Abstract
Horizontal transfer (HT) is central to the evolution of prokaryotic species. Selfish and mobile genetic elements, such as phages, plasmids, and transposons, are the primary vehicles for HT among prokaryotes. In multicellular eukaryotes, the prevalence and evolutionary significance of HT remain unclear. Here, we identified a set of DNA transposon families dubbed SPACE INVADERS (or SPIN) whose consensus sequences are ≈96% identical over their entire length (2.9 kb) in the genomes of murine rodents (rat/mouse), bushbaby (prosimian primate), little brown bat (laurasiatherian), tenrec (afrotherian), opossum (marsupial), and two non-mammalian tetrapods (anole lizard and African clawed frog). In contrast, SPIN elements were undetectable in other species represented in the sequence databases, including 19 other mammals with draft whole-genome assemblies. This patchy distribution, coupled with the extreme level of SPIN identity in widely divergent tetrapods and the overall lack of selective constraint acting on these elements, is incompatible with vertical inheritance, but strongly indicative of multiple horizontal introductions. We show that these germline infiltrations likely occurred around the same evolutionary time (15–46 mya) and spawned some of the largest bursts of DNA transposon activity ever recorded in any species lineage (nearly 100,000 SPIN copies per haploid genome in tenrec). The process also led to the emergence of a new gene in the murine lineage derived from a SPIN transposase. In summary, HT of DNA transposons has contributed significantly to shaping and diversifying the genomes of multiple mammalian and tetrapod species.
Keywords: genome evolution, lateral gene transfer, transposable elements, transposase
Lateral or horizontal transfer (HT), the transfer of genetic material between reproductively isolated species, is a frequent occurrence in prokaryotes with selfish and mobile genetic elements such as phages, plasmids, and transposons, serving as the primary vehicles for HT of prokaryotic genes (1). In contrast, reports of HT are scarce in eukaryotes and most cases of nuclear acquisition implicate transfers from prokaryotes or endosymbionts (2–6). The best documented instances of HT between the nuclear genomes of multicellular eukaryotes involve mobile genetic elements, and in particular class 2 or DNA mediated transposons (7, 8). Thus far, conspicuous cases of HT of DNA transposons have been detected among insects (8–12), fish (13) and, in one example, between plants (14). Germline invasions by retroviruses have been documented for several mammals (15–18), and there is mounting evidence supporting the horizontal introduction of a snake retroposon in ruminants (19, 20). However, to our knowledge, there has been no substantiated report of HT of DNA transposons in mammals. Here, we present unequivocal evidence for the repeated HT of a DNA transposon family named SPACE INVADERS in 7 tetrapod lineages, including 5 mammalian orders.
Results
Discovery of SPIN Transposons.
While surveying DNA transposons in the draft genome assembly of the bushbaby Otolemur garnettii, a prosimian primate, we discovered a previously uncharacterized family of elements of the hAT (hobo/Activator/Tam3) superfamily, which we dubbed SPACE INVADERS, or SPIN. Alignment of 21 full-length or nearly full-length SPIN copies allowed us to reconstruct the putative ancestral consensus sequence for this family. The consensus sequence was 2,836-bp long and contained a single long ORF encoding a 602-aa transposase with a conserved hAT dimerization domain at the C terminus (PFAM05699). In addition, each SPIN copy examined was flanked by 8-bp target site duplications, another hallmark of the hAT superfamily (21).
To determine the species distribution of SPIN, the bushbaby consensus sequence was used as a query in Blastn searches of all GenBank databases, which currently contains whole genome shotgun sequence assembly for 33 vertebrate species, including 23 placental mammals, 1 marsupial, 1 monotreme, 3 non-mammalian tetrapods, and 5 fish (see Materials and Methods). These searches yielded a large number of extremely high score hits (score >1,800, e value = 0.00) in 5 mammalian species (tenrec, little brown bat, mouse, rat, and opossum) and in 2 tetrapods (green anole lizard and African clawed frog). In addition, one lower score, but potentially still significant hit (score = 330, e value = 8 × 10−27), was returned in guinea pig that had 84% sequence identity over 325 bp of the query. No other significant hits (scores ≤102, e values ≥8 × 10−19) were found in the other 27 vertebrate genomes or in any other species represented in the GenBank databases.
