Abstract
Loss of biological diversity because of extinction is one of the most pronounced changes to the global environment. For several decades, researchers have tried to understand how changes in biodiversity might impact biomass production by examining how biomass correlates with a number of biodiversity metrics (especially the number of species and functional groups). This body of research has focused on species with the implicit assumption that they are independent entities. However, functional and ecological similarities are shaped by patterns of common ancestry, such that distantly related species might contribute more to production than close relatives, perhaps by increasing niche breadth. Here, we analyze 2 decades of experiments performed in grassland ecosystems throughout the world and examine whether the evolutionary relationships among the species comprising a community predict how biodiversity impacts plant biomass production. We show that the amount of phylogenetic diversity within communities explained significantly more variation in plant community biomass than other measures of diversity, such as the number of species or functional groups. Our results reveal how evolutionary history can provide critical information for understanding, predicting, and potentially ameliorating the effects of biodiversity loss and should serve as an impetus for new biodiversity experiments.
Keywords: community ecology, ecosystem function, phylogenetic diversity, biodiversity experiments, metaanalysis
The modern era has come to be defined as a period of rapid environmental change. One of the most prominent changes taking place globally is a reduction in the number of genes, species, and functional groups of organisms that comprise the biological diversity of natural and managed communities. Widespread loss of biodiversity has prompted scientists from an increasing number of disciplines to begin studying the social, economic, and environmental impacts of diversity change (1–5). For example, seminal experiments in the 1990s suggested that species loss might reduce the amount of biomass produced by plants (6–9), possibly translating to a loss of important ecological services such as the ability of natural habitats to absorb CO2 from the atmosphere. These experiments stimulated 2 decades of research detailing the functional role of plant diversity in ecosystems. Recent summaries of this body of research have confirmed that systems with fewer species generally produce less biomass than those with more species (10–14).
However, why changes in the number of species cause ecosystems to be less productive is still not entirely clear. Is it because less diverse communities tend to be missing genes, metabolic pathways, or traits that would otherwise allow a more complete utilization of local conditions (4, 15)? To answer this question would require that researchers quantify the biological traits that drive resource use and biomass production. However, because a multitude of traits are potentially associated with the ecological differences among species that drive patterns of resource use, knowing the evolutionary relationships of the members of a community can serve to quantify patterns of trait diversity (16).
Here, we present results from a formal metaanalysis of experiments performed in locations around the world to show that phylogenetic diversity is the single best predictor of how community biomass is altered by changes in species diversity. Our dataset is derived from 29 experiments that manipulated the number of species of terrestrial angiosperms in experimental plots, pots, or garden beds in fields or greenhouses and then measured the impacts of plant species number on the production of plant biomass [for a summary of studies used, see supporting information (SI) Table S1]. For each of the experimental units that contained more than one species for which constituent monocultures were measured, we standardized the diversity “effect size” of the biomass produced in a polyculture to the mean of the constituent species in monoculture, as the log response ratio (LRmean; see Materials and Methods). The pool of species used includes 177 taxa that span all major functional groups of grassland ecosystem plants (C3 and C4 graminoids, legumes, etc.). We calculated not only the number of species in a plot, but also the number of functional groups (for functional group definitions, see Materials and Methods) and the amount of phylogenetic diversity in a community (PDC) in a plot (Fig. 1). PDC measures the magnitude of the divergences among species that have evolved since a common ancestor, calculated as the sum of phylogenetic branch lengths separating species on a phylogeny. We estimated the phylogenetic relationships among species by using Bayesian inference with Ultrametric rate smoothing for 143 of the 177 species for which nucleotide sequences from 4 genes (5.8s, atpb, matk, and rbcl) were available in GenBank [National Center for Biotechnology Information (www.ncbi.nlm.nih.gov); for details, including support metrics, and for comparisons with other phylogenetic methods, see SI Text and Figs. S1 and S2]. We were able to estimate PDC in 78% of all experimental polycultures (i.e., 1,315 experimental units).
