Abstract
MicroRNAs (miRNAs) play an important role in posttranscriptional regulation of genes. We developed a method to predict human miRNAs without requiring cross-species conservation. We first identified lowly/moderately expressed tissue-selective genes using EST data and then identified overrepresented motifs of seven nucleotides in the 3′ UTRs of these genes. Using these motifs as potential target sites of miRNAs, we recovered more than two-thirds of the known human miRNAs. We then used those motifs that did not match any known human miRNA seed region to infer novel miRNAs. We predicted 36 new human miRNA genes with 44 mature forms and 4 novel alternative mature forms of 2 known miRNA genes when a stringent criterion was used and many more novel miRNAs when a less stringent criterion was used. We tested the expression of 11 predicted miRNAs in three human cell lines and found 5 of them expressed in all three cell lines and 1 expressed in one cell line. We selected 2 of them, P-2 and P-27–5p, to do functional validation, using their mimics and inhibitors and using both luciferase assay and Western blotting. These experiments provided strong evidence that both P-2 and P-27–5p are novel miRNAs and that CREB3L3, which encodes cAMP-responsive element binding protein 3-like 3, is a target gene of P-2, whereas LAMB3, which encodes laminin β3, is a target gene of P-27–5p.
Keywords: microRNA, target gene, frequent pattern, functional validation, mimic and inhibitor
Although many human miRNA genes are now known, it is likely that a large number of human microRNA (miRNA) genes remain to be discovered (1–3). Most current methods [e.g., (4–8)] predict novel miRNA genes directly from RNA and/or DNA sequences. In this study, we use a different approach: we first identify putative target binding sites (motifs) in 3′ UTRs and then use these sites to infer novel miRNAs. A similar method for predicting cross-species conserved motifs in 3′ UTRs has been applied to mammalian genomic sequences (9). Briefly, we try to (i) identify a set of tissue-selective genes, which are genes that are expressed in only one or a few tissues (10); (ii) exclude highly expressed tissue-selective genes and call the remaining genes low-key tissue-selective genes; (iii) find motifs of seven nucleotides that appear frequently in the 3′ UTRs of the genes in the set; (iv) consider each of these motifs as a potential miRNA target sequence and its complementary sequence as a potential miRNA seed; (v) exclude those potential seeds that match any known miRNAs; and (vi) use the remaining potential seeds to identify potential novel miRNA genes from predicted secondary structures in the human genome. Our method is based on the reasoning that the 3′ UTRs of the target genes of a miRNA should share the same or a highly similar sequence motif [i.e., target site (TS)]. To define human tissue-selective genes, we use EST data. Moreover, we use microarray gene expression data to filter out highly expressed tissue-selective genes, because a gene that is regulated by miRNAs is more likely to be lowly or moderately expressed than to be highly expressed (11–15).
Because our approach is different from all current methods, it has the potential to find miRNA genes that have not been predicted by any current method. Indeed, we identified 36 highly confident new human miRNA genes that give rise to 44 mature forms and 4 novel alternative mature forms of 2 known human miRNAs plus many less confident predictions. An expression study of 11 predicted miRNAs, using three human cell lines, and functional validation of 2 predicted miRNAs provided evidence for good performance of our method. Thus, the new method can indeed complement existing methods.
Results
Overview of Our Approach.
We developed a seven-step computational method to predict human tissue-selective binding motifs in 3′ UTRs and their potential regulatory miRNAs [see the flowchart in supporting information (SI) Fig. S1]. Below, we briefly describe these steps.
First, we download the human UniGene (Homo sapiens: Build no. 198) data of 40 tissues from the BodyMap-Xs database (16) and map these UniGene entries to Ensembl gene identification numbers. These entries belong to 18,021 human Ensembl genes.
Second, we remove those genes that are expressed in more than 5 of the 40 tissues and call the remaining 6,496 genes tissue-selective genes.
Third, we use a microarray dataset to remove highly expressed genes from the set of tissue-selective genes. We call the remaining genes low-key tissue-selective genes.
Fourth, for each tissue, we select two mutually exclusive gene sets: a set of low-key tissue-selective genes and a set of background genes. We compare the 3′ UTR sequences in the two gene sets to identify sequence motifs that appear frequently in the 3′ UTRs of genes in the first set but rarely in the background set. We use a window size of seven nucleotides to count their frequencies in the 3′ UTRs of the genes in the tissue-selective gene set and their frequencies in the background set. For each 7-mer motif, we use the two-sample proportion test (17) to examine whether the motif is significantly overrepresented in the tissue-selective gene set (i.e., P ≤ 0.01).