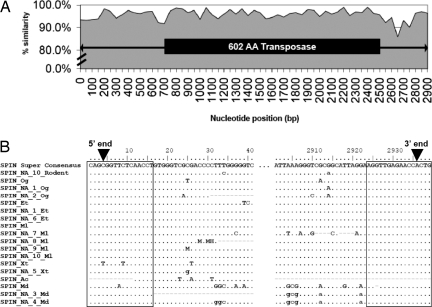
In tenrec and bat, SPIN was present in multiple full-length copies, which allowed the reconstruction of species-specific consensus sequences of 2,871 and 2,867 bp in length, respectively, both encoding a 601-aa hAT transposase. In the other species (opossum, mouse, rat, frog, and lizard), we could detect no more than two full-length and often only partial copies, precluding the reconstruction of reliable consensus sequences. Nonetheless, the longest SPIN copy from each of these species was used to construct a multiple alignment together with the consensus SPIN from bushbaby, tenrec, and bat. The alignment revealed a strikingly high level of sequence identity over the entire length of SPIN elements (≈2.9 kb), with an average of 96% pairwise nucleotide identity between any two species and 98% among the consensus sequences. The level of sequence identity was similarly elevated across both coding and noncoding regions [Fig. 1A, and supporting information (SI) Fig. S1]. Furthermore, the 16-bp terminal inverted repeats (TIR) of SPIN elements were all characterized by the same mismatch at position 4 (Fig. 1B), suggesting that these elements all descend from the same active ancestral transposon carrying imperfect TIRs.
Fig. 1.
Sequence identity and multiple alignment of SPIN elements. (A) Interspecific sequence identity across the entire length of full-length SPIN elements. The plot reflects the average sequence identity in nonoverlapping bins of 50 nt across a multiple alignment of the full-length SPIN consensus sequences from bat, tenrec, and bushbaby, along with single-copy sequences from frog, lizard, and opossum. An average of 98% pairwise nucleotide identity is observed between the bat, tenrec, and bushbaby consensus sequences and an average of 96% between any 2 sequences (range = 84–99%). The transposase ORF is depicted as a black rectangle and the terminal inverted repeats are indicated by arrowheads. (B) Multiple alignment of the 5′ and 3′ ends of full-length and MITE SPIN elements. The 16-bp TIRs are boxed. All SPIN elements share the same imperfection at position 4 in their consensus (black arrowheads). The only exception is SPIN_Xt, the full-length SPIN element in frog, for which we were able to locate only a single partial copy. However, the same TIR imperfection is found in the consensus sequence of SPIN_NA_5_Xt, a MITE subfamily from frog. [Rodent, mouse/rat; Og, Otolemur garnettii (bushbaby); Et, Echinops telfairi (tenrec); Ml, Myotis lucifugus (bat); Xt, Xenopus tropicalis (frog); Ac, Anolis carolinensis (lizard); Md, Monodelphis domestica (opossum)].
Lineage-Specific Amplification of Nonautonomous SPIN Elements.
A hallmark of DNA transposon evolution in eukaryotes is the proliferation of nonautonomous copies, also known as miniature inverted-repeat transposable elements (MITEs), which may greatly outnumber their full-length, autonomous partners (21, 22). Typically, MITEs form homogeneous subfamilies that derive from the recurring transmobilization of a particular deletion derivative of a full-length copy (21, 22). We found that each of the 8 species containing full-length SPIN copies also harbor distinct populations of related MITEs in their genome. A total of 12 major MITE subfamilies were detected (Table 1) and each subfamily was found to derive from a different deletion derivative of a full-length SPIN element (Fig. S1 and Fig. S2). The presence of SPIN MITEs identified computationally was validated experimentally by PCR using oligonucleotide primer pairs internal to the TIRs of the SPIN elements (Fig. 2 and see SI Materials and Methods). PCR products of the expected size were obtained from all species that were predicted to contain SPIN, but not from two mammalian species where SPIN was undetectable by Blast searches (human and Jamaican fruit bat). DNA sequencing of PCR products confirmed that they were identical or nearly identical to SPIN transposons found in the corresponding whole genome assemblies (GenBank accession numbers EU867495–EU667500 and FJ154100).