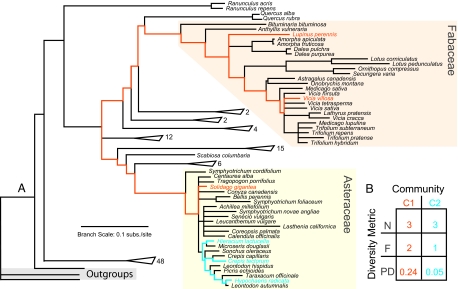
Fig. 1.
Summary of phylogenetic results. (A) Illustrated is the majority rule consensus from a Bayesian MCMC search, with several clades collapsed because of space considerations. Triangles represent collapsed clades, with numbers of species per clade indicated. Two clades (Fabaceae and Asteraceae) representing different functional groups are highlighted for illustrating different biodiversity metrics. Details of phylogenetic analyses and results, including support values, are presented in the Materials and Methods and the SI Text. (B) Biodiversity can be measured in a number of ways, including the number of species in a community (N), the number of trait-based functional groups (F), or the total phylogenetic diversity (PDC) representing the members of a community. PDC is calculated as the sum of branch lengths leading to all members of a community, and two examples are shown using orange and blue branches. The number of species and functional groups often give little information about the underlying phylogeny describing the evolutionary history of a group of species. In 2 hypothetical communities, C1 and C2, the number of species is the same, but variation exists in the number of functional groups and the calculated PDC.
Results and Discussion
Similar to prior summaries (6, 12, 17), our analyses confirm that both the number of species and the number of plant functional groups in an experiment are significant predictors of plant biomass production (Table 1, Model A, and Fig. 2 A and B). The finding of our analysis is that phylogenetic diversity is also a highly significant predictor of biomass production (Table 1, Model A, Fig. 2C). Given that we have data on the number of plant species, the number of plant functional groups, and the phylogenetic diversity in an experimental unit, it is possible to ask which of these metrics of biological diversity best explains variation in biomass production among experimental communities. Such information would potentially be useful for conservation and management where it is often beneficial to predict how community composition affects ecologically important processes. When we compared single-variable models that included the different measures of diversity as explanatory variables, PDC was a significantly better predictor of variation in plant biomass than either the number of species or functional groups (Table 1, Model A).
Table 1.
Results of linear mixed effects models predicting the log response ratio of biomass production to three measures of biodiversity
| Models and variables | F | df | P value | Rpseudo2 | Log likelihood | AIC | Akaike weight |
|---|---|---|---|---|---|---|---|
| A: Single variable with Exp as random effect | |||||||
| Spp | 90.443 | 1,262 | <0.0001 | 0.184 | −1057.728 | 2,123.456 | 3.97 × 10−6 |
| FG | 65.469 | 1,262 | <0.0001 | 0.181 | −1067.760 | 2,143.521 | 1.74 × 10−10 |
| PDC | 112.31 | 1,262 | <0.0001 | 0.206 | −1045.291 | 2,098.582 | ≈1.0 |
| B: Multivariable with Exp as random effect | |||||||
| Spp + PDc + Spp*PDc | 0.210 | −1049.218 | 2,110.436 | ||||
| Spp | 0.449 | 1,260 | 0.5028 | ||||
| PDC | 112.77 | 1,260 | <0.0001 | ||||
| Spp*PDC | 6.492 | 1,260 | 0.0110 | ||||
| C: Multivariable with FG as random effect nested in Exp | |||||||
| Spp + PDC + Spp*PDC | 0.211 | −1048.977 | 2,111.954 | ||||
| Spp | 0.402 | 1,220 | 0.5261 | ||||
| PDC | 98.154 | 1,220 | <0.0001 | ||||
| Spp*PDC | 5.905 | 1,220 | 0.0152 | ||||
Spp is the number of species in an experimental unit; FG is the number of functional groups, and PDC is phylogenetic diversity calculated from the Bayesian Inference with Ultrametric rate-smoothing phylogeny. Results Model A are for single-variable, fixed-effect models with experiment (Exp) included as a random effect. Model B is a model with Spp and PDC, and their interaction is with the Exp random effect. Model C is the two-variable model with FG as a random effect nested within Exp. Akaike weights can be interpreted as the probability that model i is the best fit to the observed data among a set of models (54).
Fig. 2.