Fifth, we filter out simple repetitive motifs such as partial poly(A) sites. One reason for the existence of such simple motifs is that alternative polyadenylation can occur in a tissue-selective manner (18). Because in ∼93% of the known human miRNAs, the nucleotide frequency entropies of their seed regions are ≥0.8, we use the threshold of 0.8 to filter out motifs of low compositional complexity. The remaining motifs are called the frequent tissue-selective motifs or simply the frequent motifs.
Sixth, we use each frequent motif identified as a seed-match region to search the miRBase (19) to remove those motifs that perfectly match any known human miRNAs. In an expanded analysis, we allow one G/U pairing in seven nucleotides. In a further expanded analysis, we also allow 1-nt left- or right-shift seed matches, because a seed region can start at the first, second, or third position from the 5′ end of a miRNA (20). For convenience, a frequent motif that matches the seed region of a known miRNA(s) is called a target motif in this paper.
Seventh, we use the remaining motifs to search the set of predicted secondary structures in the human genome as reported by Pedersen et al. (21) to select motifs that are good candidates for novel miRNAs. In the 48,476 well-conserved secondary structures in the human genome, Pedersen et al. (21) found 195 known miRNA genes and proposed 187 miRNA gene candidates, of which only 24 candidates have been experimentally confirmed. We download the remaining 163 candidates and compute their secondary structure with the minimum folding energy by the mFold software (22). Then, we use the unmatched frequent motifs to check whether the motifs are located in the stem regions in the secondary structures of the miRNA candidates under three criteria (see Methods). A candidate that passes the three criteria is taken as a putative miRNA.
For a more detailed description of the new method, see Methods. Below, we describe the main results.
Low-Key Tissue-Selective Genes in Tissues.
In the 18,021 selected human Ensembl genes, 16,790, 14,910, 14,766, and 14,042 genes are found in the EST data of the cerebrum, lung, testis, and eye, respectively, whereas only 948, 944, and 135 genes are found in the EST data of the salivary gland, esophagus, and brainstem, respectively (Fig. S2).
Among the 18,021 selected genes, there are 6,496 tissue-selective genes, each of which showed EST expression in five or fewer tissues. To remove highly expressed genes, we use Liang et al.'s data (10). These authors identified 3,919 tissue-selective genes from microarray expression data, each of which was highly expressed in some tissues. However, only 1,393 of them map to Ensembl genes and the tissues in the BodyMap-Xs database, and only 239 of these 1,393 genes overlap with our tissue-selective gene set. We exclude these 239 genes from our gene set and call the remaining 6,257 genes low-key tissue-selective genes. Among these genes, 3417, 1129, 720, 521, and 470 are expressed in one, two, three, four, and five tissues, respectively (Fig. S3A).
Tissue-Selective Motifs in the 3′ UTR.
We identified 2,819 frequent tissue-selective motifs. The number of motifs in a tissue is generally correlated with the number of expressed genes and with the number of tissue-selective genes in that tissue (both correlation coefficients are ≈0.6). There are 1703, 575, 284, 136, and 68 one-, two-, three-, four-, and five-tissue selective motifs, respectively. We found 53 frequent motifs (∼2%) that appear in >5 tissues, although each of the genes selected is expressed in ≤5 of the 40 tissues under study (Fig. S3 B and C).
The lymph node, placenta, kidney, lung, pancreas, and cerebrum each contain >400 tissue-selective motifs (833, 557, 489, 465, 440, and 406, respectively). Interestingly, for these six tissues, the number of tissue-selective motifs is negatively correlated with the number of low-key tissue-selective genes (382, 684, 1413, 609, 350, and 2030, respectively; the correlation coefficient is ≈−0.5). In the vein, corpus callosum/glia, adrenal gland, adipose tissue, thyroid/parathyroid, and stomach, the number of predicted motifs is <10, and in the brain stem, esophagus, bladder, and salivary gland, no tissue-selective motif is found (Fig. S3B).
The average ratio of the predicted tissue-selective motifs to the EST genes in a tissue is <0.08; that is, very few tissue-selective motifs are found in the genes expressed in a tissue. But the ratios of the number of predicted motifs over the number of low-key tissue-selective genes of the lymph node, artery/aorta, and pancreas are 2.18, 1.35, and 1.26, respectively, although those for the other 37 tissues are all <1.0. That is, the average number of tissue-selective motifs per low-key tissue-selective gene is ∼2 in the lymph node and >1 in the artery/aorta and pancreas.