Table 1.
Characteristics of SPIN elements
| Species | TE name* | Length, bp | Copy no. | Avg divergence, % |
|---|---|---|---|---|
| Mouse | SPIN_Rodent | NA† | <5 | NA‡ |
| SPIN_NA_10_Rodent | 225 | 34,041 | 16.1 | |
| Rat | SPIN_Rodent | NA | <5 | NA |
| SPIN_NA_10_Rodent | 225 | 32,567 | 16.5 | |
| Bushbaby | SPIN_Og | 2,836 | 7,145 | 7.2 |
| SPIN_NA_1_Og | 225 | 8,480 | 8.6 | |
| SPIN_NA_2_Og | 80 | 17,498 | 11.9 | |
| Tenrec | SPIN_Et | 2,871 | 13,963 | 9.4 |
| SPIN_NA_1_Et | 224 | 52,551 | 10.4 | |
| SPIN_NA_6_Et | 487 | 32,824 | 9.4 | |
| Bat | SPIN_Ml | 2867 | 2,806 | 3.9 |
| SPIN_NA_7_Ml | 212 | 21,198 | 3.2 | |
| SPIN_NA_8_Ml | 192 | 3,693 | 5.5 | |
| SPIN_NA_9_Ml | 311 | 10,638 | 3.1 | |
| SPIN_NA_10_Ml | 223 | 11,984 | 3.3 | |
| Opossum | SPIN_Md | NA | <5 | NA |
| SPIN_NA_3_Md | 192 | 3,671 | 3.9 | |
| SPIN_NA_4_Md | 718 | 1136 | 5.7 | |
| Frog | SPIN_Xt | NA | <5 | NA |
| SPIN_NA_5_Xt | 186 | 3,992 | 5.2 | |
| Lizard | SPIN_Ac | NA | <5 | NA |
| SPIN_NA_11_Ac§ | 273 | 12,138 | 9.1 |
For each species in which SPIN elements were discovered, the name of the element along with the length, copy number (of both full-length and fragmented elements) and average percent sequence divergence from the consensus sequence are shown. A total of 12 distinct MITE families, along with consensus sequences, could be identified in the 8 species.
*MITE families are denoted with ″NA″ for nonautonomous.
†NA indicates that a consensus sequence could not be derived because too few full-length copies could be identified.
‡NA indicates that an average percent divergence could not be determined for full-length SPIN elements due to low copy number and a high level of fragmentation.
§A complete consensus sequence for SPIN_NA_11_Ac, a lizard-specific MITE family, could not be confidently reconstructed due to uncertainty in its central region. The copy number for this family was estimated based on counts of the 5′ and 3′ terminal regions, for which a reliable consensus could be reconstructed.
Fig. 2.
Experimental verification of the presence or absence of SPIN transposons. PCR fragments of expected sizes were obtained in all species (or a close relative) where SPIN elements were identified computationally. A PCR product of the expected size (data not shown) was also obtained with DNA from the opossum, Monodelphis domestica. DNA sequencing of cloned PCR products for each species confirmed that they represent distinct SPIN family members (see SI Materials and Methods).
Although the consensus sequences for each MITE subfamily differ in size, their shared regions display a level of interspecific pairwise sequence identity (>91%) comparable to full-length SPIN elements (Table 1). They also display the same imperfect TIRs (Fig. 1B). Within each species the MITE subfamilies achieved very high copy numbers, ranging from ≈4,000 copies in frog to ≈99,000 copies in tenrec (Table 1). Thus, whereas full-length SPIN elements from different species are indistinguishable in terms of their sequence and structure, they have colonized the 7 species lineages with differential success and given rise to structurally distinct MITE subfamilies in each of these lineages.
Evidence for the Horizontal Introduction of SPIN Elements.
