Abstract
The search for target genes involved in unbalanced acquired chromosomal abnormalities has been largely unsuccessful, because the breakpoints of these rearrangements are too variable. Here, we use the example of dicentric chromosomes in B cell precursor acute lymphoblastic leukemia to show that, despite this heterogeneity, single genes are targeted through a variety of mechanisms. FISH showed that, although they were heterogeneous, breakpoints on 9p resulted in the partial or complete deletion of PAX5. Molecular copy number counting further delineated the breakpoints and facilitated cloning with long-distance inverse PCR. This approach identified 5 fusion gene partners with PAX5: LOC392027 (7p12.1), SLCO1B3 (12p12), ASXL1 (20q11.1), KIF3B (20q11.21), and C20orf112 (20q11.1). In each predicted fusion protein, the DNA-binding paired domain of PAX5 was present. Using quantitative PCR, we demonstrated that both the deletion and gene fusion events resulted in the same underexpression of PAX5, which extended to the differential expression of the PAX5 target genes, EBF1, ALDH1A1, ATP9A, and FLT3. Further molecular analysis showed deletion and mutation of the homologous PAX5 allele, providing further support for the key role of PAX5. Here, we show that specific gene loci may be the target of heterogeneous translocation breakpoints in human cancer, acting through a variety of mechanisms. This approach indicates an application for the identification of cancer genes in solid tumours, where unbalanced chromosomal rearrangements are particularly prevalent and few genes have been identified. It can be extrapolated that this strategy will reveal that the same mechanisms operate in cancer pathogenesis in general.
Keywords: ALL, breakpoint cloning, molecular copy number counting
Chromosomal rearrangements are recurrent findings in human cancer and result in aberrant restructuring of the genome (1). Reciprocal (balanced) translocations may lead to abnormal gene function by direct disruption of coding sequences, such as the formation of chimaeric fusion genes. To date, 358 fusion genes have been identified in human malignancy, the majority of which are the result of balanced chromosomal rearrangements (2). However, the majority of translocations described in human cancer are unbalanced (3), suggesting that other cancer genes remain to be identified. The analysis of unbalanced translocations has largely failed to identify target genes because of the heterogeneity of the chromosomal breakpoints and the multiplicity of partner chromosomes. Thus, it has been assumed that these rearrangements affect gene function through the loss or gain of chromosomal material. Identification of the key molecular events resulting from unbalanced rearrangements would be a significant step toward understanding their role in cancer pathogenesis.
Examples among which both breakpoint heterogeneity and multiplicity of partners are found are dicentric chromosomes: one of the cytogenetic features found in patients with B cell precursor acute lymphoblastic leukaemias (BCP-ALL). The breakpoints principally target the short arm of chromosome 9, which predominantly rearranges with chromosomes 7, 12, and 20, giving rise to the dic(7;9)(p11;p11∼13) (4), dic(9; 12)(p11∼13;p13) (5), and dic(9;20)(p11∼13;q11), respectively (6). These dicentric chromosomes can coexist with established chromosomal changes, for example, dic(7;9) is found in association with t(9;22)(q34;q11) (BCR-ABL1 fusion) (7), and dic(9;12) occurs with t(12;21)(p13;q22) (ETV6-RUNX1 fusion) (8), suggesting that they are important cooperating events. A fusion between the PAX5 and ETV6 genes on chromosomes 9 and 12, respectively, was reported to define the dic(9;12) (7). In contrast, array-based comparative genomic hybridization failed to identify consistent breakpoint within genes in small numbers of patients with the dic(7;9) and dic(9;20) (8, 9).
Using a variety of classical and innovative molecular techniques, we investigated a large cohort of BCP-ALL patients with dicentric chromosomal abnormalities. This approach identified a new subtype of BCP-ALL with genomic disruption of PAX5. This showed that specific gene loci may be the target of heterogeneous translocation breakpoints in human cancer, acting through a variety of mechanisms. The application of this strategy to the analysis of solid tumours, in which unbalanced chromosomal rearrangements are prevalent and relatively few genes have been identified, will lead to the identification of novel cancer genes, expanding our understanding of the genetic basis of cancer pathophysiology.