The proportional difference between polyculture biomass and mean biomass of species monocultures [log ratio (mean)] is positively related to: the number of species (A), the number of plant functional groups (B), and plot phylogenetic diversity based on Bayesian Inference with Ultrametric rate smoothing (C). (D) Significant relationship between the number of species and phylogenetic diversity. The linear fits come from single-variable mixed-effects models (Table 1, Model A), and the dashed lines represent the case where polyculture biomass equals mean monoculture biomass.
When productivity was modeled as a function of species number (the form of diversity directly manipulated in these studies), PDC, and their two-way interaction, we found that PDC was not only a significant predictor of plant biomass, but that there was a highly significant interaction between species number and PDC (Table 1, Model B). Thus, the impact of plant species number on biomass in past experiments is at least partially explained by the amount of phylogenetic diversity represented in an experimental unit. Further, because PDC and species number are positively correlated (Fig. 2D), we examined the impacts of PDC on production within species number treatments. We found that the effects of PDC on production were most pronounced at lower numbers of species where studies typically included many different combinations of species (2 species: F1,439 = 4.813, P = 0.029; 4 species: F1,277 = 10.435, P = 0.001), whereas the impacts of PDC became less pronounced at higher levels of diversity (6 species: F1,106 = 0.026, P = 0.873; 8 species: F1,90 = 3.077, P = 0.083), probably because researchers tended to use far fewer combinations of species, resulting in less variation in PDC.
The number of functional groups of plants was also significantly related to PDC among species within experimental units (F1,1262 = 1016.61, P < 0.0001, also see Fig. S3). This relationship raises the possibility that the explanatory value of PDC might simply be a result of its correlation to the number of functional groups. However, when we ran a mixed effects model that included a hierarchical effect of the number of functional groups used in an experiment and the effects PDC, species number and the species number–PDC interaction, PDC and the interaction were both significant predictors of plant community biomass (Table 1, Model C). Thus, given that controlling for the effect of the number of functional groups does not diminish the productivity–PDC relationship, we conclude that existing definitions of functional groups are too coarse and do not correspond well to true functional trait differences in communities (also see ref. 18).
Although the Rpseudo2 (see Materials and Methods) values from the mixed effects models ranged from 0.181 to 0.211 (Table 1), individual experiments were highly variable in the amount of variation explained. The within-experiment R2 for the effect of species number on biomass ranged from <0.001 to 0.62, and ranged from 0.01 to 0.69 for PDC and 0.03 to 0.78 for the full model, including species number, PDC, and their interaction. Although there are strong overall effects of PDC and species number on community biomass production, environmental or experimental context seems to be a major factor determining the magnitude of these effects within individual experiments. However, the explanatory power of PDC appeared unchanged even after we accounted for the fact that a small number of species are widely used in experiments, and the prevalence of just a few species could dictate the results. When we excluded species found in >10% of experimental units (n = 14 species) and reran the analyses, none of our conclusions was altered (see Table S2). The results were also robust to five commonly used methods used to estimate phylogenies (see SI Text), and regardless of which phylogeny was used to estimate PDC, phylogenetic diversity was always a superior predictor of community biomass production compared with the number of species or functional groups included in diversity experiments, indicating that our conclusions are not sensitive to minor differences in estimates of PDC caused by using different phylogenetic methods (see SI Text).
Data and analyses presented here show that when controlled experiments simulate changes in species diversity, changes in community biomass are greater for groups of plant species that have a distant common ancestor than for groups that share a recent common ancestor. However, it is not yet clear why increasing phylogenetic diversity results in increased plant community biomass (but see ref. 19). Some research suggests that phylogenetic relatedness is an indicator of the ecological uniqueness of species and a predictor of patterns of competitive coexistence (20–24; but see ref. 25). For example, it has long been assumed, and sometimes demonstrated, that within a habitat type, the amount of ecological differentiation among species is proportional to the amount of evolutionary and genetic divergence (26). Ecological differentiation can result in reduced resource use overlap between species, allowing species to stably coexist together (e.g., niche partitioning). These ecologically differentiated species could potentially complement each other in their resource use by differentially capturing resources in space and/or time. Greater niche and trait differences could, in turn, translate to higher production of biomass (4, 15, 19, 21, 27).