The level of sequence identity of SPIN transposons among such widely divergent tetrapods is exceptional, being greater than some of the most conserved protein-coding genes in vertebrates (e.g., RAG-1) (23) and comparable to the so-called ultraconserved elements (24). Such levels of sequence identity can be explained by one of two alternatives: Either SPIN elements have been vertically inherited from the last common ancestor of these species (≈360 mya (25)) and preserved by intense purifying selection in these lineages, or full-length SPIN progenitors were introduced horizontally in these lineages and subsequently spread within each genome.
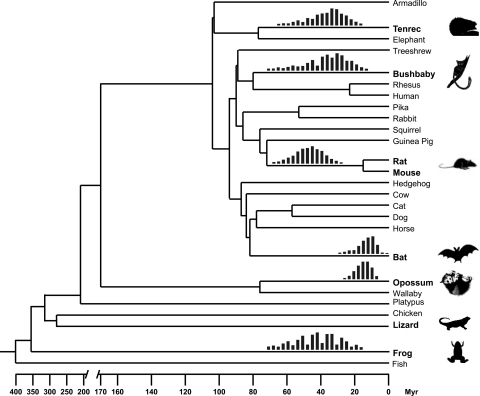
Several observations indicate that a scenario of vertical acquisition of SPIN elements is untenable. Under this hypothesis, the patchy taxonomic distribution of SPIN would require that these elements were lost repeatedly during tetrapod evolution (at least 13 times independently (Fig. 3)), while being maintained active in a subset of lineages. This scenario would also imply that some ancestral SPIN copies have been retained at orthologous genomic positions in some of these species or in the sister taxa represented in the databases. Such ancient transposon fossils are generally well preserved in the genomes of eutherians owing to their relatively slow substitution rates (26–28). However, with the exception of mouse and rat, where nearly all SPIN elements were found at orthologous genomic positions and therefore must have inserted before the divergence of these rodents (Fig. 3), none of the SPIN elements from one species were present at orthologous positions in any of the other species for which complete or nearly complete genome sequences are available. Moreover, we could readily identify SPIN copies present only in one species (bushbaby, bat, or tenrec), but precisely absent at orthologous position in species representing other mammalian orders (Fig. S3). These results strongly indicate that SPIN amplification occurred independently in each of these lineages and that it postdates the radiation of these mammalian orders. Together, these data are inconsistent with a scenario of ancient origin followed by vertical persistence of SPIN activity throughout tetrapod evolution.
Fig. 3.
Species distribution and timing of amplification of SPIN transposons. The tree depicts the phylogenetic relationship and divergence times of the vertebrate species with complete or nearly complete genome sequences currently available (23, 42). The species harboring SPIN transposons are in bold. The timing of SPIN amplification in each species lineage is shown by the red vertical bars above the corresponding branches. Each set of bars represents the age span for all SPIN MITE subfamilies found in the species with each individual bar showing the relative proportion of elements falling within the same, nonoverlapping 3-myr bin (see SI Materials and Methods). The age span is not shown for lizard because the neutral substitution rate is undetermined for this species. However, we note that the level of sequence divergence of SPIN in lizard is similar to those observed in bushbaby and tenrec (see Table 1).