Results and Discussion
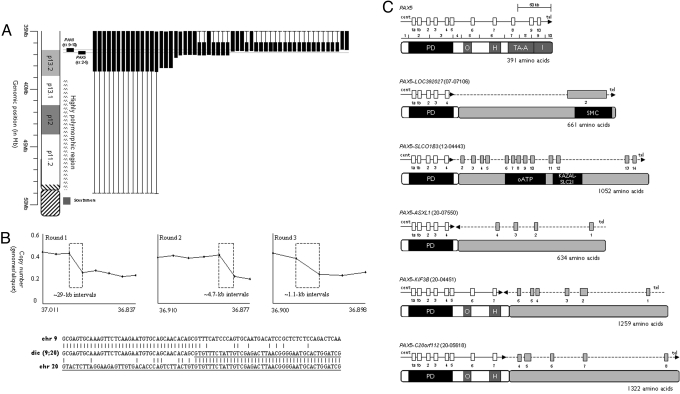
Diagnostic cytogenetic and FISH data were collected on a series of 110 BCP-ALL patients with dic(7;9) (n = 13), dic(9;12) (n = 38), and dic(9;20) (n = 59) [supporting information (SI) Table S1]. To define the breakpoints of these translocations, fluorescence in situ hybridization (FISH) was performed on fixed cell suspension from 54 of these patients, using clones identified from the National Center for Biotechnology Information map for chromosomes 9. Breakpoints on 9p were heterogeneous (Fig. 1A). Fourteen patients exhibited breakpoints within the highly polymorphic region close to the centromere (9p11), whereas the remaining 40 showed breakpoints within 9p13.2. All breakpoints resulted in loss of a large number of genes on 9p, including the tumor suppressor gene, CDKN2A (10). Apart from 2 cases with dic(9;20) (20-06897 and 20-10401), all showed either deletion of the entire PAX5 gene (n = 29, 53.7%) or a breakpoint within the gene (n = 23, 42.6%). PAX5 is essential for B-lineage commitment and maintenance (11), has been shown to be deleted in >30% of childhood ALL samples (10) and is involved in the formation of chimaeric fusion genes: Fusions to ETV6, FOXP1, ZNF521, ELN, and PML were reported in refs. 7, 10, 12, and 13. In this study, intragenic PAX5 breakpoints were shown to be involved in translocations of chromosome 9 with chromosomes 7 (n = 1), 12 (n = 19), and 20 (n = 3). Using FISH with clones specifically targeting the gene, ETV6 was implicated as the partner in 18/19 (95%) dic(9;12) cases. A nested PCR followed by direct sequencing showed that the breakpoints were located within intron 4 of PAX5 in 8 cases, whereas 7 were within ETV6 intron 2 as reported in ref. 7. A single case (12–02398) displayed a breakpoint within intron 1 (Fig. S1), which resulted in the retention of the entire N-terminal (SAM) domain of the predicted fusion protein, potentially altering its protein–protein interactions (14).
Fig. 1.
FISH and sequence analysis of BCP-ALL patients with dicentric chromosomal abnormalities. (A) FISH data for chromosome 9 from patients with dicentric chromosomes. A partial idiogram of the short arm of chromosome 9 is shown on the left with the position of the PAX5-specific FISH clone shown in black. Each column shows an individual patient with the position and extent of the deleted region represented by the vertical black box. In several cases, it was not possible to refine the position of the deletion breakpoint (shown by the thin vertical line). This was due either to the lack of uniquely binding FISH clones in the polymorphic region of 9p11–13.1 or lack of material. (B) Representative MCC and sequence analysis of PAX5 disruption in dicentric chromosome translocations. The results from 3 rounds of MCC on patient 20–4551 are shown above, where each round further delineates the position of the breakpoint. The x and y axes show the genomic position and degree of copy number change, respectively. The size and position of the area with the copy number change is shown by the dashed box. Below is the sequence data from the same patient. The fusion sequence is shown between normal sequence for chromosome 9 and 20, where the chromosome 20 sequence is underlined. (C) Schematic of PAX5 genomic breakpoints and partner genes. The exons for PAX5 and the partner genes are shown by the white and gray boxes, respectively. The gene orientation is shown by the horizontal arrow. Below each genomic schematic is the corresponding predicted protein structure and domains. PD, paired; O, octapeptide; H, homeodomain-like; TA-A, transactivation, activating; I, inhibitory; HLH, helix–loop–helix; ETS, Ets; oATP, organic anion transporter polypeptide; KAZAL-ALC21, azal-type serine protease inhibitor; SMC, structural maintenance of chromosomes.