If this interpretation is correct, then future work should be able to map variation in niche differences (or plant traits that confer such differences) onto our phylogenies and find strong correspondence. However, until the time that such datasets exist, we suggest that phylogenetic diversity may be a useful biodiversity metric for predicting the ecological consequences of modern diversity change and for scaling from organism physiology to ecosystem processes (28). This tool may prove especially useful in the world's ecosystems where organisms are too large (e.g., rainforests), the systems too vast (e.g., plankton of the open ocean and the taxa of the ocean floor), or population sizes already too small (endangered species) to allow manipulative biodiversity experiments.
Materials and Methods
Obtaining and Standardizing Data.
We used recent reviews and metaanalyses of biodiversity and ecosystem studies (4, 10, 12, 13) to identify 29 experiments that have experimentally manipulated the diversity of 3 or more terrestrial plant species in greenhouse or field settings. Most of these experiments were performed by using seasonal systems where most above-ground biomass has a yearly senescence phase. We obtained data on the species composition and community-level biomass in each experimental pot or plots used in these 29 studies by using data presented in the original publication, online data repositories, or directly from the principal investigators of the experiments (refs. 7, 17, 29–37; see Table S1 and Fig. S4 for a summary of experiments). Our dataset included a total of 177 species of angiosperms that are inhabitants of ecosystems found throughout the world (see SI Text).
We used two complementary metrics to characterize diversity effects on the production of biomass in each plot or pot (10, 12, 38). The first metric characterizes the net diversity effect size. It is estimated as the log ratio: LRmean = ln(yip/ym̄), which gives the proportional difference between biomass production (y) of a polyculture (p) and the mean biomass of those same species in monoculture (m̄) in experiment i. The second metric gives the proportional difference in biomass production (y) of a polyculture (p) and the biomass of the maximum producing monoculture (m̂) in experiment i, LRmax = ln(yip/ym̂). This metric characterizes the amount of “transgressive overyielding” in a community, which occurs when a diverse polyculture produces more biomass than even its single most productive species. The main text of this article focuses on the net diversity effect size, LRmean, whereas results for LRmax are reported in the SI Text, Table S3, and Fig. S6.
Because these two metrics require information about the biomass that species achieve individually in monoculture, a number of polyculture plots could not be included in our analyses. This was true for two principal reasons. First, some experiments did not include monocultures of all species in the experimental design, which meant that only a subset of polycultures could be included in our analyses (e.g., 7, 30, 36). Second, in a subset of experiments, unintentional species were sown into plots (37). These additions created 2-species polycultures in place of the intended single-species monocultures. In either case, adequate monoculture estimates could not be calculated for all species in 310 plots, which were excluded from our analyses.
Constructing the Phylogeny.
The 177 species recorded in these experiments included mainly herbaceous angiosperms (both monocots and eudicots), with experimenters explicitly focusing on species that mimic herbaceous grassland-type communities. We pooled all 177 species together to construct a master phylogeny. We used two methods to construct this phylogeny: (i) by using the angiosperm supertree (39); and (ii) from molecular data where we estimated a phylogeny from either (i) maximum likelihood or (ii) Bayesian inference analyses, and for the final two phylogenies we further used Ultrametric rate smoothing on the two molecular phylogenies.
Angiosperm supertree.
We constructed this phylogeny by using the Davies et al. (39) supertree and generated a phylogeny for our species list by using Phylomatic (40). We then used the BLADJ procedure in Phylocom 3.40 (41) to scale branch lengths by using known node ages. For angiosperms, we used the divergence times estimated by Wikstrom et al. (42). Therefore, our estimates of phylogenetic distance from the supertree are in millions of years.
Molecular phylogenies.
For each of the 177 species, we searched GenBank (43) for 4 gene sequences commonly used in published angiosperm phylogenies: atpb, matk, rbcl, and 5.8s. Of the 177 species, 110 had at least 1 gene represented in GenBank. For a further 33 species, we used gene sequences for a congeneric relative only if there were not any other congeners used in the experimental plots. We also included 3 representatives of early diverging lineages as outgroup species, including Amborella trichopoda, Magnolia grandiflora, and Nymphaea odorata. For these 148 species we aligned sequences by using MUSCLE (44). We then selected best-fit models of nucleotide substitution for each gene by using the Akaike Information Criterion, as implemented in Modeltest and MrModeltest (45, 46).
