Abstract
The transforming growth factor type β-1 (TGF-β) signaling pathway is a major tumor suppressor during early carcinogenesis, and its growth-suppressive activity is commonly lost during early tumor progression. IκB kinase α (IKKα) also acts as a tumor suppressor in stratified epithelia, and its expression and nuclear localization are progressively down-regulated during malignant progression of squamous cell carcinoma (SCC) and acquisition of an invasive phenotype. A critical role for IKKα in TGF-β signaling in stratified epithelia was identified recently during normal keratinocyte differentiation, and both IKKα and components of the TGF-β signaling pathway are required for induction of antiproliferative Myc antagonists in such cells. We now describe that the interaction between IKKα and the TGF-β signaling pathway is also important in a subset of SCCs. In SCCs that are unable to shuttle IKKα to the nucleus, defective TGF-β-induced growth arrest was rescued by introduction of a constitutively nuclear IKKα variant. These results suggest that the tumor-suppressive activity of IKKα in stratified epithelia may be exerted in part via the TGF-β signaling pathway.
Keywords: squamous cell carcinoma, Myc, skin cancer
The pathogenesis of squamous cell carcinomas (SCCs) involves activating H-Ras mutations as well as activation of c-Myc, loss of p53 function, and expression of mitogenic and inflammatory cytokines (1, 2). In addition, transforming growth factor type β-1 (TGF-β) was detected in both in situ and invasive SCCs at levels that correlate with malignancy (3–5). TGF-β is a pleiotropic molecule acting as a potent growth suppressor on epithelial cells (6). However, experimental evidence suggests that in SCCs TGF-β is required for progression from carcinoma in situ to invasive cancer and for the epithelial to mesenchymal transition that results in the genesis of spindle cell carcinomas and a metastatic phenotype (3–5, 7). TGF-β overproduced by epithelial cells acts on the tumor microenvironment to induce release of inflammatory cytokines, metalloproteinases, and angiogenic factors that contribute to the progression and invasiveness of carcinoma cells, which become resistant to TGF-β-mediated growth arrest (7). The growth-suppressive activity of TGF-β on normal epithelial cells is mediated by the coordinate transcriptional activation of a subset of genes, including inhibitors of cyclin-dependent kinases and the early down-regulation of c-Myc transcription and activity (8, 9).
IκB kinase α (IKKα) is a component of the IKK complex, which is activated by a variety of inflammatory stimuli and has a key role in NF-κB activation (10). IKKα, however, also exhibits a kinase-independent but critical function in the development of skin and other stratified epithelia (11, 12). During keratinocyte differentiation, IKKα accumulates in the nucleus, where it orchestrates cell cycle exit and terminal differentiation (13). This antiproliferative and prodifferentiative effect of IKKα is mediated in part through interaction with TGF-β-regulated SMAD transcription factors (14). Furthermore, TGF-β induces nuclear accumulation of IKKα, and IKKα and SMAD3 coregulate the expression of several antiproliferative Myc antagonists (14). A tumor-suppressive role for IKKα in SCC was recently identified in a mouse model of chemically induced SCC and a small number of human samples (15). More recent evaluation of IKKα expression in SCC revealed its loss in about 30% of such cancers, especially in those that develop a highly invasive phenotype (16). It is not clear, however, how IKKα exerts its tumor-suppressive activity in human SCC, and different possible modes of action were recently discussed (17).
We now confirm that IKKα is down-regulated and loses its nuclear localization in a subset of human SCC derived from stratified epithelia, such as skin, lung, esophagus, and larynx/pharynx. Furthermore, IKKα appears to act as a tumor suppressor by modulating sensitivity to TGF-β-dependent growth arrest through selective regulation of c-Myc antagonists and TGF-β-dependent proinvasive genes.
Results
IKKα Is Down-Regulated and Delocalized in SCC.
