Abstract
B lymphocytes are an integral part of the adaptive immune system. On antigen binding to the B-cell receptor (BCR), B cells rapidly proliferate and differentiate into antibody-secreting plasma cells. The p38 mitogen-activated protein kinase (MAPK) pathway functions downstream of the BCR to control cell proliferation, but the transcriptional effectors of this pathway in B cells have remained elusive. In the present study, we inactivated Mef2c exclusively in B cells by conditional gene targeting in mice. Loss of MEF2C function resulted in a reduced immune response to antigen, defective germinal center formation, and a severe defect in B-cell proliferation, and we show that MEF2C regulates proliferation in response to BCR stimulation via the p38 MAPK pathway. p38 directly phosphorylates MEF2C via three residues in the C-terminal transactivation domain, establishing MEF2C as a direct transcriptional effector of BCR signaling via p38 MAPK.
Keywords: MEF2, knockout, mouse, germinal center, B cell receptor
B and T lymphocytes are the main effectors of the adaptive immune system and function to eliminate specific pathogens, to discriminate between self and foreign antigens, and to develop immunological memory (1). Mature follicular B (Fo B) cells play a critical role in an effective immune response by producing soluble antibodies that bind to pathogens at specific epitopes, which leads to their degradation (2). Antigen binding to the B-cell receptor (BCR) results in rapid proliferation of Fo B cells in germinal centers (GCs) of the spleen and lymph node (3). BCR stimulation leads to the initiation of a host of signaling cascades, including the p38 mitogen-activated protein kinase (MAPK) pathway, which has been demonstrated to control cell proliferation (4), but the transcription factors responsible for controlling cell cycle genes downstream of p38 MAPK in B cells have not been clearly defined.
MEF2C is a MADS domain transcription factor that regulates the development and differentiation of many tissue types (5). Underscoring its importance, inactivation of Mef2c in mice results in embryonic lethality at E10 because of profound cardiovascular defects (6–8). MEF2C functions as a signal-dependent transcriptional switch, which allows it to function as an integrator of a variety of upstream signals. It has been shown to be phosphorylated by p38 MAPK in myocytes and macrophages, which leads to increased expression of known MEF2-dependent targets (9–11). The Mef2c gene is highly expressed in B cells of the spleen and lymph node (12), but the function of MEF2C in B cells has not been determined previously.
In the present study, we inactivated MEF2C exclusively in B cells by conditional gene targeting in mice. Loss of MEF2C function resulted in a profound decrease in peak IgG1 titers on immunization with a T-dependent antigen because of a reduced GC response and severely diminished proliferative capacity. Gene expression profiling of Mef2c-null B cells demonstrated a decrease in many cell cycle genes and suggested a defect in the p38 MAPK pathway. We show that MEF2 activity is strongly stimulated by BCR stimulation and that this activation is dependent on direct p38 MAPK phosphorylation of MEF2C. These data suggest a pathway in which MEF2C acts downstream of BCR signaling via the p38 MAPK cascade to drive B-cell proliferation in response to antigen stimulation.
Results
Mef2c Is Required for an Efficient Humoral Immune Response.
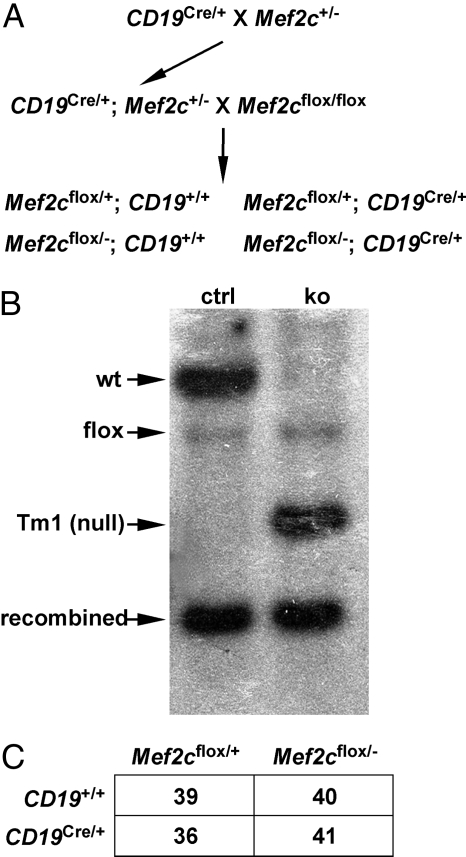
To determine the function of Mef2c in B cells in vivo, we used a conditional knockout (KO) strategy by crossing Mef2c+/−; CD19Cre/+ mice with Mef2cflox/flox mice (Fig. 1A). CD19Cre is expressed exclusively in B cells from early in development (13), such that this cross resulted in specific inactivation of Mef2c in B cells (Fig. 1B). Mef2c B-cell KO mice were born alive and in Mendelian ratios (Fig. 1C). The total number of B cells in the spleen and lymph nodes was similar between Mef2c KO and control mice (data not shown). All surface markers analyzed appeared to be expressed normally in Mef2c-deficient splenic B cells, with the exception of CD23, which exhibited a broader range of expression (Supplemental Material, supporting information (SI) Fig. S1).
Fig. 1.