Maximum likelihood phylogeny from gene sequences.
Using the aligned sequences and the estimated models of nucleotide substitution, we estimated a maximum likelihood phylogeny by using the PHYML algorithm with a BIONJ starting tree (47, 48). To assess nodal support on maximum likelihood phylogenies, we report Approximate Likelihood Ratio Test (aLRT) scores, which have been shown to correlate with ML bootstrap scores but require much less computational time (48).
Bayesian phylogeny from gene sequences.
We conducted partitioned Bayesian Inference, estimating the posterior probability distribution of all possible phylogenies by using Metropolis Coupled Markov Chain Monte Carlo (MCMCMC), as implemented in MrBayes (49, 50). Four independent Markov Chains were run, each with 3 heated chains for 100 million generations. To monitor possible convergence of the separate MCMC runs, we tracked the standard deviation of split frequencies (SDSF), which was 0.022 at the end of the analysis. We sampled the runs every 10,000 generations and used a burnin of 70 million steps to generate a majority rule consensus tree that was used to calculate PDC. The Bayesian phylogeny is shown in Fig. S1.
Maximum likelihood and Bayesian trees with Ultrametric rate smoothing.
We created two additional phylogenies that represent estimated divergence times based on Ultrametric transformations of the maximum likelihood and Bayesian phylogenetic analyses described above. We performed nonparametric rate smoothing (51) on both phylogenies.
Calculating Phylogenetic Diversity.
We calculated the phylogenetic diversity of plant species sown together in each experimental unit as total phylogenetic branch lengths connecting species together by using script provided by T. Jonathan Davies run in R 2.6.2 (R Development Core Team, www.R-project.org). The results are provided in the SI Text.
Another PD metric, Faith's PD (52) or phylogenetic diversity, is identical to ours except that it calculates PD including the root node of a larger regional phylogeny. Faith's PD then is a measure of the proportion of evolutionary history represented within a local community. Our measure, PDC, simply calculates the phylogenetic distance connecting all members of a community together without considering a larger regional phylogeny. We calculated PDC for plots that had all resident species included in all phylogenies, to compare results among the different phylogenetic methods; therefore, the species included in the supertree phylogeny mirror those for the molecular phylogeny.
In the main text, we present only the results from one phylogenetic analysis (Bayesian inference with Ultrametric rate smoothing; see SI Text for results from other phylogenetic methods) because estimates of PDC depended very little on the specific method of phylogenetic analysis. This finding is evidenced by a high correlation of PDC values calculated with trees resulting from five different phylogenetic methods (for all, r > 0.912, P < 0.01; see Fig. S2). We chose to present the Bayesian tree in particular because Bayesian search algorithms represent a more thorough exploration of parameter space than maximum-likelihood methods, likely optimizing tree topology and branch lengths better. We also chose to use the Ultrametric version of the Bayesian tree because present-day taxa are assumed to be equally divergent from the shared ancestor.
Assigning Functional Groups.
To assign species to plant functional groups we used the classifications provided by individual researchers for their own experiments (17, 34, 36, 37), which generally organized plants into nitrogen fixers (Fabaceae), woody species, C3 graminoids, C4 graminoids, and nonnitrogen-fixing forbs. To standardize the classification of graminoids, we used a single source (53) to determine C3–C4 status. We then enumerated the number of functional groups within experimental plots.
Statistical Analysis.
Our analyses focused on comparing the relative importance of seven potential predictors of LRmean, including species number, the number of plant functional groups, and the five PDC estimates from different phylogenetic methods. For each predictor we ran single variable mixed effects models of the general form:
where yi is the LRmean value in a plot in experiment i, βo is the intercept, β1 is the coefficient associated with the fixed effect variable xi (either species number, PDC, or functional group number), bi is the coefficient of the random effect (experiment), and the error term, εi, is the remaining variation. Parameters in all mixed-effects models were estimated by using restricted likelihood estimation (54). Individual models were compared by using log-likelihood values, Akaike Information Criterion (AIC), and the Akaike weight, which gives the likelihood that model i explains the most variation in an observed data given a set of candidate models (55). We also calculated pseudo-R2 values by regressing model-fitted response values to the observed response variable. Twenty-one statistical outliers (of 1,315) were excluded from our analyses. These were identified from Bonferroni 2-sided tests on Studentized residuals.