In addition, several lines of evidence allowed us to rule out that SPIN elements, as a whole, have evolved under purifying selection in the lineages examined. First, neighbor-joining phylogenies of SPIN elements mined from each species revealed a star-like topology indicative of a single burst of transposition followed by accumulation of discrete mutations along each branch (Fig. S4). This idiosyncratic pattern of evolution is consistent with neutral evolution and is typical of DNA transposons (9, 10, 21). Furthermore, we found no significant difference in the level of sequence identity among full-length SPIN copies at the first, second, or third codon positions of the transposase ORF (P > 0.05, χ2 test) and no signature of purifying selection acting on SPIN transposase sequences since their divergence from their ancestral consensus sequence (i.e., Ka/Ks not significantly <1, see Materials and Methods). The single exception to this pattern is a peculiar SPIN transposase ORF found at orthologous position in mouse and rat (chr5:130,377,377–130,379,669 in the mm8 mouse genome assembly, see Fig. S5) that has apparently evolved under strong evolutionary constraint since the divergence of the two murine species (Ka/Ks = 0.10; significantly <1, P < 0.0001). The noncoding regions of the corresponding SPIN element are still recognizable, but they have suffered secondary transposable element insertions and extensive mutational decay (Fig. S5). Based on mouse transcriptome data (e.g., full-length cDNA clones BC137610 and AK010551, see Fig. S5), this SPIN transposase appears to be expressed from a far-upstream promoter in fusion with flanking exons encoding a CHCH domain (PFAM06747). The resulting putative chimeric protein (756 aa) might have been “exapted” for a cellular function in the murine lineage. This is similar to the SETMAR protein of primates, which results from the fusion of a mariner transposase with flanking exons of a SET domain gene (29). Notwithstanding this possible case of transposase exaptation, we can reject the hypothesis that the extreme level of SPIN sequence identity among widely divergent tetrapods reflects the systematic action of purifying selection. Thus, the only plausible scenario is that active and nearly identical SPIN elements were introduced horizontally, and relatively recently, into several tetrapod species and subsequently spawned different waves of SPIN amplification along these species lineages.
Timing of SPIN Colonizations.
To gain further insight into the timing of the transfers, we derived an estimate of the time elapsed since the amplification of SPIN families in each of the host species lineages. Because the elements have evolved neutrally since their insertion, the age of individual insertions can be approximated by measuring the sequence divergence from the ancestral consensus sequence and by applying a neutral substitution rate characteristic of the species lineage (26–28). To estimate the neutral substitution rate in the lineages of bushbaby, murine rodents, bat, and tenrec, we retrieved, aligned, and compared a large and diverse set of ancient transposable elements present at orthologous genomic positions in human, which is known to have diverged from each of these species ≈80, 89, 94, and 104 mya, respectively (see SI Materials and Methods). For the opossum lineage, we used ancient repeats found at orthologous positions in opossum and wallaby, which diverged ≈76 mya. Based on the number of substitutions per million years, we infer that both full-length and MITE SPIN families amplified within a fairly narrow evolutionary window (≈31–46 mya) in the lineages of rodents, bushbaby, tenrec, and frog, consistent with a single wave of germ-line infection of diverse tetrapods (Fig. 3). The burst of SPIN amplification seems more recent in the bat and opossum lineages, ≈15 mya (Fig. 3), which may indicate more recent SPIN transfers in these lineages or a delay in their amplification following the initial horizontal introduction.
Discussion
Our analysis provides unequivocal evidence for the HT of a DNA transposon into distant tetrapods, including species from five distinct mammalian orders. These findings are startling given the barriers apparently opposing the entry of exogenous DNA into the sequestered germline of animals (8). The few convincing cases of HT that have been documented generally involve transfers between fairly closely related taxa, such as P and copia elements among drosophilids (8, 30), Mutator-like element transposons between grasses (14), or the concurrent germline infiltrations of Pan troglodytes endogenous retroviruses in chimpanzee and gorilla (17). Some DNA transposons, such as mariner elements, are apparently able to cross wider evolutionary distances, in part because their transposition does not seem to require specific host factors (8–11). SPIN is remarkable in that it entered the germline and reached high copy numbers in placental, marsupial, reptilian, and amphibian species. This is consistent with the ability of hAT superfamily transposons to jump efficiently in a wide range of heterologous species and conditions (31–33).