Molecular copy number counting (MCC) was carried out on DNA from 5 patients (dic(7;9) (n = 1), dic(9;12) (n = 1), and dic(9;20) (n = 3]) (Fig. 1B). MCC allows the progressive delineation of unbalanced copy number breakpoints to within a few hundred base-pairs and facilitates rapid sequence analysis. With the use of subgenome quantities of DNA distributed into a 96-well plate, the frequency of PCR well positivity for any given primer pair is a direct reflection of the genomic copy number at that site (15). Based on the final round of MCC, PCR primers were selected that amplified the translocation breakpoint from chromosome 9 into the unknown sequence. Using this approach, 5 fusion sequences were identified in which PAX5 partnered the LOC392027 (7p12.1), SLCO1B3 (12p12), ASXL1 (20q11.1), KIF3B (20q11.21), and C20ORF112 (20q11.1) loci (Fig. 1C and Fig. S1). The PAX5-ASXL1 and PAX5-KIF3B fusion sequences were in opposing orientation; the remaining 3 fusions were in the same orientation but out of frame, suggesting that the biological consequence of these fusion events was loss of gene function (Fig. 1C). The identification of the PAX5-SLCO1B3 fusion indicated that the dic(9;12) is not universally defined by the PAX5-ETV6 fusion described in ref. 7. In agreement with the reports in refs. 7, 10, 12, and 13, all of the predicted fusion proteins retained the DNA-binding paired domain of PAX5. In contrast, a number of the partner genes were predicted to lose the capacity to bind DNA, providing further evidence for the importance of the disruption of normal PAX5, rather than the gain of functional elements to be the consequence of the fusion event.
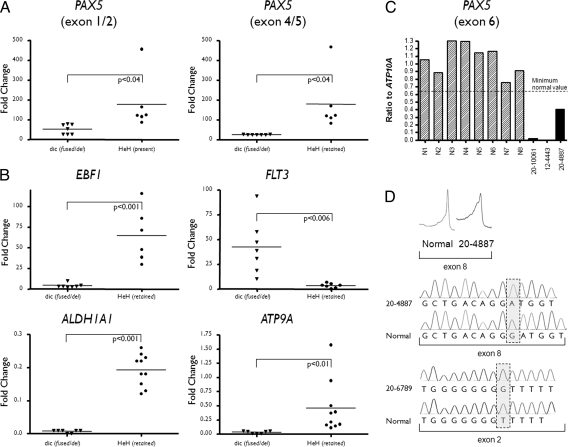
However, in those cases without a PAX5 fusion, alternative genes may be involved. FISH and molecular evidence implicates PAX5 disruption through deletion as the key mechanism in these patients. To confirm the importance of PAX5 disruption, quantitative PCR (qRT-PCR) was used to assess the expression of PAX5 in cases with dicentric chromosomes compared with a control cohort. Patients with high hyperdiploid ALL (HeH) were used as the control cohort, because PAX5 inactivation events are rare in this cytogenetic subtype (10). FISH analyses confirmed the presence of 2 copies of chromosome 9 in all patients from the HeH control cohort. Initial qRT-PCR analysis showed that the expression of exons 1 and 2 (P < 0.04), and 4 and 5 (P < 0.04) were significantly underexpressed in the patients with dicentric chromosomes compared with the control cohort (Fig. 2A and Table S2), clearly demonstrating that both deletion and gene fusion result in the same reduced expression of wild-type PAX5. The same approach was applied to measure the expression of the PAX5 downstream target genes, EBF1, FLT3, ALDH1A1, and ATP9A (10). Differential inactivation of gene expression in the presence of PAX5 alterations was apparent for EBF1 (P < 0.001), ALDH1A1 (P < 0.001), and ATP9A (P < 0.01), whereas FLT3 (P < 0.006) showed the reverse: gene activation in the presence of PAX5 alteration (Fig. 2B and Table S2). These results further supported the hypothesis that PAX5 was the target in patients with dicentric chromosomes, even in the absence of a PAX5 fusion gene.