Species number, PDC, and their interaction were then combined into the single mixed-effects model:
With this model we tested the assumption that the predictor variables were fixed effects, as in a standard mixed-effects model, vs. whether heterogeneous results necessitate allowing β estimates to vary among experiments. To test this assumption, we fit models with random effects for both the intercept and the slope estimates for either species number or PDC nested within experiment and compared these models with the corresponding one where the intercept and slope were modeled as fixed effects (54). Fixed independent-variable models (i.e., single β) were most parsimonious for both richness and PDC (likelihood ratios: 2.995, P = 0.2236; 4.091, P = 0.1293, respectively).
Although we know that functional groups are predictors of LRmean, we also know that functional groupings generally have a phylogenetic signal (28). Also, from the data it is apparent that the number of functional groups in a plot is correlated with the average PDC in that plot (see Fig. S3). Therefore, because not all experiments explicitly manipulated the number of plant functional groups, we included the number of functional groups, j, as a random effect nested hierarchically within experiment. The model is:
 |
where yij is the LRmean value in a plot in experiment i with j functional groups, βo is the intercept, βsp is the slope of the effect of species number, βPC is the slope of the PDC effect, and βsp×PD is the slope of the effect of the interaction between species number and PDC. bi is the coefficient of the random effect of experiment i, bij is the coefficient of the random effect of j number of functional groups nested with experiment i, and the error term, εi, is the remaining variation. Because of the apparent colinearity between predictors, we preformed a ridge regression (e.g., 56) on the full model and found no significant change in parameter estimation with an estimated Hoerl–Kennard–Baldwin parameter of 2.315.
Finally, species varied in the number of plots in which they occurred (see Fig. S5), and commonly used species could have a disproportionate effect on the results. Therefore, we created n data subsets corresponding to the n species found in >10% of plots. In each data subset, all plots containing common species ni were removed. We reran the statistical analyses and compared these results with the full dataset to determine whether species ni had a disproportionate effect on the results (see SI Text). All analyses were run by using R 2.6.2 (R Development Core Team, www.R-project.org).
Supplementary Material
Acknowledgments.
We thank T. Jonathan Davies for providing R code that calculates PDC and for discussions; D. D. Ackerly, T. J. Davies, M. J. Donoghue, E. J. Edwards, A. Hector, S. Kembel, S. Mazer, D. Tilman, and an anonymous reviewer for providing comments on an earlier version of the manuscript; and researchers for supplying raw datasets. This work was supported by National Science Foundation Grant DEB-0614428 (to B.J.C.). A portion of this work was conducted while M.W.C. was a Postdoctoral Associate at the National Center for Ecological Analysis and Synthesis, a Center funded by National Science Foundation Grant DEB-0553768, the University of California, Santa Barbara, and the State of California.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805962105/DCSupplemental.