Tissue-Selective Motifs That Match Known Human miRNAs.
We compared the identified frequent motifs with all known human miRNAs in the miRBase (release 11.0). First, we found a total of 98 tissue-selective motifs that perfectly match the 133 mature sequences of known human miRNA. (The latter number is larger than the former because the seed regions for different miRNAs can be the same.) Second, we allowed 1-nt left- or right-shift seed matches. The total number of matched miRNAs increased to 267. Finally, when one G/U pairing is also allowed in the seed match, the total number increases to 814 frequent motifs (≈29% of all identified motifs).
In total, there are 483 mature sequences of known human miRNAs that match our predicted frequent tissue-selective motifs (Fig. S4), whereas only 194 known human miRNAs do not match any of our predicted frequent motifs. Thus, our method can indeed recover more than two-thirds of the known human miRNAs.
When G/U pairing and left- or right-shift imperfect seed matches are allowed, the mappings between miRNAs and their target motifs may not be one-to-one but can be one-to-many, many-to-one, or many-to-many (see the examples in Fig. 1). Possibly, even one nucleotide change in the seed region of a miRNA can lead to completely different target genes that are expressed in different tissues.
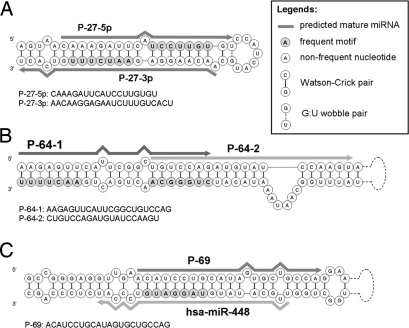
Fig. 1.
Two examples of the frequent tissue-selective motifs that match known human miRNAs. (A) The seed region of miR-203 is UGAAAUG. If we do not allow G/U pairing or left- or right-shift matches, the target motif for prefect matching is ACUUUAC, which is found only in lymph node and lung tissues. However, when the one-nucleotide shift and one G/U pairing are allowed, we find seven other motifs as their putative target motifs in 12 tissues, including the lymph node and lung. (B) miR-206a and miR-206b are two members of the miR-206 family that differ by only one base in the seed region, but their tissue-selective motifs are completely different and the target genes are also expressed in different tissues. In each example, the seed matches include (i) perfect matches and (ii) imperfect matches allowing one G/U pairing, and matches allowing a left or right shift.
Secondary Structure of Predicted Novel miRNA Genes.
Because the unmatched motifs may be potential TSs of unknown miRNAs, we check each of them to see whether it is located in any of the stem regions in the secondary structures of the miRNA candidates predicted by Pedersen et al. (21).
Because the locations of seed regions in the predicted secondary structures are unknown, we consider three possible binding situations. First, two different motifs match the two primes of the stem part of a miRNA gene candidate (Fig. 2A). Second, two motifs are mapped onto the same stem of a miRNA gene candidate but at two different locations (Fig. 2B). The predicted mature sequences are two potential alternative forms of this candidate miRNA. Third, a motif matches a region of a known miRNA gene, but our predicted mature miRNA is different from the known mature sequence in miRBase (Fig. 2C and additional examples in Table S1 and Table S2).
Fig. 2.
Three cases of frequent motifs that match predicted secondary structures of mature miRNA candidates. (A) AAUCUUU and UCCUUGU. (B) AACUUUU and CUGGGCA. (C) UAGGAUG.
When G/U pairing is not allowed, 60 frequent motifs that do not match any known miRNA seed regions give rise to 48 mature miRNA candidates (Table S1). However, P-9–3p and -5p are two novel alternative mature forms of hsa-miR-652; that is, these two sequences overlap with that of hsa-miR-652 but have different seed regions. Similarly, P-27–3p and -5p are two novel alternative mature forms of hsa-miR-802 (Table S1). When one G/U pairing is allowed in the seed match, 116 frequent motifs give rise to 93 mature miRNA candidates (Table S2). P-62, P-63–1 and -2, P-64–1 and -2, P-65, P-66, P-67, P-68–1 and -2, and P-69 are alternative mature forms of has-miR-544, hsa-miR-1264, has-miR-1298, hsa-miR-873, hsa-miR-376b, hsa-miR-381, hsa-miR-365, and hsa-miR-448, respectively (Table S2).