Conditional inactivation of Mef2c in B cells. (A) A conditional KO strategy, using Cre recombinase driven by the CD19 locus, was used to excise Mef2c flanked by LoxP sites (Mef2cflox/flox). All mice were backcrossed into a C57J/BL6 background for nine generations. For all experiments, CD19Cre/+; Mef2cflox/+ littermates were used to control for the effect of CD19 heterozygosity. (B) Deletion of the second coding exon of Mef2c in sorted Fo B cells was confirmed by Southern blot analysis. Conditional KO (ko) mice had the null allele (Tm1) but not the wild type (wt) allele, which was present in control littermates (ctrl). Note that the conversion of the “floxed” allele (flox) to the recombined form occurred in both control and KO Fo B cells with high efficiency because of the presence of the CD19Cre allele. (C) Mef2c B cell KO animals (CD19Cre/+; Mef2cflox/−) were born at the expected 1:4 ratio.
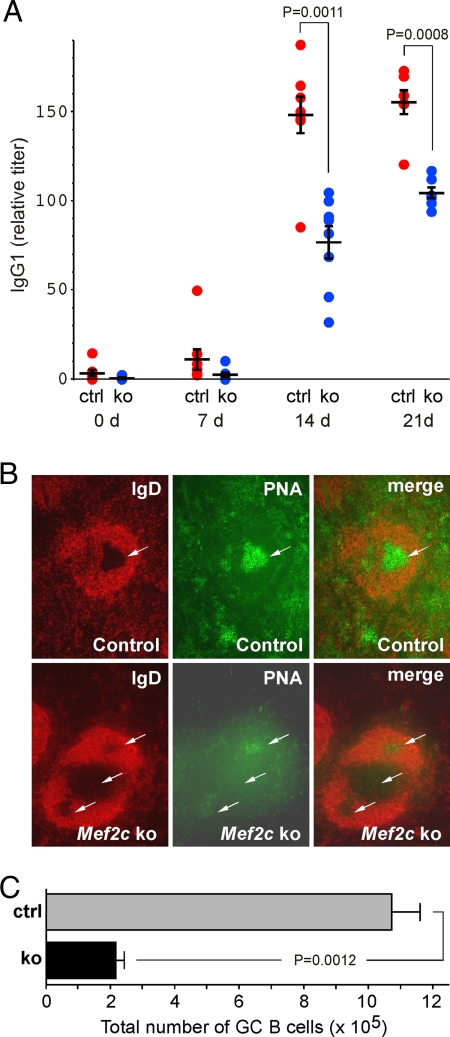
Because Mef2c expression was highest in Fo B cells (Supplemental Material, Fig. S2), we tested the function of these cells by immunizing 8-week-old Mef2c B-cell KO mice and littermate controls with a T-cell–dependent antigen and examined IgG1 titers at 0, 7, 14, and 21 days after immunization. Peak response for IgG1 at 14 days was reduced by 50% in Mef2c B-cell KO mice and remained low at 21 days (Fig. 2A). To determine whether the lower IgG1 titers in Mef2c B-cell KO mice were attributable to a defect in GC formation, we examined lymph nodes from B-cell KO and littermate control mice histologically at 14 days after immunization. IgD and peanut agglutinin (PNA) were used to highlight the lymphoid follicle and GC, respectively. IgD expression was similar in Mef2c B-cell KO and control lymph nodes, suggesting that follicle structure did not depend on MEF2C (Fig. 2B). By contrast, Mef2c B-cell KO mice showed a reduction in the robustness of GC formation in response to antigen, as evidenced by staining with PNA (Fig. 2B). Consistent with the reduced PNA staining by histology, Mef2c B-cell KO mice had fewer GC B cells (PNA+, FAS+, B220+) than control mice (Fig. 2C). These data indicate that Mef2c is important for the humoral response to immunization and GC formation.
Fig. 2.
Mef2c is required for efficient humoral immune response against T-dependent antigens. (A) Mef2c B-cell conditional KO (ko, blue circles) mice exhibit reduced NP-specific IgG1 titers compared with control (ctrl, red circles) mice at 14 and 21 days after immunization with NP-chicken gamma globulin. (B) Immunohistochemistry on sections from lymph nodes depicts a reduced GC response (PNA, green) at 14 days after immunization in Mef2c B-cell KO (bottom row) mice compared with control (top row) mice (white arrows indicate GCs). Lymphoid follicle structure (IgD, red) remained intact in both groups. (C) Quantification of the GC defect by flow cytometry shows a ≈80% reduction in total PNA+, FAS+, B220+ GC B cells in lymph nodes of Mef2c B-cell KO mice compared with control mice.
MEF2C Regulates B-Cell Proliferation in Response to BCR Stimulation.