References
- 1.Sala OE, et al. Biodiversity: Global biodiversity scenarios for the year 2100. Science. 2000;287:1770–1774. doi: 10.1126/science.287.5459.1770. [DOI] [PubMed] [Google Scholar]
- 2.Vitousek PM, Mooney HA, Lubchenco J, Melillo JM. Human domination of Earth's ecosystems. Science. 1997;277:494–499. [Google Scholar]
- 3.Chapin SI, et al. Ecosystem consequences of changing biodiversity. Bioscience. 1998;48:45–52. [Google Scholar]
- 4.Hooper DU, et al. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol Monogr. 2005;75:3–35. [Google Scholar]
- 5.Tilman D, Downing JA. Biodiversity and stability in grasslands. Nature. 1994;367:363–365. [Google Scholar]
- 6.Tilman D, et al. Diversity and productivity in a long-term grassland experiment. Science. 2001;294:843–845. doi: 10.1126/science.1060391. [DOI] [PubMed] [Google Scholar]
- 7.Tilman D, Wedin D, Knops J. Productivity and sustainability influenced by biodiversity in grassland ecosystems. Nature. 1996;379:718–720. [Google Scholar]
- 8.Naeem S, Thompson LJ, Lawler SP, Lawton JH, Woodfin RM. Declining biodiversity can alter the performance of ecosystems. Nature. 1994;368:734–737. [Google Scholar]
- 9.Hector A, et al. Plant diversity and productivity experiments in European grasslands. Science. 1999;286:1123–1127. doi: 10.1126/science.286.5442.1123. [DOI] [PubMed] [Google Scholar]
- 10.Cardinale BJ, et al. Impacts of plant diversity on biomass production increase through time due to complementary resource use: A meta-analysis. Proc Natl Acad Sci USA. 2007;104:18123–18128. doi: 10.1073/pnas.0709069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stachowicz J, Bruno JF, Duffy JE. Understanding the effects of marine biodiversity on communities and ecosystems. Annu Rev Ecol Evol Syst. 2007;38:739–766. [Google Scholar]
- 12.Cardinale BJ, et al. Effects of biodiversity on the functioning of trophic groups and ecosystems. Nature. 2006;443:989–992. doi: 10.1038/nature05202. [DOI] [PubMed] [Google Scholar]
- 13.Balvanera P, et al. Quantifying the evidence for biodiversity effects on ecosystem functioning and services. Ecol Lett. 2006;9:1146–1156. doi: 10.1111/j.1461-0248.2006.00963.x. [DOI] [PubMed] [Google Scholar]
- 14.Schmid B, et al. The design and analysis of biodiversity experiments. In: Loreau M, Naeem S, Inchausti P, editors. Biodiversity and Ecosystem Functioning: Synthesis and Perspectives. Oxford: Oxford Univ Press; 2002. p. xii.p. 294. [Google Scholar]
- 15.Petchey OL, Hector A, Gaston KJ. How do different measures of functional diversity perform? Ecology. 2004;85:847–857. [Google Scholar]
- 16.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- 17.Lanta V, Leps J. Effect of functional group richness and species richness in manipulated productivity–diversity studies: A glasshouse pot experiment. Acta Oecolog Int J Ecol. 2006;29:85–96. [Google Scholar]
- 18.Wright JP, et al. Conventional functional classification schemes underestimate the relationship with ecosystem functioning. Ecol Lett. 2006;9:111–120. doi: 10.1111/j.1461-0248.2005.00850.x. [DOI] [PubMed] [Google Scholar]
- 19.Venail PA, et al. Diversity and productivity peak at intermediate dispersal rate in evolving metacommunities. Nature. 2008;452:210–257. doi: 10.1038/nature06554. [DOI] [PubMed] [Google Scholar]
- 20.Cavender-Bares J, Wilczek A. Integrating micro- and macroevolutionary processes in community ecology. Ecology. 2003;84:592–597. [Google Scholar]
- 21.Maherali H, Klironomos JN. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science. 2007;316:1746–1748. doi: 10.1126/science.1143082. [DOI] [PubMed] [Google Scholar]
- 22.Silvertown J, Dodd M, Gowing D, Lawson C, McConway K. Phylogeny and the hierarchical organization of plant diversity. Ecology. 2006;87:S39–S49. doi: 10.1890/0012-9658(2006)87[39:pathoo]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 23.Webb CO. Exploring the phylogenetic structure of ecological communities: An example for rain forest trees. Am Nat. 2000;156:145–155. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- 24.Valiente-Banuet A, Verdu M. Facilitation can increase the phylogenetic diversity of plant communities. Ecol Lett. 2007;10:1029–1036. doi: 10.1111/j.1461-0248.2007.01100.x. [DOI] [PubMed] [Google Scholar]
- 25.Cahill JF, Kembel SW, Lamb EG, Keddy PA. Does phylogenetic relatedness influence the strength of competition among vascular plants? Perspect Plant Ecol Evol Syst. 2008;10:41–50. [Google Scholar]