Expression Tests of Predicted Novel miRNAs.
We selected 11 predicted mature miRNA candidates to test whether they are indeed expressed in breast and lung, using three cell lines: the MCF-7 breast cancer cells, the MCF-10A mammary epithelial cells, and the IMR-90 lung fibroblasts. We used real-time PCR to measure the expression level of a miRNA candidate and checked the sizes of PCR amplicons by gel electrophoresis to exclude nonspecific amplifications such as primer dimer formation. As shown in Fig. S5A, 7 novel miRNA candidates were amplified in all three cell lines and one (P-15–5p) was amplified only in MCF-7 cells. The eight PCR-amplified fragments were cloned and sequenced, and six fragments were found to have the correct sequences. The results indicate that 6 of the 11 selected miRNA candidates are expressed in one or more types of human cells (Table 1).
Table 1.
Expression tests and sequencing validation of 11 predicted miRNAs in breast and lung
| Predicted novel miRNA | Experimental validation |
||
|---|---|---|---|
| PCR | DNA electrophoresis | Cloning and sequencing | |
| P-2 | V | V | V |
| P-27–3p | V | V | V |
| P-11–3p | V | V | |
| P-15–5p | V | V | V |
| P-17 | V | V | V |
| P-21 | V | V | V |
| P-23 | |||
| P-24–3p | V | V | |
| P-25 | |||
| P-27–5p | V | V | V |
| P-36 | |||
P-17, P-21, and P-24-3p have been registered as hsa-miR-2052, hsa-miR-2054, and hsa-miR-2053, respectively. V, positive.
We then focused on two predicted mature miRNAs, P-2 and P-27–5p, which were expressed in all three cell lines. We used the U6 snRNA gene as an internal standard because U6 is known to be expressed stably with a significant amount under different biological conditions. We found P-2 to be expressed at a higher level than U6 snRNA in all three cell lines, whereas P-27–5p was expressed at a lower level than U6 snRNA in all three cell lines (Fig. S5B).
Functional Validation of Novel miRNAs by Luciferase Assay and Immunoblotting.
The 3′ UTRs of CREB3L3 (cAMP-responsive element binding protein 3-like 3) and LAMB3 (laminin β3) were screened for complementarities to the seed sequences of P-2 and P-27–5p because they appeared to be, respectively, good candidate target genes of P-2 and P-27–5p. To find out whether CREB3L3 and LAMB3 are indeed, respectively, the targets of P-2 and P-27–5p, we used the luciferase reporter construct to determine the effects of miRNA mimics, which are small partially double-stranded RNAs that mimic endogenous precursor miRNAs, and inhibitors, which inhibit the action of specific miRNAs, on expression of the luciferase gene.
The reporter plasmids bearing the P-2 TS on CREB3L3 3′ UTR (or the P-27–5p TS on LAMB3 3′ UTR) were cotransfected with P-2 (or P-27–5p) mimics (or inhibitors), and the luciferase activity was then measured using the Dual-Light (Applied Biosystems) luciferase and β-galactosidase reporter gene assay system at 48 h after transfection. Fig. 3A shows that P-2 can target to four potential TSs on CREB3L3 3′ UTR (Fig. S6) in the three cell lines. Similarly, the luciferase activity in the P-27–5p mimic transfected cells was lower than that in the negative control transfected cells, and the activity in the anti-P-27–5p transfected cells was higher than that in the negative control transfected cells (Fig. 3B). Thus, P-2 and P-27–5p can bind specifically to CREB3L3 and LAMB3, respectively (Fig. 3C).
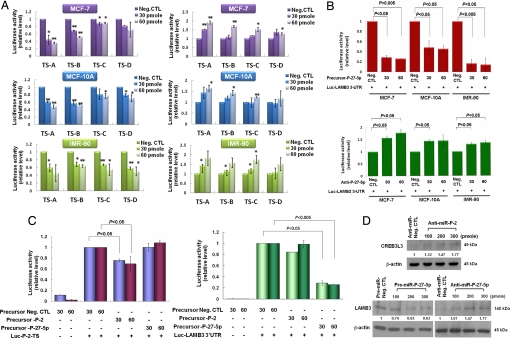
Fig. 3.