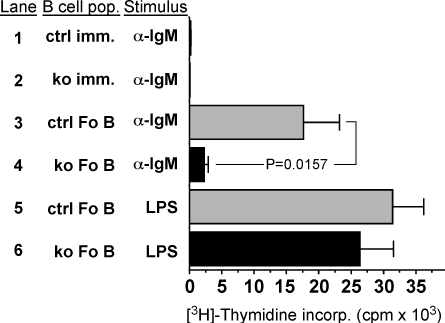
To test the hypothesis that the reduced GC response in Mef2c B-cell KO mice might be attributable to a defect in Fo B cell proliferation on antigen stimulation, we measured [3H]-thymidine incorporation into B cells from Mef2c B-cell KO and control animals (Fig. 3). An equal number of immature transitional splenocytes (total B220+, AA4.1+) and Fo B cells (B220+, AA4.1−, CD21+, CD23+) with equivalent levels of CD21 and CD23 expression between KO and control animals (Supplemental Material, Fig. S1) were isolated and allowed to proliferate ex vivo in response to BCR stimulation. Fo B cells from control mice exhibited 8-fold greater proliferation than B cells lacking MEF2C function when the BCR was stimulated by addition of α-IgM F(ab′)2 (Fig. 3, lanes 3, 4). The difference in proliferation between KO and control animals was slightly reduced on exposure to higher concentrations of α-IgM F(ab′)2 (data not shown), suggesting that strong BCR stimulation could partially overcome the proliferative defect.
Fig. 3.
MEF2C regulates B-cell proliferation in response to BCR stimulation. [3H]-thymidine incorporation assay shows a nearly 90% reduction in proliferation of sorted Fo B cells from KO (ko) mice compared with controls (ctrl) on stimulation of the BCR with α-IgM (lanes 3, 4) but not LPS (lanes 5, 6). Immature B cells (imm.) from either population failed to proliferate on BCR stimulation (lanes 1, 2). Data represent the mean plus SEM for three independent assays.
To determine if the requirement of MEF2C for proliferation was specific to BCR stimulation, we induced proliferation using bacterial LPS, which stimulates Toll-like receptors and associated signaling pathways, and therefore initiates B-cell proliferation via a non–BCR-dependent pathway (14). Importantly, B cells lacking MEF2C function proliferated as well as control B cells in response to LPS stimulation (Fig. 3, lanes 5, 6), indicating that MEF2C was not required for B-cell proliferation in general. As expected, immature AA4.1+ B cells from both groups failed to proliferate on BCR stimulation (Fig. 3, lanes 1, 2). These data demonstrate that MEF2C is a critical effector of B-cell proliferation in response to antigen stimulation of the BCR.
Activation of MEF2C by BCR Stimulation Requires p38 MAPK.
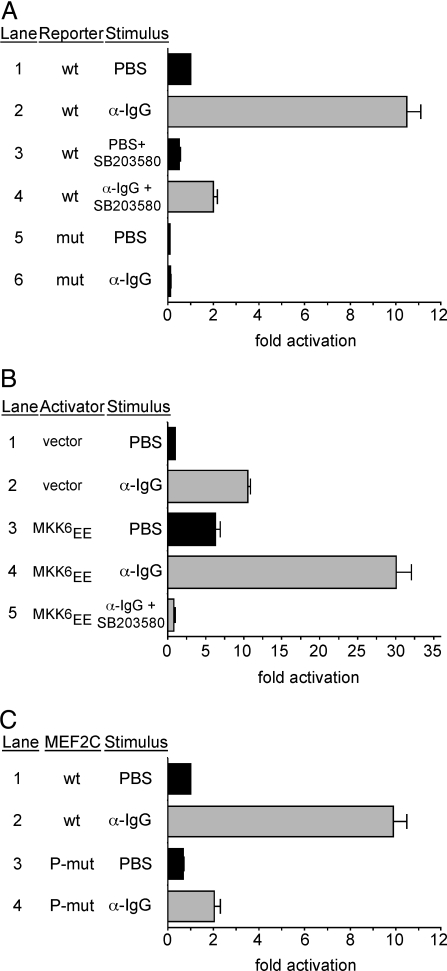
BCR stimulation results in activation of p38 MAPK signaling and subsequent proliferation (15). Previous studies performed in myocytes and macrophages have demonstrated that MEF2C is a direct target of p38 MAPK via phosphorylation of three residues in the C-terminal transactivation domain (10, 11, 16). Therefore, we reasoned that MEF2C might be an effector of p38 MAPK signaling in B cells in response to BCR stimulation. To test this notion, we transfected the 2PK3 B-cell line with an MEF2-dependent reporter, composed of four MEF2 sites directing luciferase expression, and measured the activity of this reporter in response to BCR stimulation. BCR stimulation resulted in a ≈10-fold increase in MEF2-dependent activation (Fig. 4A, lanes 1, 2). This activation was dependent on MEF2 binding to the reporter, because transfection with a mutant MEF2 reporter showed no increase in luciferase activity in response to BCR signaling (Fig. 4A, lanes 5, 6). Addition of SB203580, a p38-specific inhibitor (17), blocked the activation of the reporter (Fig. 4A, lanes 3, 4), indicating that p38 activity was critical for BCR-induced activation of MEF2.
Fig. 4.