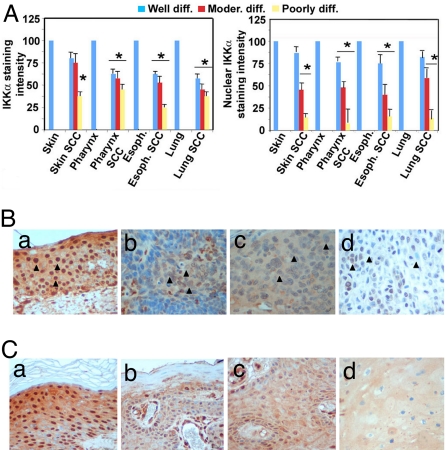
We analyzed IKKα expression and subcellular localization by immunohistochemistry (IHC) in a panel of 245 SCC sections from various tissues and 39 sections of normal noncancerous tissues. IKKα was down-regulated in 78% of skin SCCs and in 82% of SCCs originated from other stratified epithelia, such as lung, esophagus, and oral cavity/larynx (Fig. 1A). The amount of IKKα correlated with clinical stage, being highest in well-differentiated tumors and lowest in high-grade, poorly differentiated, primary SCCs (Fig. 1A Left). Interestingly, although in normal stratified epithelia strong nuclear localization of IKKα was detected primarily in postmitotic cells of the suprabasal layers, in 65% of the examined SCCs IKKα appeared stranded in the cytoplasm with little nuclear staining [Fig. 1B and supporting information (SI) Fig. S1A]. By analyzing skin SCCs together with adjacent normal tissue, we observed that nuclear IKKα was progressively lost at the boundary zone where the hyperplastic/dysplastic epidermis (actinic keratose) and in situ SCC convert into an invasive SCC (Fig. 1B and Fig. S1A), suggesting that IKKα down-regulation and delocalization may be associated with this critical step in tumor progression.
Fig. 1.
IKKα is down-regulated and delocalized in SCC arising from stratified epithelia. (A) Anti-IKKα staining intensity was quantified in 3 microscopic fields for each tissue section analyzed (245 SCCs and 39 controls) by ImageJ software (National Institutes of Health). Mean staining intensity of normal tissue was set as 100%, and the number of samples for each SCC group is indicated. Error bars represent 1 standard deviation (Left). Mean staining intensity of IKKα-positive nuclei was calculated from tissue arrays and SCC samples with adjacent normal skin stained with anti-IKKα using ImageJ software after setting the mean cytoplasmic staining intensity as background. Error bars represent 1 standard deviation (Right). Asterisk (*) represents statistical significance (P < 0,05) with respect to normal tissue as determined by χ2 test. (B) A series of 245 tumors and 39 normal tissue samples was subjected to IHC with IKKα-specific antibody. The panels shown here are representative of IKKα staining in normal skin (a), and poorly differentiated, invasive SCC from skin, esophagus, and lung (b, c, and d, respectively). IKKα nuclear staining is strongly reduced in tumors vs. normal epithelium (compare nuclei indicated by arrowheads in a with those in b, c, and d). (Magnification: 40×.) (C) A biopsy from skin SCC with adjacent normal skin was subjected to anti-IKKα IHC; a–d are different regions of the same section, representative of normal skin (a), hyperplastic skin/actinic keratosis (b), in situ SCC (c), and invasive SCC (d). (Magnification: 40×.)
Down-Regulation of IKKα in Primary Keratinocytes Alters Expression of TGF-β-Regulated Genes.