Functional target validation of novel mature miRNAs by luciferase assay and immunoblotting using their mimics and inhibitors. The MCF-7, MCF-10A, and IMR-90 cell lines each were transfected with 30–60 pmol PremiR or anti-miR miRNAs of P-2 or P-27–5p. The three different cell lines were cotransfected with luciferase reporter constructs bearing a P-2 target sequence on CREB3L3 3′ UTR or a P-27–5p target sequence on LAMB3 3′ UTRs. The cells were assayed for luciferase activity after transfection for 48 h. (A) Use of P-2 mimics (Left) or inhibitors (Right) showed that P-2 can bind to four possible TSs (TS-A, TS-B, TS-C, and TS-D in Fig. S6) in CREB3L3 3′ UTR in three cell lines (*, P < 0.05, **, P < 0.001). (B) The luciferase activity of cells transfected with P-27–5p mimic was decreased and that transfected with anti-P-27–5p was increased in the three types of cells. (C) Luciferase reporter-based assay was also used to determine miRNA specificity. P-2 and P-27–5p could target specifically to CREB3L3 and LAMB3, respectively. (D) MCF-7 cells were transfected with 100–300 pmol mimic or inhibitor. Protein expression of LAMB3 was decreased after treatment with P-27–5p mimic and increased after treatment with P-27–5p inhibitor for 48 h. CREB3L3 was increased after treatment with P-2 inhibitor.
The immunoblotting showed that the protein expression of LAMB3 was decreased after treatment with P-27–5p mimic but increased after treatment with P-27–5p inhibitor (Fig. 3D). CREB3L3 was increased after treatment with P-2 inhibitor. These immunoblotting data also indicate that CREB3L3 and LAMB3 are the targets of P-2 and P-27–5p, respectively.
Discussion
At the time when we selected P-27–5p for functional validation, we were not aware that it is an alternative mature form of hsa-miR-802. Our experimental study showed that P-27–5P is indeed a miRNA. However, to our knowledge, the only experimental studies on miR-802 were an expression and sequencing study in mice (23) and a miRNA RT-PCR and in situ hybridization study using human fetal brain and heart specimens (24). Thus, it remains to be shown that hsa-miR-802 is functional.
Some of our identified genes may not be tissue selective because we used EST data in only 40 tissues. However, our objective is to predict novel tissue-selective motifs rather than genes. If our identified genes have a high proportion of truly tissue-selective genes, the series of statistical tests proposed in this paper should be powerful enough to exclude those patterns that occur by chance. At the end, the number of tissue-selective motifs we predicted was only 2,819, although the number of the low-key tissue-selective genes with the motifs was 4,721.
A computational approach that is based on cross-species conservation would not be able to predict “young” or species-specific miRNAs and their targets. We therefore proposed a method that requires no cross-species conservation. Actually, methods without this requirement [e.g., REDUCE (regulatory element detection using correlation with expression) (25)], have already been proposed but have not been applied to search for novel miRNAs and their targets.
Counting the numbers of expressed tissues for the target genes predicted by TargetScanS (7) according to the BodyMap-Xs database, we found that ∼72% of the genes were expressed in ≥20 tissues, whereas only ∼5% of the genes were expressed in ≤5 tissues; the mean is 28 tissues. Thus, most currently predicted target genes are not tissue selective. The observation that currently predicted miRNA targets tend to be broadly expressed has been made earlier by Liang and Li (26). In our study, we used only genes expressed in ≤5 tissues to detect the frequent motifs. Thus, only a few of our predicted tissue-selective genes overlap with the target genes predicted by TargetScanS.
Alternative splicing is widely found in higher eukaryotes to generate different mRNA isoforms in specific cell or tissue types (27). In this study, we downloaded all 3′ UTR sequences of each tissue-selective gene from Ensembl but only selected the longest one as the input 3′ UTR sequence. However, the longest one may not be the major form in the selected tissue. Different transcripts spliced from the same gene can be targets of different miRNAs (14). Zhang et al. (18) have analyzed the differences in ploy(A) sites involved in regulating alternative polyadenylation in different tissues. Moreover, in a recent analysis of alternative 3′ UTR isoforms during T-cell proliferation, Sandberg et al. (28) found an increase in the expression of mRNAs terminating at upstream polyadenylation sites (i.e., shorter 3′ UTRs) and that a reduction in protein expression could be reversed by deletion of predicted miRNA TSs in the 3′ UTR region. Thus, alternative polyadenylation can play an important role in mRNA regulation by miRNAs, and it should be taken into consideration in the prediction of potential miRNA binding motifs in future studies.
Materials and Methods
Collection of Human Gene Expression Data.