Fig. 2.
Quantitative PCR analysis of expression and copy number and sequence analysis of BCP-ALL patients with dicentric chromosomes. (A) Quantitative RT-PCR for PAX5 expression exons 1/2 and 4/5. The patient subgroup and fold change in expression are shown in the x and y axes, respectively. Patients with a dicentric chromosome resulting in either fusion or deletion of PAX5 [dic (fused/del)] and with a high-hyperdiploidy karyotype [HeH (retained)]. (B) Quantitative RT-PCR expression of PAX5 target genes. (C) Quantitative genomic PCR for exon 6 of PAX5. Based on a series of control patients (n = 180) with 2 normal copies of PAX5 a minimal ratio for a normal copy number was defined and is shown by the horizontal dashed line. Each patient is shown as a vertical bar on the x axis, where N1-N8 are patients with 2 copies of PAX5, 12-4443 and 20-10061, have homozygous, and 20-4887 has heterozygous loss of PAX5. (D) PAX5 mutation analysis in patients with dicentric chromosomes. The DHPLC traces are shown above for a normal and patient 20-4887, where the x and y axes show retention time and spectrophotometrical detection at 253 nM, respectively. Below are the sequence traces for 2 patients with mutations of PAX5. The box shows the mutation in each case. Patient 20-4887 contained a DelG1450 frame-shift mutation in exon 8, which translates to a truncated protein composed of the original 334 AA of PAX5 then a sequence of 19 AA followed by a premature stop. Patient 20–6789 harboured an insG525 frame-shift mutation in exon 2, which translates to a truncated protein with 25 intact AA followed by a sequence of 48 AA then a premature stop.
The intact PAX5 allele was examined for deletions and mutations. Sixteen patients were screened for submicroscopic deletions, using genomic quantitative PCR. The heterozygous loss of PAX5 as a result of dicentric chromosome formation was confirmed, whereas homozygous deletions of PAX5 were observed in 2 patients (Fig. 2c). The same exons were screened for mutations by denaturing high-performance liquid chromatography (DHPLC) in 20 patients. Direct sequencing revealed that cases 20–4887 and 20–6789 harbored a single base pair deletion of exon 8 (delG1450 frame-shift mutation) and a single base pair insertion in exon 2 (insG525 frame-shift mutation), respectively. Both mutations would translate into a truncated PAX5 protein (Fig. 2D). This suggested that the formation of the dicentric chromosome provided leukemic potential by abrogating normal PAX5 function in these cases.