Activation of MEF2C by BCR stimulation requires p38 MAPK. (A) BCR stimulation (α-IgG) of 2PK3 B cells transfected with an MEF2-dependent reporter plasmid (wt) showed a 10-fold increase in reporter activity compared with PBS-treated cells (lanes 1, 2), whereas mutant reporter plasmid (mut) did not (lanes 5, 6). Addition of a p38-specific inhibitor (SB203580) blocked BCR-induced reporter activity (lanes 3, 4). (B) Cotransfection with MKK6EE and stimulation with α-IgG showed a synergistic increase in MEF2-dependent reporter activity (lane 4) compared with transfection of MKK6EE without stimulation (lane 3) or reporter alone stimulated with α-IgG (lane 2). Addition of p38 inhibitor blocked this activation (lane 5). (C) p38 phosphorylation sites on MEF2C are necessary for its activity. Addition of α-IgG to cells transfected with P-mut MEF2C failed to activate the MEF2-dependent reporter (lanes 2, 4). Data in all panels represent the mean plus SEM for five independent transfections and analyses.
p38 MAPK functions via a downstream phosphorylation cascade that includes the MAPK kinase, MKK6 (18). Consistent with a role for the p38 pathway in MEF2 activation in B cells, MKK6EE, a constitutively active form of MKK6, induced MEF2 reporter activity in 2PK3 B cells by ≈5-fold (Fig. 4B, lanes 1, 3). Interestingly, BCR stimulation of cells expressing MKK6EE resulted in a strong synergistic activation of the MEF2-dependent reporter, which was blocked by addition of SB203580 (Fig. 4B, lanes 3–5). These results further support the notion that BCR stimulation works through the p38 pathway to stimulate MEF2C activity.
To test the role of the p38 MAPK pathway further in the activation of MEF2C in response to BCR stimulation, we mutated the three p38 phosphorylation sites (T293, T300, and S387) in MEF2C to alanines and tested the activity of the MEF2-dependent reporter when coexpressed with this phosphomutant (P-mut) form of MEF2C (Fig. 4C). MEF2C is expressed endogenously in 2PK3 B cells (data not shown), and expression of additional wild type MEF2C did not have an obvious influence on the ability of BCR stimulation to activate the MEF2-dependent reporter (compare ∼10-fold activation in Fig. 4C, lane 2, with similar activation in Fig. 4A, lane 2). By contrast, expression of MEF2C(P-mut) in 2PK3 B cells inhibited BCR-dependent activation of the MEF2-dependent reporter (Fig. 4C, lane 4), suggesting that the P-mut form of MEF2C functioned as a dominant negative with regard to endogenous MEF2 activity and indicating that the p38 phosphorylation sites on MEF2C are important for the response to BCR stimulation.
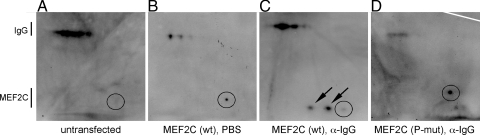
To examine whether BCR stimulation results in the direct phosphorylation of MEF2C, we examined MEF2C and MEF2C(P-mut) by two-dimensional (2D) immunoblot in response to BCR stimulation (Fig. 5). Transfection of B cells with a plasmid encoding a FLAG-tagged version of MEF2C resulted in the presence of a single spot on the 2D immunoblot (Fig. 5, compare A and B). Stimulation of the BCR resulted in phosphorylation of MEF2C, which could be seen by the presence of two additional spots on the 2D immunoblot (Fig. 5C). Detection of the phosphorylated forms of MEF2C was dependent on the p38 phosphorylation sites, because FLAG-tagged MEF2C(P-mut) did not exhibit a change in isoelectric point in response to BCR stimulation (Fig. 5D). These observations establish a link between BCR stimulation and the activation of the MEF2C transcription factor via p38 phosphorylation.
Fig. 5.
BCR stimulation results in phosphorylation of MEF2C. 2D immunoblot demonstrates that stimulation of BCR by α-IgG resulted in a shift in the isoelectric point of MEF2C (B, C), whereas P-mut MEF2C did not (D). Encircled is MEF2C; the arrows point to shifts in the isoelectric point of MEF2C. Spots were identified by molecular weight (MEF2C-FLAG = ∼53 kDa, IgG = ∼150 kDa) compared with a standard protein ladder run in nondenaturing conditions.
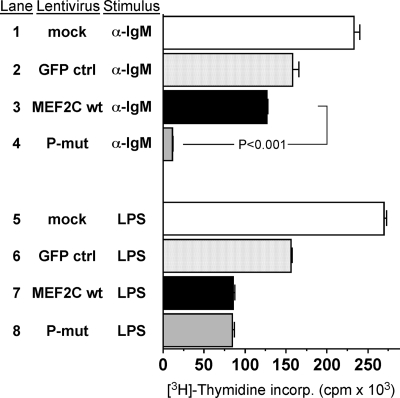
The dominant negative effect of MEF2C(P-mut) observed in Fig. 4C suggested that phosphorylation of MEF2C by p38 is essential for MEF2C to elicit a response downstream of the BCR. If this were the case, then MEF2C(P-mut) might inhibit B-cell proliferation in response to BCR stimulation. To test this notion, we used lentivirus to transduce MEF2C and MEF2C(P-mut) into primary Fo B cells isolated from wild type C57J/BL6 mice and measured [3H]-thymidine incorporation in response to BCR stimulation (Fig. 6). On stimulation with α-IgM, primary Fo B cells expressing MEF2C(P-mut) showed greater than 90% reduction in proliferation compared with untransfected cells or cells overexpressing either wild type MEF2C or GFP (Fig. 6, lanes 1–4). By contrast, LPS stimulation of primary Fo B cells resulted in no difference in proliferation between cells overexpressing wild type MEF2C versus MEF2C(P-mut) (Fig. 6, lanes 7, 8). Interestingly, there was a slight reduction in [3H]-thymidine incorporation when either the wild type or P-mut form of MEF2C was introduced in primary Fo B cells stimulated with LPS (Fig. 6, lanes 5–8), suggesting that overexpression of unphosphorylated MEF2C might interfere with activation of cell cycle genes in response to Toll-like receptor activation.