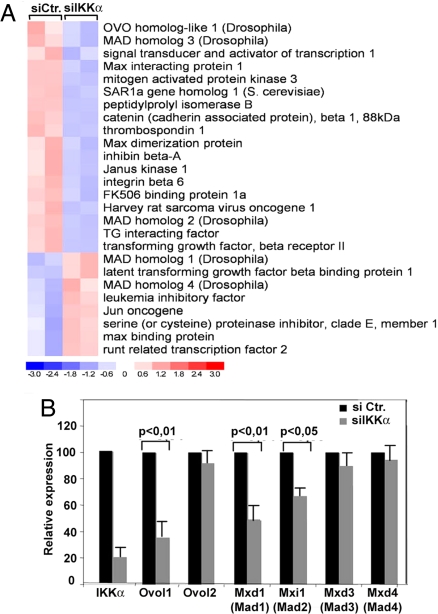
To identify potential mediators of IKKα tumor suppressor activity in stratified epithelia, we performed expression profiling on primary mouse keratinocytes silenced for IKKα by RNAi (Fig. S1B). Knockdown of IKKα in primary mouse keratinocytes significantly (P < 0.05) reduced expression of 981 genes and up-regulated 410 other genes (Fig. 2A). As expected, and in agreement with the role of IKKα in skin development (11, 13, 14, 18), genes down-regulated in IKKα-depleted cells included genes involved in epidermal development, proliferation, and differentiation/keratinization, such as keratins, stratifin, small proline-rich proteins (Sprr2A, Sprr2J), and PERP (Fig. S1C). In addition, we identified a group of genes belonging to or regulated by the TGF-β pathway that are differentially expressed in IKKα knockdown cells relative to controls. As shown in the heatmap and gene list (Fig. 2A), we observed a down-regulation of TGF-βRII, SMAD3, β-catenin, and SMAD-specific E3 ubiquitin ligase 2, and up-regulation of latent TGF-β-binding protein, SMAD4, and c-Jun, suggesting that the TGF-β pathway is altered in IKKα-silenced cells, as previously found in the epidermis of Ikkα−/− mice (14).
Fig. 2.
IKKα down-regulation in primary keratinocytes interferes with expression of TGF-β-regulated genes. (A) Total RNA from control or IKKα-silenced primary keratinocytes was analyzed using Mouse Genome 420 microarrays. Data were analyzed by dChip software (Department of Biostatistics, Dana Farber Institute, Harvard, MA). The heatmap of genes belonging to the TGF-β pathway that were significantly up- or down-regulated in IKKα-silenced cells is shown. (B) Primary mouse keratinocytes were transfected with IKKα-specific or control siRNA, and expression of IKKα, Ovol1, Ovol2, Mad1, Mad2, Mad3, and Mad4 mRNAs was examined 24 h after transfection by quantitative RT-qPCR. The results shown are representative of 3 independent experiments. Statistical significance is indicated. Histogram is representative of 3 experiments performed in triplicate. Error bars represent 1 standard deviation.
TGF-β is a major tumor suppressor during early carcinogenesis because of its prominent antiproliferative activity on epithelial cells, which is exerted mainly by dampening of c-Myc function and expression (18, 19), which also requires p53 family members (20). Interestingly, and as found by analysis of Ikkα−/− epidermis (14), our expression analysis showed marked down-regulation of Ovol1, Max dimerization protein 1 (Mad1, Mxd1), and Mxi1 (Mad2), known as TGF-β-induced inhibitors of c-Myc transcription and activity (21–23), but not of Ovol2, Mad3, and Mad4, suggesting that the hyperproliferation observed in Ikkα-deficient keratinocytes (Fig. 3 and ref. 13) and in SCCs may at least in part be due to loss of TGF-β-mediated restraint of c-Myc activity and expression.
Fig. 3.