- 26.Stephens PR, Wiens JJ. Convergence, divergence, and homogenization in the ecological structure of emydid turtle communities: The effects of phylogeny and dispersal. Am Nat. 2004;164:244–254. doi: 10.1086/422342. [DOI] [PubMed] [Google Scholar]
- 27.Heemsbergen DA, et al. Biodiversity effects on soil processes explained by interspecific functional dissimilarity. Science. 2004;306:1019–1020. doi: 10.1126/science.1101865. [DOI] [PubMed] [Google Scholar]
- 28.Edwards EJ, Still CJ, Donoghue MJ. The relevance of phylogeny to studies of global change. Trends Ecol Evol. 2007;22:243–249. doi: 10.1016/j.tree.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Dimitrakopoulos PG, Schmid B. Biodiversity effects increase linearly with biotrope space. Ecol Lett. 2004;7:574–583. [Google Scholar]
- 30.Dukes JS. Productivity and complementarity in grassland microcosms of varying diversity. Oikos. 2001;94:468–480. [Google Scholar]
- 31.Fridley JD. Resource availability dominates and alters the relationship between species diversity and ecosystem productivity in experimental plant communities. Oecologia. 2002;132:271–277. doi: 10.1007/s00442-002-0965-x. [DOI] [PubMed] [Google Scholar]
- 32.Fridley JD. Diversity effects on production in different light and fertility environments: An experiment with communities of annual plants. J Ecol. 2003;91:396–406. [Google Scholar]
- 33.Naeem S, Hakansson K, Lawton JH, Crawley MJ, Thompson LJ. Biodiversity and plant productivity in a model assemblage of plant species. Oikos. 1996;76:259–264. [Google Scholar]
- 34.Naeem S, Tjossem SF, Byers D, Bristow C, Li SB. Plant neighborhood diversity and production. Ecoscience. 1999;6:355–365. [Google Scholar]
- 35.Reich PB, et al. Plant diversity enhances ecosystem responses to elevated CO2 and nitrogen deposition. Nature. 2001;410:809–812. doi: 10.1038/35071062. [DOI] [PubMed] [Google Scholar]
- 36.Spehn EM, et al. Ecosystem effects of biodiversity manipulations in European grasslands. Ecol Monogr. 2005;75:37–63. [Google Scholar]
- 37.Tilman D, et al. The influence of functional diversity and composition on ecosystem processes. Science. 1997;277:1300–1302. [Google Scholar]
- 38.Hector A, Bazeley-White E, Loreau M, Otway S, Schmid B. Overyielding in grassland communities: Testing the sampling effect hypothesis with replicated biodiversity experiments. Ecol Lett. 2002;5:502–511. [Google Scholar]
- 39.Davies TJ, et al. Darwin's abominable mystery: Insights from a supertree of the angiosperms. Proc Natl Acad Sci USA. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webb CO, Donoghue MJ. Phylomatic: Tree assembly for applied phylogenetics. Mol Ecol Notes. 2005;5:181–183. [Google Scholar]
- 41.Webb CO, Ackerly DD, Kembel SW. Phylocom: Software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics. 2008;24:2098–2100. doi: 10.1093/bioinformatics/btn358. [DOI] [PubMed] [Google Scholar]
- 42.Wikstrom N, Savolainen V, Chase MW. Evolution of the angiosperms: Calibrating the family tree. Proc R Soc London Ser B. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 46.Posada D, Crandall KA. Selecting the best-fit model of nucleotide substitution. Syst Biol. 2001;50:580–601. [PubMed] [Google Scholar]
- 47.Anisimova M, Gascuel O. Approximate likelihood ration test for branches: A fast accurate and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 48.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 49.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 50.Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 51.Sanderson MJ. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol Biol Evol. 1997;14:1218–1231. [Google Scholar]
- 52.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 53.Waller SS, Lewis JK. Occurrence of C3 and C4 photosynthetic pathways in North American grasses. J Range Man. 1979;32:12–28. [Google Scholar]
- 54.Pinheiro JC, Bates DM. Mixed-Effects Models in S and S-PLUS. New York: Springer; 2004. p. 528. [Google Scholar]
- 55.Johnson JB, Omland KS. Model selection in ecology and evolution. Trends Ecol Evol. 2004;19:101–108. doi: 10.1016/j.tree.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 56.Byrtek M, O'Sullivan F, Muzi M, Spence AM. An adaptation of ridge regression for improved estimation of kinetic model parameters from PET studies. IEEE Trans Nucl Sci. 2005;52:63–68. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.