Fig. 6.
Expression of a P-mut form of MEF2C severely reduces the proliferative capacity of primary Fo B cells. Wild type primary Fo B cells were infected with lentivirus expressing either MEF2C wild type (wt), MEF2C(P-mut), or GFP control (ctrl) and were stimulated to proliferate with either α-IgM (BCR stimulation) (lanes 1–4) or LPS (Toll-like receptor stimulation) (lanes 5–8). Expression of MEF2C(P-mut) severely inhibited proliferation compared with expression of MEF2C wt (lanes 3, 4; P < 0.001). No difference in proliferation was observed between cells infected with MEF2C wt versus MEF2C(P-mut) when stimulated with LPS (lanes 7, 8). Data represent the mean plus SEM for three independent assays of primary Fo B cells isolated from three different wild type C57J/BL6 mice.
The observation that MEF2C controls B-cell proliferation in response to p38 phosphorylation downstream of the BCR strongly suggested that MEF2C might regulate the expression of cell cycle genes downstream of p38 MAPK signaling. To test this hypothesis, we examined the expression of an array of genes involved in the MAPK pathway, including numerous cell cycle genes, by Taqman real-time PCR (Table S1). Consistent with an MEF2C-dependent cell cycle program downstream of the BCR, we observed that the expression of many cell cycle genes was significantly reduced in unstimulated Fo B cells lacking MEF2C compared with controls, including Cyclin D3 (fold change = −6.1, P = 0.013), Cyclin B1 (fold change = −5.7, P = 0.030), Cdk inhibitor 1b (fold change = −7.8, P = 0.018), and Cdk inhibitor 1a (fold change = −9.6, P = 0.050). These results highlight the requirement for MEF2C for normal expression of cell cycle genes in Fo B cells and suggest a coordinated p38 MAPK-MEF2C–dependent program for proliferation on BCR stimulation.
Discussion
Distinct Transcriptional Programs Control Unique Aspects of B-Cell Proliferation.
B-cell proliferation and differentiation in response to antigen are critical to a robust immune response and must occur rapidly during infection to limit tissue damage (3). Given the importance of B cells in the immune response, it is not surprising that multiple B-cell intrinsic receptor pathways regulate B-cell proliferation, including Toll-like receptors in response to LPS and the BCR on specific antigen stimulation (15, 19). LPS/Toll-like receptor–induced B-cell proliferation has been suggested to represent a more evolutionarily ancient pathway, characteristic of the innate immune system, whereas BCR-directed proliferation functions as part of a more versatile adaptive immunity (20).
There are two distinct classes of mature B cells involved in humoral immunity, and they function differentially in immune responses. Marginal zone (MZ) B cells function in a more innate-like fashion compared with the classical Fo B cells (20). Interestingly, we observed that Mef2c expression is highest in Fo B cells and lowest in MZ B cells (Supplemental Material, Fig. S2), supporting a role for MEF2C specifically in a classical humoral immune response by Fo B cells. By contrast, NF-κB is expressed in all classes of B cells, and inactivation of NF-κB results in a broad proliferation defect on stimulation with either LPS or α-IgM (21, 22). Inactivation of another set of transcription factors, OCT-2 and OCA-B, causes a defect in B-cell proliferation only on LPS stimulation but not on α-IgM stimulation (23). Together, these observations suggest that distinct transcriptional programs may function coordinately to control B-cell proliferation in response to unique stimuli with some pathways activating both classes of B cells, whereas others promote MZ or Fo B cell proliferation specifically. Our data demonstrate that MEF2C is a critical regulator of the adaptive immune response specifically downstream of BCR stimulation.
MEF2 Factors as Regulators of B-Cell Development and Function.
MEF2 transcription factors are widely appreciated for their roles in the development and function of muscle lineages, but Mef2 genes are broadly expressed, suggesting roles in numerous other tissues (5, 24). Indeed, recent studies have highlighted roles for Mef2 genes in skeletal and craniofacial development and in the development and function of the central nervous system (5). Here, we demonstrate an essential role for MEF2C in B-cell proliferation, IgG1 production, and GC formation in response to BCR stimulation. While this paper was in final preparation, Wilker et al. (25) reported similar findings, and our studies are generally in close agreement. The authors of that study also show that MEF2C is an important regulator of IgG1 production and GC formation in response to antigen stimulation, and they demonstrate an important role for MEF2C in proliferation downstream of the BCR (25). Wilker et al. (25) further show that the expression of several cell cycle and cell survival genes was reduced in the absence of Mef2c on BCR stimulation. However, the intracellular signaling molecules linking the BCR to MEF2C were not identified. Our work confirms and extends the report by Wilker et al. (25) by establishing that BCR stimulation results in activation of MEF2C transcriptional activity and that this occurs through direct phosphorylation of MEF2 by p38 MAPK.