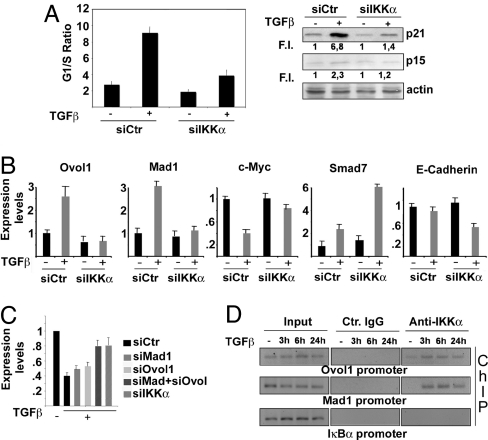
IKKα is part of the TGF-β-activated antiproliferative pathway in human keratinocytes. (A) HaCaT cells were transfected with either control or IKKα-specific siRNA. The transfected cells were treated with TGF-β for 24 h or left untreated and stained with propidium iodide to determine cell cycle profile. Results from 3 independent experiments performed in duplicate are expressed as G1/S ratio. Parallel samples were used to detect p21 and p15-INKB expression in total cell lysates. Numbers under each lane indicate fold-induction relative to controls. Band intensities were quantified by Kodak image software. (B) HaCaT cells were transfected with either control or IKKα siRNAs and treated with TGF-β as described above. RT-qPCR was performed using primers specific to the indicated genes. Results are mean expression values of 3 different experiments performed in triplicate. Error bars are standard deviations. (C) HaCaT cells were transfected with either control siRNA or siRNAs specific for Mad1, Ovol1, Mad1 plus Ovol1, or IKKα, and the ability of TGF-β to down-regulate c-Myc was evaluated by RT-qPCR. Results are the mean expression values of 3 different experiments performed in triplicate. Error bars indicate standard deviation. Expression values in untreated cells were set as 1. (D) HaCaT cells were treated with TGF-β for the indicated times. IKKα-containing protein–DNA complexes were immunoprecipitated with IKKα-specific antibody. PCR was performed on the recovered DNA using specific primers for a region spanning positions −1337 to −1170 of the Mad1 gene and −747 to −555 of the Ovol1 gene. Control IgG and PCR on IkBα promoter served as negative controls. PCR primers were designed based on location of potential TGF-β-responsive elements (SMAD-binding sites) identified by MatInspectorSoftware (Genomatix GmbH).
Expression of both Ovol1 and Mad1 mRNAs, and to a lesser extent Mad2, was inhibited in IKKα -ilenced cells (Fig. 2B), thus confirming the gene profiling results. Mad1 and Ovol1 are up-regulated in nuclei of suprabasal keratinocytes, and their expression correlates spatially and temporally with exit of keratinocytes from the cell cycle and expression of differentiation markers (22, 24). Mad1 and IKKα had similar expression patterns in human skin because they were both expressed in the nuclei of suprabasal layers (Fig. S2A) and were progressively lost in invasive SCCs (Fig. 1 Ca–Cd and Fig. S2 Ab and Ac). In well-differentiated SCCs that still express nuclear IKKα, we detected an accented nuclear staining for Mad1 in the differentiated portions of the tumor (Fig. S2 Ad and Ae), supporting the contention that Mad1 may be a relevant antiproliferative target of IKKα also in human epidermis, in addition to the previously established relationship between the two in mouse skin (14).
IKKα Regulates TGF-β Antiproliferative Activity.
The identification of a gene set belonging to the TGF-β signaling pathway in IKKα-silenced cells suggests that down-regulation of nuclear IKKα in SCC may cause unresponsiveness to TGF-β-induced growth-inhibitory signals. We thus examined the impact of RNAi-mediated IKKα knockdown (Fig. S3A) on TGF-β-induced growth arrest of TGF-β1-sensitive human keratinocyte HaCaT cells. TGF-β induced cell cycle arrest in control cells but had a weaker antiproliferative effect in IKKα-down-regulated cells (Fig. 3A).
The failure of IKKα-down-regulated cells to undergo a complete cell cycle arrest correlated with reduced expression of the cyclin-dependent kinase inhibitors p21 and p15INK4B (Fig. 3A), and of Mad1 and Ovol1, as well as with failure to down-regulate c-Myc (Fig. 3B). IKKα depletion enhanced expression of SMAD7 (Fig. 3B), a potent inhibitor of TGF-β signaling, whose elevated expression was detected in aggressive SCCs (25), and resulted in inhibition of E-cadherin, which correlates with tumor invasiveness (Fig. 3B).