The apparent overlap in the timing of SPIN amplification in the different lineages (Fig. 3) and the fact that the SPIN consensus sequences are phylogenetically equidistant to each other (Fig. 1, Fig. S1, and Fig. S6) suggest that different tetrapod species were infected at close evolutionary time points by essentially the same active element. Presumably, this element originated from the same ancestral source species. At the moment, the taxonomic identity of the source species cannot be identified because SPIN-encoded transposases revealed no more than ≈45% amino acid similarity with other eukaryotic hAT transposases found in the databases (Fig. S6). It is also not possible to distinguish whether each tetrapod species acquired SPIN independently from the same exogenous source (e.g., a common prey or parasite) or whether SPIN was acquired once by a tetrapod species from an exogenous source and then spread by HT between tetrapod species. The mechanisms underlying such recurrent HTs remain to be clarified, but we note that DNA viruses are increasingly recognized as potential intermediates for HT of mobile elements between widely divergent animals (20, 34–36). Recently Piskurek and Okada (20) reported on the transfer of a snake retroposon into the genome of taterapox virus, a poxvirus that infects West African rodents. Vertebrate poxviruses are good candidates as SPIN vectors because many have the ability to infect a broad range of species and cell types and some, such as smallpox virus, have been implicated in zoonosis (37). Interestingly, several of the mammals harboring SPIN (i.e., bats, opossum, murine rodents) are notorious reservoir species for diverse viruses, including poxviruses. However, our current view of the taxonomic distribution of SPIN is heavily biased by the sample of species available in the databases. To gain insights into the origin and evolutionary history of SPIN, future experiments shall focus on a systematic exploration of the taxonomic distribution of these elements.
Despite these outstanding questions, our data provide evidence that HT has contributed significantly to diversifying and shaping the genomes of mammals and other tetrapods. With copy numbers per haploid genome ranging from 4,000 to nearly 100,000 copies, to the best of our knowledge SPIN ranks as the most successful DNA transposon family ever reported in any species. DNA transposons have been shown to cause both large- and small-scale genomic rearrangements because of ectopic recombination or aberrant transposition events, and they have also contributed to the creation of new genes (21). Here, we discovered an apparently functional, murine-specific chimeric gene derived from a SPIN transposase (Fig. S5). Thus, there is little doubt that SPIN amplification not only added megabases of DNA to the genomes, but it also promoted lineage-specific changes in chromosomal architecture and fueled evolutionary innovation.
Materials and Methods
Identification of SPIN Elements.
The sequence of SPIN_NA_2_Og, also known as PMER1 in Repbase (38), was used as a query in local Blastn searches (v. 2.2.14) (39) against the bushbaby whole-genome shotgun (WGS) assembly (otoGar1) downloaded from the UCSC Genome Browser (http://genome.ucsc.edu) (40). Sequences matching the 5′ and 3′ terminal regions of SPIN_NA_2_Og separated by >2,000 bp were extracted and used to reconstruct the consensus sequence for the full-length SPIN_Og element. The SPIN_Og consensus sequence was used as a query in Blastn searches against all National Center for Biotechnology Information Blast databases (including trace archives) to determine the presence or absence of SPIN elements in other species. The following Blastn parameters were used: Gap existence penalty, 6; gap extension penalty, 5; penalty for nucleotide mismatch, −5; and reward for nucleotide match, 4. SPIN elements were considered present within a species if the Blastn score (bits) for a high-scoring pair (HSP) was ≥1,718; a value corresponding to 80% of the query sequence matching a sequence within the species database with 90% identity. We chose to use scores rather than e values because the later are dependent on the size of the queried database, which varies between species. However, we note that in all species where an HSP with a score >1,718 was obtained, at least one such HSP had an e value of 0.00. SPIN elements were considered to be absent from a species if the best HSP had a score ≤158, corresponding to 25% of the query sequence matching a sequence within the species database with 90% identity. When SPIN elements were found within a genome, consensus sequences were constructed for each family by using a simple majority rule based on a multiple alignment of at least 20 copies. SPIN consensus sequences have been deposited in Repbase Update (38). We also derived a SPIN “superconsensus” by creating a multiple alignment of full-length SPIN_Ml, SPIN_Et, and SPIN_Og consensus sequences. To determine copy numbers and average percentage divergence of individual SPIN families, we used the respective consensus sequences to mask the corresponding genome sequence by using RepeatMasker v. 3.1.5 (41). We note that some of the MITE consensus sequences used have been deposited previously in Repbase. These include: SPIN_NA_10_Rodent (URR1A), SPIN_NA_2_Og (PMER1), SPIN_NA_5_Xt (URR1_Xt), SPIN_NA_3_Md (URR1A_Mdo), SPIN_NA_4_Md (URR1B_Mdo), SPIN_NA_7_Ml (nhAT4a_ML), SPIN_NA_8_Ml (nhAT4b_ML), SPIN_NA_9_Ml (nhAT5a_ML), and SPIN_NA_10_Ml (nhAT5b_ML). All other consensus sequences were derived during this work.