Here, we have shown that specific gene loci may be the target of heterogeneous breakpoints in human cancer, acting through a variety of mechanisms. Although several small investigations had failed to identify the key molecular events in patients with dicentric chromosomes (8, 9), this large study, using comprehensive molecular analysis, identified PAX5 as the key target gene on chromosome 9 as a consequence of its involvement in multiple fusion genes and by deletion. This approach has considerable application to the identification of cancer genes in solid tumours, where unbalanced chromosomal rearrangements are particularly prevalent and relatively few genes have been identified. It has wider application in cases with deletions and amplifications, in which target genes are often involved through the formation of fusion genes (16). Unbalanced derivative chromosomes may result from an unstable balanced translocation which occurs at much higher frequency (14). Cytogenetic data from the Mitelman Database of Chromosome Aberrations in Cancer (http://cgap.nci.nih.gov/chromosomes/mitelman) showed >80 recurrent unbalanced translocations in breast and lung cancer alone. This number is certainly an underestimation of the true frequency of unbalanced translocations, because many remain unidentified. This was indicated by the frequent cytogenetic description, using “add.” To conclude, we have shown an efficient strategy for the identification of cancer genes, supported by the identification of PAX5 as a key genetic target of dicentric chromosomes in patients with ALL. In the absence of a dicentric chromosome, PAX5 is also targeted by interstitial deletions and copy number neutral LOH events (10, 17), further supporting the importance of investigating the underlying molecular basis of unbalanced translocations. There are now exciting high-throughput sequencing strategies emerging that will surely enhance the discovery of aberrant genes in cancer cells (18, 19). However, the approach described here offers a simple, cost-effective and accessible approach to the identification of cancer genes. From the expansion of this approach into other tumor types, a large number of novel genes will surely emerge, expanding our understanding of carcinogenesis and ultimately leading to improved management of patients with cancer.
Materials and Methods
Patients.
All patients had a confirmed diagnosis of BCP ALL. They were entered onto a Medical Research Council/National Cancer Research Institute Childhood or Adult ALL treatment trial and registered on the Leukaemia Research United Kingdom Cancer Cytogenetics Group Karyotype Database in Acute Leukaemia (20). Informed consent was obtained in accordance with the Declaration of Helsinki. Diagnostic cytogenetic and FISH analysis of bone marrow or peripheral blood was carried out in the U.K. regional genetic laboratories and described according to the International System for Human Cytogenetic Nomenclature (21). The presence of a dicentric chromosome was confirmed with chromosome painting and locus-specific probes according to the studies in ref. 22. In total, 110 patients harboured a dicentric chromosome involving the short arm of chromosome 9 with 3 different partner chromosomes; dic(7;9)(p11;p11∼13) (n = 13), dic(9;12)(p11∼13;p13) (n = 38) and dic(9;20)(p11∼13;q11) (n = 59) (Table S1).
FISH.
The position of deletion breakpoints were investigated with clones from the National Center for Biotechnology Information map for chromosomes 7, 9, 12, and 20. DNA was extracted, labeled, hybridized, and visualized using standard methodologies. Probes located to the long arm of each chromosome were used as controls. The involvement of PAX5 was determined with a dual-color break-apart probe constructed from clones position proximal (RP11–297B17) and distal (RP11–344B23) to the gene.
Molecular Copy-Number Counting (MCC) and Long-Distance Inverse PCR (LDI-PCR) Cloning.
MCC was carried out as described in refs. 15 and 23, using markers within PAX5 (SI Materials and Methods). Three PCR primers (external forward, internal forward, and common reverse) were designed to amplify each marker (Table S3). LDI-PCR was carried out as described in ref. 23. Briefly, 1 μg of genomic DNA was digested with the relevant restriction enzyme (New England BioLabs), ligated at 4 °C, and purified using QIAquick PCR Purification Kit (Qiagen). For the first round of PCR, LDI-PCR primers were used to amplify the ligated DNA, whereas the product from the second round nested PCR was gel purified for direct sequence analysis. Further experimental detail and the sequences of MCC and LDI-PCR primers along with their genomic positions are shown in SI Materials and Methods and Table S4.
Genomic Breakpoint Cloning with PCR.
A seminested PCR was carried out to amplify the fusion sequences in 5 dic(9;12) cases with good quality genomic DNA available (cases 2616, 3742, 4662, 8952, and 10630) (SI Materials and Methods). Two oligonucleotides spanning PAX5 exon 4 and the beginning of intron 4 were chosen as the common first-(AGCCACCCAACCAACCAG) and second-forward (GTCACAGCATAGGTAAGAGG) primers. A set of 4 scattering oligonucleotides in ETV6 intron 2 (base pairs 11833587–11859330) were chosen as reverse primers. The sequences of reverse primers are as follows: GAGGAGAGTGAGGCAGG (base pairs 11833587–11833603), CTTACAGGAATCTTTATGG (base pairs 11835721–11835739), GCACCCTCCATACCTAAG (base pairs 11854626–11854643) and CACTAAGTCCTAAGTAGG (base pairs 11859313–11859330). PCR was performed using GoTaq Flexi DNA polymerase (Promega) according to the manufacturer's recommendation, and the PCR products were purified for direct sequencing.