We downloaded the human UniGene (Homo sapiens: Build no. 198) data and the tissue expression information from the BodyMap-Xs database (16). Then, we mapped the UniGene data to the Ensembl gene identification number and downloaded the transcript data from Ensembl. Finally, we downloaded the 3,919 tissue-selective genes predicted from microarray expression data by Liang et al. (10).
Identification of Tissue-Selective Motifs in 3′ UTRs.
For each tissue-selective gene, we downloaded the 3′ UTR sequence of its longest transcript from Ensembl. For each tissue, we selected two mutually exclusive gene sets: a set of low-key tissue-selective genes (F) and a set of background genes (B). For 7-mer motif mk, let fk and bk be its frequencies in F and B, respectively. We used the one-sided two-sample proportion test (17, 29) to examine whether fk is significantly higher than bk. If so, mk is overrepresented in F. In this case, motif mk is significantly overrepresented in the tissue with a P value ≤0.01.
Matching Frequent Motifs to the Seed Regions of Known Mature miRNAs.
We downloaded the 678 known human mature miRNA sequences from miRBase (release 11.0) (19) and then examined whether a frequent motif that we predicted perfectly matches any of the seed regions of mature miRNAs. We also allowed one G/U pairing, and 1-nt left or right shift in the matching comparison.
Secondary Structures of Potential Novel miRNAs.
We downloaded the 169 human sequences that were predicted to be miRNA genes from the viewpoint of secondary structure (21), but they have not been experimentally validated. For each of these miRNA candidates, we predicted its secondary structure using the mFold software (22). Then, we checked whether the unmatched motifs are located at the stem regions of the secondary structures of some candidates under three criteria: (i) matching should be exact or include only one G/U pair between the motif and its complement; (ii) the length of predicted mature miRNA is at least 17 nucleotides; and (iii) the predicted mature miRNA should not extend to the opposite prime of the stem part.
Cell Culture.
Human breast cancer MCF-7, mammary epithelial MCF-10A, and human lung fibroblast IMR-90 cell lines were purchased from the Bioresource Collection and Research Center, Taiwan. The MCF-7 cell line was maintained in DMEM (Atlanta Biologicals) supplemented with 10% FBS (Gibco). The normal mammary epithelial MCF10A cell line was cultured in growth medium, DMEM, 10 μg/ml insulin (Sigma), 20 ng/ml human epidermal growth factor (Invitrogen Corporation), and 500 ng/ml hydrocortisone (Sigma), supplemented with 5% FBS. The human lung fibroblast IMR-90 cell line was grown in MEM (Sigma) supplemented with 10% FBS. Cells at the logarithmic growth phase were used for our experiments. Cultures were maintained at 37 °C in a humidified atmosphere with 5% CO2.
Designing Stem-Loop RT Primers and PCR Primers.
To test the expression of a predicted novel miRNA gene, we designed PCR primers according to the stem-loop RT-PCR method (30). This method includes two steps: stem-loop RT and real-time PCR. A stem-loop RT primer binds to the 3′ portion of miRNA molecules and forms a spatial constraint structure, which prevents it from binding double-strand genomic DNA molecules. Therefore, stem-loop RT-PCR is the preferred method for rapid and accurate detection of miRNAs. All the novel miRNA sequences studied and the PCR primers are listed in Table S3.
Small RNA Isolation and Real-Time PCR Confirmation.
Total RNA was isolated from cultured cells using the mirVana miRNA Isolation Kit (Ambion, Inc.) according to the manufacturer's instructions. The concentration of RNA was determined by a NanoDrop ND-1000 Spectrophotometer (NanoDrop Tech.). The miRNA cDNA was prepared by the specific miRNA stem loop primer using the Taqman miRNA Reverse Transcription kit (Applied Biosystems). The RT reaction was done starting from 20 ng of total RNA and using the looped primers. The quantitative real-time PCR of each sample was performed with the iCycler iQ Real-Time detection system (BioRad) using the iQ SYBR Green Supermix kit (BioRad). The 25-μl PCR included a 5-μl RT product, 1×SYBR Green Supermix, 0.5 μl of 10-mM forward primer, and 0.5 μl of 10-mM reverse primer. The reactions were incubated in a 96-well plate at 95 °C for 3 min followed by 50 cycles of 95 °C for 10 sec and 60 °C for 30 sec. All reactions were done in triplicate and included no template controls. PCR amplicons were also checked using gel electrophoresis. The PCR amplicons of predicted novel miRNAs P-2, P-11–3p, P-15–5p, P-24–3p, and P-27–5p were checked with 5% agarose gel; P-17, P-27–3p, and P-21 were checked with 12% acrylamide gel. The U6 small nuclear RNA was used as an internal control for cross-experimental normalization. The cycle number at which the reaction crossed an arbitrary threshold (CT) was determined for each gene, and the relative amount of each miRNA to U6 RNA was described using the equation 2−ΔCT, where ΔCT = (CT miRNA − CT U6RNA). To confirm the predicted novel miRNA sequence, the amplified DNA fragment with 3′-A tailing was cloned into the pGEM-T Easy Vector System (Promega) and then sequenced (Mission Biotech Co. Ltd).
