fukunaga R, Yokoyama S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNA-Leu reveal two modes of discriminator-base recognition. Nat Struct Mol Biol. 2005;12:915–922. doi: 10.1038/nsmb985. [DOI] [PubMed] [Google Scholar]
- 15.Sharma D, Southworth DR, Green R. EF-G-independent reactivity of a pretranslocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HD, Puglisi JD, Chu S. Fluctuations of transfer RNAs between classical and hybrid states. Biophys J. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci USA. 2007;104:4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Semenkov Y, Shapkina T, Makhno V, Kirillov S. Puromycin reaction for the A site-bound peptidyl-tRNA. FEBS Lett. 1992;296:207–210. doi: 10.1016/0014-5793(92)80380-y. [DOI] [PubMed] [Google Scholar]
- 19.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 20.Savelsbergh A, et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 21.Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodnina MV, Wintermeyer W. GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc Natl Acad Sci USA. 1995;92:1945–1949. doi: 10.1073/pnas.92.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milon P, et al. Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Methods Enzymol. 2007;430:1–30. doi: 10.1016/S0076-6879(07)30001-3. [DOI] [PubMed] [Google Scholar]
- 24.Konevega AL, et al. Spontaneous reverse movement of mRNA-bound tRNA through the ribosome. Nat Struct Mol Biol. 2007;14:318–324. doi: 10.1038/nsmb1221. [DOI] [PubMed] [Google Scholar]
- 25.Konevega AL, et al. Purine bases at position 37 of tRNA stabilize codon–anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA. 2004;10:90–101. doi: 10.1261/rna.5142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baxter WT, Leith A, Frank J. SPIRE: The SPIDER reconstruction engine. J Struct Biol. 2007;157:56–63. doi: 10.1016/j.jsb.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 27.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 28.Gabashvili IS, et al. Solution structure of the E. coli 70S ribosome at 11.5-Å resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- 29.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]