Fusion Protein Sequences Prediction.
Sequences of chimeric proteins were predicted using the online program GENSCAN (http://genes.mit.edu/genscan).
Quantitative Analysis of PAX5 and Target Gene Expression.
qRT-PCR was carried out to assess the expression of PAX5 (exon boundaries 1/2 and 4/5) and PAX5-target genes EBF1, FLT3, ALDH1A1, and ATP9A genes, using the Taqman gene expression assays (Applied Biosystems) as described in ref. 24 (SI Materials and Methods). Seven patients with dicentric chromosomes were compared with 6 cases with a high-hyperdiploidy karyotype (where chromosome 9 was diploid). The comparative Ct method was used for quantitation of relative gene expression. The average Ct value of the endogenous control gene, GAPDH, was subtracted from the average experimental gene Ct value to give the ΔCt value. Differences between control and test were carried out by using ΔΔCt. Differences in gene expression between the 2 groups was performed using a standard t test.
PAX5 Copy Number Analysis with Quantitative Genomic PCR.
This assay was performed as described in ref. 10 but using ATP10A as the control gene (SI Materials and Methods). Briefly, 5-point standard curves ranging from 150 ng to 1 ng/reaction were constructed using normal human genomic DNA (Roche) and amplified for the 3 target PAX5 exons (exons 3, 6, and 8) and control ATP10 gene (Table S5). Assays were performed in duplicate on 2 separate occasions with 50 ng of sample DNA per 20 μL of reaction using a Taqman 7500 Real-Time PCR System (Applied Biosystems). PAX5 gene dosage for each exon was calculated by dividing the value obtained for PAX5 by the corresponding value for ATP10A.
Mutation Analysis of PAX5.
PAX5 exons, previously identified to house mutations in childhood ALL, were amplified by PCR using genomic or whole genome amplified DNA from dicentric cases (SI Materials and Methods). The amplicons were then screened for mutations by DHPLC using a transgenomic wave machine (Crewe). Primer sequences were those previously cited (10), and the DHPLC parameters used are available on request. For those amplicons with chromatographic profiles that differed from wild type, direct sequencing was performed by purifying 100 μL of PCR product using a Qiaquick PCR purification kit (Qiagen) with a final elution volume of 30 μL and then sequenced using both forward and reverse primers with the ABI Version 3 BigDye terminator cycle sequencing kit and analyzed on an ABI prism DNA sequencer (Applied Biosystems). Sequence alignments were carried out using DS gene software (Accelrys).
Supplementary Material
Acknowledgments.
We thank for their dedication the National Cancer Research Institute Childhood and Adult Leukaemia Working Parties and their members, who have designed and coordinated the clinical trials through which these patients were identified and treated and the U.K. Cancer Cytogenetics Group laboratories for the contribution of cytogenetic and FISH data and fixed cell suspensions. This work was supported by Leukaemia Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803494105/DCSupplemental.