To determine whether Ovol1 and Mad1 are involved in c-Myc mRNA down-regulation in response to TGF-β, we selectively inhibited their expression in HaCaT cells by RNAi (Fig. S3B). Whereas TGF-β-mediated down-regulation of c-Myc mRNA was only partially inhibited in HaCaT cells silenced for either Ovol1 or Mad1, silencing of both genes almost completely prevented TGF-β-induced c-Myc down-regulation, an effect comparable to that observed in IKKα-silenced cells (Fig. 3C). These results suggest that Mad1 and Ovol1 act downstream of IKKα to restrain c-Myc expression and function. To assess the direct involvement of IKKα in activation of these genes upon TGF-β treatment, we conducted ChIP experiments. IKKα was recruited to the Ovol1 and Mad1 promoters in TGF-β-stimulated HaCaT cells with similar kinetics, suggesting that IKKα may act as a direct transcriptional coactivator (Fig. 3D). IKKα binding to the Mad1 and Ovol1 promoters correlates with induction of transcription from a Mad1-luc reporter that was potentiated by exogenously expressed IKKα and required the presence of a SMAD-binding site (Fig. S2B and ref. 14).
Rescue of TGF-β Sensitivity in SCC Cell Lines by Nuclear IKKα.
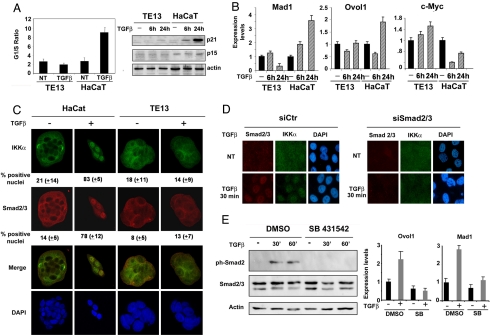
To substantiate the role of IKKα in SCC susceptibility to TGF-β-mediated cell growth arrest, we analyzed the TGF-β sensitivity of TE13 cells, an SCC cell line derived from an esophageal cancer (26). In contrast with HaCaT cells, the TE13 cell cycle profile was only marginally altered by TGF-β, correlating with reduced p21 and p15 expression (Fig. 4A). TE13 cells also failed to up-regulate Ovol1 or Mad1 transcription and down-regulate c-Myc in response to TGF-β (Fig. 4B). Similar results were obtained with 2 other SCC cell lines (Fig. S4 A and B).
Fig. 4.
Impaired TGF-β response correlates with defective nuclear accumulation of IKKα and Ovol1/Mad1 activation in SCC cells. (A) Esophageal SCC TE13 and HaCaT cells were exposed to TGF-β for 24 h or left untreated. Cells then were stained with propidium iodide, and their DNA content was analyzed by flow cytometry to determine cell cycle distribution. The histogram (Left) shows the G1/S ratio and is representative of 3 independent experiments. Lysates from sister cultures were analyzed by immunoblotting for p21 and p15-INKB expression (Right); actin was used as loading control. (B) TE13 and HaCaT cells were treated with TGF-β as described above or left untreated. Total RNA was analyzed by RT-qPCR to determine expression of indicated genes. (C) Control and TGF-β-treated TE13 and HaCaT cells cultured on coverslides were fixed, and subcellular distribution of IKKα, Smad2/3, and Smad4 was examined by indirect immunofluorescence. The percentage of cells displaying a strong nuclear signal was determined by counting at least 200 cells per field in 3 separate experiments done in duplicate (Magnification 40×). (D) HaCaT cells were transfected with scrambled siRNA (siCtr) or with a mixture of SMAD2 and SMAD3 siRNAs (siSMAD2/3). After 24 h, the cells were treated with TGF-β, and localization of SMAD2/3 and IKKα was determined as above (Magnification 20×). (E) HaCaT cells were left untreated or were treated with TGF-β for the indicated times in the presence or absence of 10 μM SB 431542 (TGF-βRI/II kinase inhibitor). Cells were lysed, and expression of phospho-SMAD2, SMAD2/3, and actin was evaluated by immunoblotting. In a parallel experiment, cells were treated with SB 431542 in the presence or absence of TGF-β for 24 h, and total RNA was extracted and subjected to real-time qPCR to detect expression of Ovol1 and Mad1. Histograms are representative of 2 independent experiments performed in triplicate. Error bars indicate standard deviation.