Other Mef2 genes are also expressed in lymphocytes (12) (Fig. S3). Interestingly, Mef2a and Mef2d expression increased significantly when Mef2c was inactivated in B cells (Supplemental Material, Fig. S3), suggesting possible compensatory roles for other MEF2 family members in B-cell development and function. It is also interesting to note that other MEF2 proteins are regulated by p38 MAPK signaling in macrophages and in muscle and neural lineages (10, 26, 27) and that multiple isoforms of p38, including p38α, p38γ, and p38δ, are abundant in B and T lymphocytes (28). Together, these observations suggest that a wide array of p38 MAPK-MEF2 signaling pathways may function in various aspects of B- and T-cell development, responsiveness, and function. It will be important in future studies to define the requirement for different MEF2 family members and their posttranslational modifications in adaptive immunity.
Implications for MEF2C in B-Cell–Related Immune Diseases.
Dysregulation of B-cell proliferation can cause inadequate immune response, immunodeficiency, or leukemia (29, 30). Our results highlight an important role for MEF2C in regulating B-cell proliferation. In other tissues, MEF2C is known to function as a phosphorylation-dependent switch; as such, it can serve as either an activator or repressor of transcription (31–33). In this regard, it is attractive to speculate that MEF2C may regulate B-cell proliferation both negatively and positively, depending on its phosphorylation state. Interestingly, deregulated Mef2c expression has been shown recently to accelerate Sox4-induced myeloid leukemia in a population of cells that share a common progenitor with B cells (34). Our studies suggest that modulation of the p38 MAPK-MEF2C pathway may be an important target for controlling proliferation in leukemia and other diseases involving aberrant B-cell growth and function.
Materials and Methods
Transgenic Mice and Genotyping.
CD19Cre/+, Mef2c+/−, and Mef2cflox/flox mice have been described previously (13, 35). For our studies, we backcrossed each of these strains for nine generations into a pure C57J/BL6 background. Genotyping was performed as reported previously (13, 35).
Immunization and ELISA.
Eight-week-old animals were immunized with 50 μg of sterile 4-Hydroxy-3-nitrophenylacetyl-chicken gamma globulin (NP-CGG) precipitated in 9% AlK(SO4)2 (pH 7.25) by intraperitoneal injection. For endpoint ELISA assays, plates were coated with 200 μg of NP-BSA overnight at 4 °C. For detection of NP-specific IgG1 titers, a 1:2000 dilution of goat anti-mouse IgM-AP or goat anti-mouse IgG1-alkaline phosphatase (AP) (Southern Biotech) was used. p-Nitrophenyl Phosphatase (Southern Biotech) was used as a substrate for the AP-conjugated antibodies. Relative titer concentrations were calculated by comparison to a standard curve from serial dilutions of sera from hyperimmunized wild type C57J/BL6 mice.
FACS.
Lymphoid organs were harvested and dissociated using a 70-μm cell strainer into Hank's buffered salt solution with 4% FBS to create a single cell suspension. Cells were stained for 30 min with the following antibodies at a 1:40 dilution: B220-Peridinin-chlorophyll Protein complex (BD Biosciences no. 553093), CD21-FITC (BD Biosciences no. 553818), CD23-phycoerythrin (BD Biosciences no. 553139), and AA4.1-Allophycocyanin (APC) (eBiosciences no. 17–5892-82). Cell viability was measured by addition of propidium iodide (5 μg/ml). Sorting was performed on a BD Biosciences FacAria cell sorter.
[3H]-Thymidine Incorporation Assay.
Immature (B220+, AA4.1+) and Fo (B220+, AA4.1−, CD21+, CD23+) B cells were isolated by FACS as described previously. Next, 2 × 105 cells were added in duplicate to a 96-well plate in RPMI-1640 plus 10% FBS. Cells were stimulated by addition of α-IgM F(ab′)2 (7.5 μg/ml; Jackson Immunoresearch no. 115–006-006) or LPS (5 μg/ml) to each well and were incubated for 44 h at 37 °C before addition of 1 μCi of [3H]-thymidine to each well. Cells were then allowed to proliferate for another 4 h, when they were harvested, washed, and counted on a scintillation counter.
Plasmids and Tissue Culture.
The MEF2 reporter plasmid, pMEF2 × 4-E1b-luc was constructed by cloning four MEF2 consensus binding sites (GGGTTATTTTTAGAGCGATCC) into a modified pGL2-Basic vector (Promega) that contains the adenovirus E1b minimal promoter. pMEF2 × 4-E1b-luc (mut) contains mutations in each of the consensus binding sites (TTACCGGTAG). PRK5-MEF2C-VP16 has been described previously (36). MEF2C(P-mut) was created by mutating the Mef2c cDNA at the regions encoding amino acid residues T293, T300, and S387 in the mouse protein (accession no. AAH26841) to alanines via PCR mutagenesis. The MKK6EE expression plasmid has been described previously (18). Plasmids pRK5-MEF2C-FLAG and pRK5-MEF2C(P-mut)-FLAG encode FLAG-tagged versions of MEF2C, which contain the influenza virus M2 FLAG epitope fused in-frame at the C-terminus of MEF2C.
For Amaxa transfections, 1 × 106 2PK3 B cells were resuspended in 100 μl of Amaxa solution V (VCA-1003), and 1 μg of reporter plasmid and 1 μg of activator plasmid were added to the cells, which were then subjected to electroporation using program X-001 on an Amaxa Nucleofector II machine. Cells were then plated in 1 ml of growth media in a 12-well plate, harvested 24 h after transfection, and assayed for luciferase activity. Fifteen micrograms of α-IgG F(ab′)2 (Jackson Immunoresearch) was added to cells for BCR stimulation studies. The p38 inhibitor SB203580 (Sigma) was used at a final concentration of 10 μM in DMSO.