Construction of Luciferase Reporter Vector with miRNA-Binding Region/Site.
Plasmids were constructed using standard techniques. The full-length 3′ UTR of LAMB3 was amplified from MCF-10A cDNA (primers: 5′-TTCATACTAGTCAGCAGGGGGCAGAGGCAGTCC-3′, where the SpeI site is underlined, and 5′-AGTCTAAGCTTCTCGTTGAACCTCCAGGCTCTTT-3′, where the HindIII site is underlined). After restriction enzyme reaction, the PCR fragment was then directly ligated into the SpeI and HindIII cloning sites of the pMIR-REPORT luciferase expression vector (Ambion). Clones were selected after colony PCR and restriction enzyme digestion. The clones were verified by sequencing (Mission Biotech Co. Ltd).
Given that we were not able to amplify the 3′ UTR of CREB3L3 from the three cell lines directly, we used synthetic oligonucleotides bearing the P-2 target sequence. The TSs sites are TS-A sense and anti-sense: 5′-AATGCGAGCTCAATGGGGGAGGCAGCTCAGCAAGCTTAATGC-3′ and 5′-GCATTAAGCTTGCTGAGCTGCCTCCCCCATTGAGCTCGCATT-3′; TS-B sense and anti-sense: 5′-AATGCGAGCTCAAACAGACCCGGACAGACAGCTCAGCAAGCTTAATGC -3′ and 5′-GCATTAAGCTTGCTGAGCTGTCTGTCCGGGTCTGTTTGAGCTCGCATT-3′; TS-C sense and anti-sense: 5′-AATGCGAGCTCAAACAGATCCGGACAGACAGCTCAGCAAGCTTAATGC -3′ and 5′-GCATTAAGCTTGCTGAGCTGTCTGTCCGGATCTGTTTGAGCTCGCATT-3′; and TS-D sense and anti-sense: 5′-AATGCGAGCTCAAACAGACCTGGACAGACAGCTCAGCAAGCTTAATGC -3′ and 5′-GCATTAAGCTTGCTGAGCTGTCTGTCCAGGTCTGTTTGAGCTCGCATT-3′. To anneal the oligonucleotides, 2 μg of each strand was added to 46 μl of DNA annealing buffer [30 mM Hepes (pH 7.4), 100 nM potassium acetate, 2 mM magnesium acetate] for a final volume of 50 μl and incubated at 90 °C for 3 min and then at 37 °C for 1 h. The annealed oligonucleotides were digested with HindIII and SacI and used to ligate into HindIII and SacI of the pMIR-REPORT luciferase expression vector (Ambion). Positive clones were selected after screening by restriction digestion with BlpI and were verified by sequencing (Mission Biotech Co. Ltd). All the restriction enzymes were purchased from New England Biolabs.
Transfection.
All transfections were carried out in triplicate. To transfect the miRNA inhibitors or precursors (Ambion) with reporter vectors, we preplated 5 × 104 cells 24 h before transfection in 24-well plates. On the following day, 200 ng of each vector (control and experimental vector) and 30–60 pmol of inhibitor were diluted into 50 μl of Opti-MEM (Gibco) into round-bottom polystyrene tubes. Next, 4 μl of lipofectamine 2000 (Invitrogen, Inc.) was diluted into 50 μl of Opti-MEM and incubated at room temperature for 5 min. The diluted DNA/inhibitor or precursor was next added into the transfection agent complex and incubated at room temperature for 20 min. The growth medium was changed into new growth medium without serum and antibiotics, and 100 μl of the complex was added to the cells. Luciferase reporter activity was measured at 48 h after transfection.
Luciferase Reporter Assays.