We next examined the potential role of IKKα nuclear accumulation in TGF-β responsiveness. TGF-β induced nuclear accumulation of IKKα along with SMAD2/3 in HaCaT cells, but was unable to do so in TE13 cells (Fig. 4C), which also show defective activating receptor-regulated (R) SMAD (SMAD2/3) phosphorylation upon TGF-β stimulation (Fig. S3C). This is in line with recent evidence for direct physical interaction between R-SMAD and IKKα (14) and suggests that the lack of TGF-β responsiveness may be linked to the prevalent cytoplasmic localization of R-SMAD and IKKα in these cells. SMAD4 nuclear localization remained intact in these cells, indicating that it is insufficient for TGF-β-induced growth arrest (Fig. S3D). Previously, IKKα was found to participate in a SMAD4-independent TGF-β signaling, and SMAD4 was shown to have no role in keratinocyte differentiation (14).
To determine the role of SMAD2/3 in IKKα translocation and activation of antiproliferative genes, we inhibited R-SMAD function either by siRNAs or by a specific inhibitor of R-SMAD phosphorylation. We observed that IKKα translocation following TGF-β treatment is dependent on expression of SMADs 2 and 3, because their selective down-regulation (Fig. S3E) also inhibited IKKα translocation (Fig. 4D). Treatment of HaCaT cells with TGF-βRI/II kinase activity inhibitor, SB431542, blocked SMAD2 phosphorylation (Fig. 4E Left); the transcription of IKKα-dependent, antiproliferative genes Mad1 and Ovol1 (Fig. 4E Right); and IKKα nuclear translocation (Fig. S3F), indicating that R-SMAD phosphorylation is required for IKKα to be translocated to the nucleus and for transcriptional regulation of Myc inhibitors.
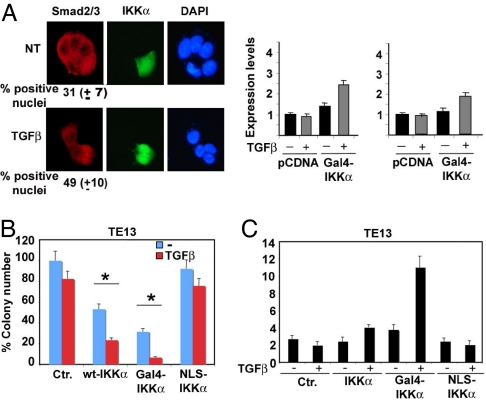
We tried to restore TGF-β sensitivity by reconstituting nuclear IKKα in TE13 cells. To this end, a Gal4-IKKα chimera was generated by fusing IKKα to the Gal4 DNA-binding domain, which contains a constitutive nuclear localization signal (NLS). As expected, Gal4-IKKα was strongly expressed in the nuclei of TE13 cells, whereas WT-IKKα was both cytoplasmic and nuclear, and an IKKα construct bearing point mutations in its endogenous NLS (NLS-IKKα) was only cytoplasmic (Fig. S5A). Gal4-IKKα expression led to constitutive SMAD2/3 nuclear localization that was enhanced modestly by TGF-β stimulation (Fig. 5A) and resulted in reinduction of Mad1 and Ovol1 transcription (Fig. 5A).
Fig. 5.