Real-Time PCR.
The Applied Biosystems Assays on Demand primer plus probe for Mef2c (assay no. Mm01344729_m1) and HPRT (assay no. Mm00446968_m1) were used to detect Mef2c expression. RNA was isolated from sorted B cells from 10 wild type C57J/BL6 mice. B-cell stages were defined using surface markers as follows: pro-B cell (B220+, CD43+, IgM−), pre-B cell (B220+, CD43−, IgM−), immature B cell (B220+, CD43+, IgM+, IgD−), transitional 1 (B220+, AA4.1+, IgM+, CD23−), transitional 2 (B220+, AA4.1+, IgM+, CD23+), Fo B cell (B220+, AA4.1−, CD21+, CD23+), and MZ B cell (B220+, AA4.1−, CD21+, CD23−). Taqman-PCR was performed on an ABI 7500 Real-Time PCR machine. For RT-PCR array experiments, Fo B cells were harvested and sorted from KO and control animals as described previously. One microgram of RNA was then used to generate cDNA with the SuperArray RT2 First Strand Kit. cDNA template and SuperArray 2 × PCR master mix were then applied to the SuperArray Mouse MAPK signaling RT-PCR array and run on a Stratagene Mx3005p RT-PCR machine.
Two-Dimensional Immunoblot.
B-cell lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton-X 100, 10 μg/ml aprotinin, 2 μg/ml leupeptin, 100 μM PMSF, 5 mM DTT, 1 mg/ml lysozyme, 5 mM EDTA, and 100 μM Na3VO4) was used to lyse transfected 2PK3 B cells for 30 min on ice. Lysates were pelleted, and resulting supernatants were subjected to the BioRad Ready-Prep 2-D cleanup kit and allowed to adsorb overnight in BioRad immobilized pH gradient (IPG) (pH 3–10) 11-cm strip. Isoelectric focusing was performed for 20,000 volt-hours, and IPG strips were then run in 10% BioRad Criterion XT native gel and transferred to Immobilon PVDF membranes. MEF2C-FLAG protein was detected using α-FLAG M2 primary (Sigma F3165) and goat anti-mouse IgG peroxidase conjugate (Sigma A4416) secondary antibodies. Membranes were then developed using the Amersham ECL kit.
Lentiviral Transduction.
MEF2C and MEF2C(P-mut) were amplified from their respective pCDNA1/amp (Invitrogen) expression plasmids using the following primers, which added a PacI site at the 5′ end and an AscI site at the 3′ end—Mef2c-PacIF: 5′-CGGCTTAATTAAATGGGGAGAAAAAGATTC-3′ and Mef2c-AscIR: 5′-GGCGCGCCCTATTAAGTAATAATGTGATCA-3′. The PCR product was then ligated into the lentiviral packaging vector FuPw at the unique PacI and AscI sites. As described previously (36), 293T cells were used to package the lentivirus. Following lipofectamine transfection into 293T cells, virus was produced for 48 h. A total of 6 × 105 sorted primary Fo B cells were then added to a 12-well plate and infected with 2 ml of lentivirus-containing 293T supernatant. Cells were then transferred to a 96-well plate and subjected to [3H]-thymidine incorporation assay.
Supplementary Material
Acknowledgments.
We thank Robin Lesley, Guy Cinamon, Larry Shiow, Chris Allen, Nicole Haynes, and other members of the Cyster laboratory for technical assistance. We thank Tod Gulick for MKK6 expression plasmids and Beth and Anthony Firulli for advice about 2D gels. We also thank Ryan Swenerton for help with 2D immunoblotting and Maha Abdulla, Zachary Mackey, and James McKerrow for assistance with immunizations. This work was supported by grants from the National Institutes of Health (B.L.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804868105/DCSupplemental.