Tests for Purifying Selection.
To examine the pattern of evolution of SPIN coding sequences, we retrieved the least degraded transposase sequences from bat, tenrec, and bushbaby, aligned them to their respective consensus sequences (which offers a theoretical approximation of their ancestral sequence), removed codons containing obvious nonsense and frameshift mutations, and computed pairwise Ka/Ks ratios with Mega 3.1 by using the model “Codon: Nei-Gojobori method with the Jukes-Cantor correction”. P values were calculated by using the codon-based Z test with the “Codon: Modified Nei-Gojobori method with the Jukes-Cantor correction” model and 500 bootstrap replications.
Supplementary Material
Acknowledgments.
We thank E. Betrán, C. Casola, J. Demuth, J. Fondon, E. Pritham, and members of the Feschotte lab for critical comments and useful suggestions during the preparation of this manuscript; M. Batzer (Louisiana State University, Baton Rouge, LA), E. de la Casa Esperon (University of Texas, Arlington, TX), P. Chippindale (University of Texas, Arlington, TX), S. Goodman (Field Museum of Natural History, Chicago, IL), P. Michalak (University of Texas, Arlington, TX), D. Ray (West Virginia University, Morgantown, WV), T. Robinson (University of Stellenbosch, Stellenbosch, South Africa), and A. Ropiquet (Univerity of Stellenbosch, Stellenbosch, South Africa) for the generous gift of DNA or tissue samples used in this study; and Patrick McGuigan of the UTA Distributed and Parallel Computing Cluster for assistance with computing resources, whose work was supported in part by the National Science Foundation Grant EIA-0216500. This work was supported by National Institutes of Health Grant R01GM77582 (to C.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data Deposition: Consensus sequences for the SPIN families described in this study have been deposited in the Repbase database, www.girinst.org. All sequences reported in this paper have been depostied in the GenBank database (accession nos. EU867495–EU867500).
See Commentary on page 16827.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806548105/DCSupplemental.
References
- 1.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 2.Hotopp JC, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- 3.Andersson JO. Lateral gene transfer in eukaryotes. Cell Mol Life Sci. 2005;62:1182–1197. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loftus B, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 5.Morrison HG, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 6.Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in Bdelloid rotifers. Science. 2008;320:1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- 7.Syvanen M, Kado C, editors. Horizontal Gene Transfer. London: Academic; 2002. [Google Scholar]
- 8.Silva JC, Loreto EL, Clark JB. Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol. 2004;6:57–71. [PubMed] [Google Scholar]
- 9.Robertson HM. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington DC: ASM Press; 2002. pp. 1093–1110. [Google Scholar]
- 10.Hartl DL, Lohe AR, Lozovskaya ER. Modern thoughts on an ancyent marinere: Function, evolution, regulation. Annu Rev Genet. 1997;31:337–358. doi: 10.1146/annurev.genet.31.1.337. [DOI] [PubMed] [Google Scholar]
- 11.Yoshiyama M, et al. Possible horizontal transfer of a transposable element from host to parasitoid. Mol Biol Evol. 2001;18:1952–1958. doi: 10.1093/oxfordjournals.molbev.a003735. [DOI] [PubMed] [Google Scholar]
- 12.Loreto EL, Carareto CM, Capy P. Revisiting horizontal transfer of transposable elements in Drosophila. Heredity. 2008;100:545–554. doi: 10.1038/sj.hdy.6801094. [DOI] [PubMed] [Google Scholar]
- 13.de Boer JG, Yazawa R, Davidson WS, Koop BF. Bursts and horizontal evolution of DNA transposons in the speciation of pseudotetraploid salmonids. BMC Genomics. 2007;8:422. doi: 10.1186/1471-2164-8-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diao X, Freeling M, Lisch D. Horizontal transfer of a plant transposon. PLoS Biol. 2006;4:e5. doi: 10.1371/journal.pbio.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han K, et al. Mobile DNA in old world monkeys: A glimpse through the rhesus macaque genome. Science. 2007;316:238–240. doi: 10.1126/science.1139462. [DOI] [PubMed] [Google Scholar]