Reporter activity was assayed using the Dual-Light system (Applied Biosystems) and was normalized to β-galactosidase activity to control for transfection efficiency variation among different wells according to the manufacturer's instructions. Luminescent signal was quantified by the Spectramax M5 ELISA reader (Molecular Devices). All reporter assays shown in this study are based on data averaged from at least three independent transfections.
Immunoblotting Analysis.
MCF-7 cells were transfected in a 10 cm-dish with 100–300 pmol of mimic or inhibitor and negative control. After 48 h, cells were collected and analyzed by Western blot analysis to assess CREB3L3 or LAMB3 expression. Cells were washed with PBS twice, and pellets were solubilized in lysis buffer containing 7 M urea, 4% CHAPS, 2 M thiourea, and 0.002% bromophenol blue. Lysates were centrifuged at 13,200 × g for 30 min. Proteins were loaded into 10% SDS/PAGE and transferred onto PVDF membranes (Millipore) at 150 V for 1.5 h. After blocking in 5% skim-milk in Phosphate Buffered Saline Tween-20 containing 0.05% Tween 20 at room temperature with gentle rocking for 1 h, membranes were probed with antibodies. Primary antibody was CREB3L3 (LifeSpan Biosciences, Inc.) or LAMB3 (Santa-Cruz Biotechnology, Inc.) at the recommended concentration incubated at 4 °C overnight and then incubated with secondary antibodies (anti-rabbit IgG-HRP; Abcam). Then, immunoblots were visualized with the ECL detection kit (Pierce Biotechnology, Inc.) and exposed to Fuji medical x-ray film. β-actin (Cell Signaling Technology) was used as an internal loading control.
Supplementary Material
Acknowledgments.
We thank Robert Friedman, Yitzhak Pilpel, Li-Ching Hsieh, Hsuan-Cheng Huang, Han Liang, and Henry Lu for suggestions. This work was supported by Academia Sinica, Taiwan; National Science Council of Taiwan; Institute of Information Science and Biodiversity Research Center and Genomics Research Center, Academia Sinica, Taiwan; National Taiwan University Frontier and Innovative Research Projects; Technology Commons, College of Life Science, National Taiwan University and Department of Medical Research, National Taiwan University Hospital; and National Institutes of Health Grants GM30998 and GM081724.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809151105/DCSupplemental.
References
- 1.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 2.Bentwich I, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 3.Hammond SM. RNAi, microRNAs, and human disease. Cancer Chemother Pharmacol. 2006;58(Suppl 1):s63–s68. doi: 10.1007/s00280-006-0318-2. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Kiriakidou M, et al. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Cora D, Di Cunto F, Caselle M, Provero P. Identification of candidate regulatory sequences in mammalian 3′ UTRs by statistical analysis of oligonucleotide distributions. BMC Bioinformatics. 2007;8:174. doi: 10.1186/1471-2105-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang S, Li Y, Be X, Howes S, Liu W. Detecting and profiling tissue-selective genes. Physiol Genomics. 2006;26:158–162. doi: 10.1152/physiolgenomics.00313.2005. [DOI] [PubMed] [Google Scholar]
- 11.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 12.Farh KK, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 13.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 14.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 15.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogasawara O, et al. BodyMap-Xs: Anatomical breakdown of 17 million animal ESTs for cross-species comparison of gene expression. Nucleic Acids Res. 2006;34:D628–D631. doi: 10.1093/nar/gkj137. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glantz SA. Primer of Biostatistics. New York: McGraw–Hill; 2002. [Google Scholar]
- 18.Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 21.Pedersen JS, et al. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takada S, et al. Mouse microRNA profiles determined with a new and sensitive cloning method. Nucleic Acids Res. 2006;34:e115. doi: 10.1093/nar/gkl653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn DE, et al. Human chromosome 21-derived miRNAs are overexpressed in Down syndrome brains and hearts. Biochem Biophys Res Commun. 2008;370:473–477. doi: 10.1016/j.bbrc.2008.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Bussemaker HJ, Li H, Siggia ED. Regulatory element detection using correlation with expression. Nat Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- 26.Liang H, Li WH. MicroRNA regulation of human protein protein interaction network. RNA. 2007;13:1402–1408. doi: 10.1261/rna.634607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai HK, Huang GT, Chou MY, Lu HH, Li WH. Method for identifying transcription factor binding sites in yeast. Bioinformatics. 2006;22:1675–1681. doi: 10.1093/bioinformatics/btl160. [DOI] [PubMed] [Google Scholar]
- 30.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.