References
- 1.Mitelman F, Mertens F, Johansson B. A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat Genet. 1997;15:417–474. doi: 10.1038/ng0497supp-417. [DOI] [PubMed] [Google Scholar]
- 2.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 3.Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat Genet. 2004;36:331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- 4.Raimondi SC, et al. New recurring chromosomal translocations in childhood acute lymphoblastic leukemia. Blood. 1991;77:2016–2022. [PubMed] [Google Scholar]
- 5.Carroll AJ, et al. tdic(9;12): A nonrandom chromosome abnormality in childhood B-cell precursor acute lymphoblastic leukemia: A Pediatric Oncology Group study. Blood. 1987;70:1962–1965. [PubMed] [Google Scholar]
- 6.Rieder H, et al. dic(9;20): A new recurrent chromosome abnormality in adult acute lymphoblastic leukemia. Genes Chromosomes Cancer. 1995;13:54–61. doi: 10.1002/gcc.2870130109. [DOI] [PubMed] [Google Scholar]
- 7.Strehl S, Konig M, Dworzak MN, Kalwak K, Haas OA. PAX5/ETV6 fusion defines cytogenetic entity dic(9;12)(p13;p13) Leukemia. 2003;17:1121–1123. doi: 10.1038/sj.leu.2402923. [DOI] [PubMed] [Google Scholar]
- 8.Schoumans J, et al. Characterisation of dic(9;20)(p11–13;q11) in childhood B-cell precursor acute lymphoblastic leukaemia by tiling resolution array-based comparative genomic hybridisation reveals clustered breakpoints at 9p13.2 and 20q11.2. Br J Haematol. 2006;135:492–499. doi: 10.1111/j.1365-2141.2006.06328.x. [DOI] [PubMed] [Google Scholar]
- 9.Lundin C, et al. Tiling resolution array CGH of dic(7;9)(p11 approximately 13;p11 approximately 13) in B-cell precursor acute lymphoblastic leukemia reveals clustered breakpoints at 7p11.2 approximately 12.1 and 9p13.1. Cytogenet Genome Res. 2007;118:13–18. doi: 10.1159/000106436. [DOI] [PubMed] [Google Scholar]
- 10.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 11.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 12.Bousquet M, et al. A novel PAX5-ELN fusion protein identified in B-cell acute lymphoblastic leukemia acts as a dominant negative on wild-type PAX5. Blood. 2007;109:3417–3423. doi: 10.1182/blood-2006-05-025221. [DOI] [PubMed] [Google Scholar]
- 13.Nebral K, et al. Identification of PML as novel PAX5 fusion partner in childhood acute lymphoblastic leukaemia. Br J Haematol. 2007;139:269–274. doi: 10.1111/j.1365-2141.2007.06731.x. [DOI] [PubMed] [Google Scholar]
- 14.Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet. 1998;18:184–187. doi: 10.1038/ng0298-184. [DOI] [PubMed] [Google Scholar]
- 15.Daser A, et al. Interrogation of genomes by molecular copy-number counting (MCC) Nat Methods. 2006;3:447–453. doi: 10.1038/nmeth880. [DOI] [PubMed] [Google Scholar]
- 16.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 17.Kawamata N, et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008;111:776–784. doi: 10.1182/blood-2007-05-088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bignell GR, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–1303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison CJ, Martineau M, Secker-Walker LM. The Leukaemia Research Fund/United Kingdom Cancer Cytogenetics Group Karyotype Database in acute lymphoblastic leukaemia: A valuable resource for patient management. Br J Haematol. 2001;113:3–10. doi: 10.1046/j.1365-2141.2001.02643.x. [DOI] [PubMed] [Google Scholar]
- 21.ISCN, editor. An International System for Human Cytogenetic Nomenclature. Basel, Switzerland: S. Karger; 2005. [Google Scholar]
- 22.Clark R, et al. Monosomy 20 as a pointer to dicentric (9;20) in acute lymphoblastic leukemia. Leukemia. 2000;14:241–246. doi: 10.1038/sj.leu.2401654. [DOI] [PubMed] [Google Scholar]
- 23.Jalali GR, et al. Disruption of ETV6 in intron 2 results in upregulatory and insertional events in childhood acute lymphoblastic leukaemia. Leukemia. 2008;22:114–123. doi: 10.1038/sj.leu.2404994. [DOI] [PubMed] [Google Scholar]
- 24.Strefford JC, et al. Complex genomic alterations and gene expression in acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21. Proc Natl Acad Sci USA. 2006;103:8167–8172. doi: 10.1073/pnas.0602360103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.