- 16.Gifford R, Tristem M. The evolution, distribution, and diversity of endogenous retroviruses. Virus Genes. 2003;26:291–315. doi: 10.1023/a:1024455415443. [DOI] [PubMed] [Google Scholar]
- 17.Yohn CT, et al. Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol. 2005;3:e110. doi: 10.1371/journal.pbio.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarlinton RE, Meers J, Young PR. Retroviral invasion of the koala genome. Nature. 2006;442:79–81. doi: 10.1038/nature04841. [DOI] [PubMed] [Google Scholar]
- 19.Kordis D, Gubensek F. Unusual horizontal transfer of a long interspersed nuclear element between distant vertebrate classes. Proc Natl Acad Sci USA. 1998;95:10704–10709. doi: 10.1073/pnas.95.18.10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piskurek O, Okada N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc Natl Acad Sci USA. 2007;104:12046–12051. doi: 10.1073/pnas.0700531104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feschotte C, Zhang X, Wessler SR. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz A, editors. Washington, DC: ASM; 2002. pp. 1147–1158. [Google Scholar]
- 23.Hugall AF, Foster R, Lee MS. Calibration choice, rate smoothing, and the pattern of tetrapod diversification according to the long nuclear gene RAG-1. Syst Biol. 2007;56:543–563. doi: 10.1080/10635150701477825. [DOI] [PubMed] [Google Scholar]
- 24.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 25.Hedges SB, Kumar S. Precision of molecular time estimates. Trends Genet. 2004;20:242–247. doi: 10.1016/j.tig.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 27.Yang S, et al. Patterns of insertions and their covariation with substitutions in the rat, mouse, and human genomes. Genome Res. 2004;14:517–527. doi: 10.1101/gr.1984404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pace JK, II, Feschotte C. The evolutionary history of human DNA transposons: Evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci USA. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jordan IK, Matyunina LV, McDonald JF. Evidence for the recent horizontal transfer of long terminal repeat retrotransposon. Proc Natl Acad Sci USA. 1999;96:12621–12625. doi: 10.1073/pnas.96.22.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weil CF, Kunze R. Transposition of maize Ac/Ds transposable elements in the yeast Saccharomyces cerevisiae. Nat Genet. 2000;26:187–190. doi: 10.1038/82827. [DOI] [PubMed] [Google Scholar]
- 32.Evertts AG, Plymire C, Craig NL, Levin HL. The hermes transposon of Musca domestica is an efficient tool for the mutagenesis of Schizosaccharomyces pombe. Genetics. 2007;177:2519–2523. doi: 10.1534/genetics.107.081075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawakami K. Tol2: A versatile gene transfer vector in vertebrates. Genome Biol 8 Suppl. 2007;1:S7. doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraser MJ, Smith GE, Summers MD. Acquisition of host cell DNA sequences by baculoviruses: Relationship between host DNA insertions and FP mutants of Autographa californica and Galleria mellonella nuclear polyhedrosis viruses. J Virol. 1983;47:287–300. doi: 10.1128/jvi.47.2.287-300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friesen PD, Nissen MS. Gene organization and transcription of TED, a lepidopteran retrotransposon integrated within the baculovirus genome. Mol Cell Biol. 1990;10:3067–3077. doi: 10.1128/mcb.10.6.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jehle JA, Nickel A, Vlak JM, Backhaus H. Horizontal escape of the novel Tc1-like lepidopteran transposon TCp3.2 into Cydia pomonella granulovirus. J Mol Evol. 1998;46:215–224. doi: 10.1007/pl00006296. [DOI] [PubMed] [Google Scholar]
- 37.McFadden G. Poxvirus tropism. Nat Rev Microbiol. 2005;3:201–213. doi: 10.1038/nrmicro1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 39.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smit A, Hubley R, Green P. RepeatMasker Open-3.0. 2004 Available at www.repeatmasker.org.
- 42.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.