References
- 1.Cooper MD. Exploring lymphocyte differentiation pathways. Immunol Rev. 2002;185:175–185. doi: 10.1034/j.1600-065x.2002.18515.x. [DOI] [PubMed] [Google Scholar]
- 2.DeFranco AL. Molecular aspects of B-lymphocyte activation. Annu Rev Cell Biol. 1987;3:143–178. doi: 10.1146/annurev.cb.03.110187.001043. [DOI] [PubMed] [Google Scholar]
- 3.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 5.Potthoff MJ, Olson EN. MEF2: A central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 6.Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Q, et al. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development. 1998;125:4565–4574. doi: 10.1242/dev.125.22.4565. [DOI] [PubMed] [Google Scholar]
- 8.Bi W, Drake CJ, Schwarz JJ. The transcription factor MEF2C-null mouse exhibits complex vascular malformations and reduced cardiac expression of angiopoietin 1 and VEGF. Dev Biol. 1999;211:255–267. doi: 10.1006/dbio.1999.9307. [DOI] [PubMed] [Google Scholar]
- 9.McKinsey TA, Zhang CL, Olson EN. MEF2: A calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- 10.Han J, Jiang Y, Li Z, Kravchenko VV, Ulevitch RJ. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 11.de Angelis L, et al. Regulation of vertebrate myotome development by the p38 MAP kinase-MEF2 signaling pathway. Dev Biol. 2005;283:171–179. doi: 10.1016/j.ydbio.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Swanson BJ, Jack HM, Lyons GE. Characterization of myocyte enhancer factor 2 (MEF2) expression in B and T cells: MEF2C is a B cell-restricted transcription factor in lymphocytes. Mol Immunol. 1998;35:445–458. doi: 10.1016/s0161-5890(98)00058-3. [DOI] [PubMed] [Google Scholar]
- 13.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genestier L, et al. TLR agonists selectively promote terminal plasma cell differentiation of B cell subsets specialized in thymus-independent responses. J Immunol. 2007;178:7779–7786. doi: 10.4049/jimmunol.178.12.7779. [DOI] [PubMed] [Google Scholar]
- 15.Campbell KS. Signal transduction from the B cell antigen-receptor. Curr Opin Immunol. 1999;11:256–264. doi: 10.1016/s0952-7915(99)80042-9. [DOI] [PubMed] [Google Scholar]
- 16.Yang SH, Galanis A, Sharrocks AD. Targeting of p38 mitogen-activated protein kinases to MEF2 transcription factors. Mol Cell Biol. 1999;19:4028–4038. doi: 10.1128/mcb.19.6.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuenda A, et al. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 18.Zhu B, Gulick T. Phosphorylation and alternative pre-mRNA splicing converge to regulate myocyte enhancer factor 2C activity. Mol Cell Biol. 2004;24:8264–8275. doi: 10.1128/MCB.24.18.8264-8275.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerondakis S, Grumont RJ, Banerjee A. Regulating B-cell activation and survival in response to TLR signals. Immunol Cell Biol. 2007;85:471–475. doi: 10.1038/sj.icb.7100097. [DOI] [PubMed] [Google Scholar]
- 20.Lopes-Carvalho T, Kearney JF. Development and selection of marginal zone B cells. Immunol Rev. 2004;197:192–205. doi: 10.1111/j.0105-2896.2004.0112.x. [DOI] [PubMed] [Google Scholar]
- 21.Grumont RJ, et al. B lymphocytes differentially use the Rel and nuclear factor kappaB1 (NF-kappaB1) transcription factors to regulate cell cycle progression and apoptosis in quiescent and mitogen-activated cells. J Exp Med. 1998;187:663–674. doi: 10.1084/jem.187.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siebenlist U, Brown K, Claudio E. Control of lymphocyte development by nuclear factor-kappaB. Nat Rev Immunol. 2005;5:435–445. doi: 10.1038/nri1629. [DOI] [PubMed] [Google Scholar]
- 23.Schubart K, et al. B cell development and immunoglobulin gene transcription in the absence of Oct-2 and OBF-1. Nat Immunol. 2001;2:69–74. doi: 10.1038/83190. [DOI] [PubMed] [Google Scholar]
- 24.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 25.Wilker PR, et al. Transcription factor Mef2c is required for B cell proliferation and survival after antigen receptor stimulation. Nat Immunol. 2008;9:603–612. doi: 10.1038/ni.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox DM, et al. Phosphorylation motifs regulating the stability and function of myocyte enhancer factor 2A. J Biol Chem. 2003;278:15297–15303. doi: 10.1074/jbc.M211312200. [DOI] [PubMed] [Google Scholar]
- 27.Han J, Molkentin JD. Regulation of MEF2 by p38 MAPK and its implication in cardiomyocyte biology. Trends Cardiovasc Med. 2000;10:19–22. doi: 10.1016/s1050-1738(00)00039-6. [DOI] [PubMed] [Google Scholar]
- 28.Cook R, Wu CC, Kang YJ, Han J. The role of the p38 pathway in adaptive immunity. Cell Mol Immunol. 2007;4:253–259. [PubMed] [Google Scholar]
- 29.Ehrlich M, et al. DNA methyltransferase 3B mutations linked to the ICF syndrome cause dysregulation of lymphogenesis genes. Hum Mol Genet. 2001;10:2917–2931. doi: 10.1093/hmg/10.25.2917. [DOI] [PubMed] [Google Scholar]
- 30.Danilov AV, Danilova OV, Klein AK, Huber BT. Molecular pathogenesis of chronic lymphocytic leukemia. Curr Mol Med. 2006;6:665–675. doi: 10.2174/156652406778195008. [DOI] [PubMed] [Google Scholar]
- 31.McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 32.Miska EA, et al. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma K, Chan JK, Zhu G, Wu Z. Myocyte enhancer factor 2 acetylation by p300 enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. Mol Cell Biol. 2005;25:3575–3582. doi: 10.1128/MCB.25.9.3575-3582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du Y, Spence SE, Jenkins NA, Copeland NG. Cooperating cancer-gene identification through oncogenic-retrovirus-induced insertional mutagenesis. Blood. 2005;106:2498–2505. doi: 10.1182/blood-2004-12-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verzi MP, et al. The transcription factor MEF2C is required for craniofacial development. Dev Cell. 2007;12:645–652. doi: 10.1016/j.devcel.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong K, Pertz O, Hahn K, Bourne H. Neutrophil polarization: Spatiotemporal dynamics of RhoA activity support a self-organizing mechanism. Proc Natl Acad Sci USA. 2006;103:3639–3644. doi: 10.1073/pnas.0600092103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.