TGF-β-induced growth arrest in SCC cells is restored by forced nuclear accumulation of IKKα. (A) TE13 cells were transiently transfected with the Gal4-IKKα construct and incubated or not with TGF-β. Cells were fixed and examined by immunofluorescence for distribution of Gal4-IKKα and Smad2/3 (Left) (Magnification 40×). In parallel experiments, transfected cells were lysed and analyzed by real-time PCR for expression of Mad1 and Ovol1 mRNAs (Right). (B) TE13 cells were transfected with WT-IKKα, Gal4-IKKα, and NLS-IKKα expression vectors and selected in G418 in the presence or absence of TGF-β. After staining with crystal violet, colonies >1 mm in diameter were counted using ImageJ software. The histogram represents mean colony numbers + SD in 3 independent experiments performed in duplicate. Asterisk (*) represents statistical significance (P < 0.05) with respect to control as determined by χ2 test. (C) Polyclonal cell lines expressing WT-IKKα, Gal4-IKKα, and NLS-IKKα obtained after 1 week of G418 selection were left untreated or treated with TGF-β for 24 h, stained with propidium iodide, and cell cycle distribution was determined. The histograms represent G1/S ratio of 3 experiments performed in duplicate. Error bars represent 1 standard deviation.
Colony formation assays revealed that only Gal4-IKKα restored full TGF-β sensitivity, whereas overexpression of WT-IKKα provided a partial, although statistically significant, response to TGF-β, and NLS-IKKα overexpression had no effect (Fig. 5B). It should be noted, however, that Gal4-IKKα inhibited clonogenic growth even in the absence of TGF-β. Cell cycle analysis produced similar results (Fig. 5C). Similar results were obtained in TE1 cells (Fig. S6A). Together, these experiments strongly suggest that nuclear IKKα is a crucial component of the TGF-β antiproliferative signaling network in epithelial cells.
The Tumor Suppressor Activity of IKKα Is Dependent on Its Nuclear Localization and Correlates with Myc Inhibition.
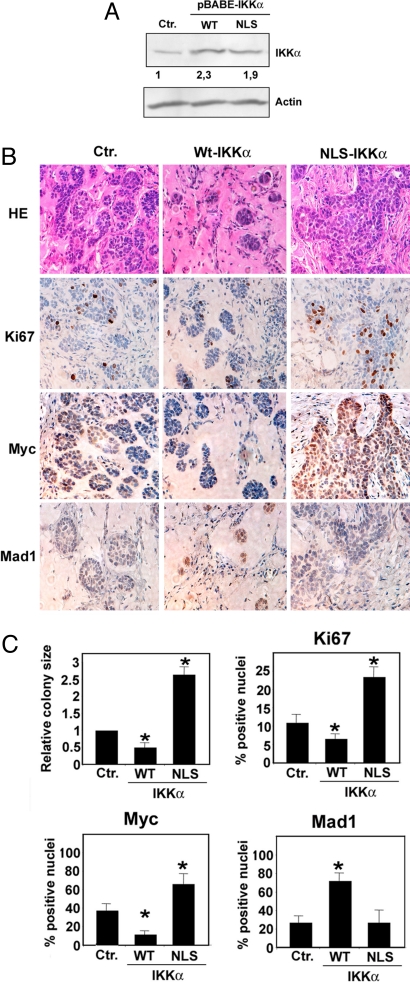
To confirm the role of the nuclear component of IKKα as a tumor suppressor in vivo, we infected TE13 cells with retroviruses expressing WT-IKKα, NLS-IKKα, or control retroviruses, generated stable cell lines (Fig. 6A), and observed their growth after s.c. transplantation into SCID mice. To facilitate tumorigenic growth, we injected TE13 cells together with Matrigel, a solubilized basement membrane preparation. Macroscopic examination of tumors 15 days after cell transplantation showed significant reduction in the size of tumors produced by WT-IKKα-transduced cells and increased vascularization of tumors formed by NLS-IKKα-transduced cultures (data not shown). Histological examination revealed the presence of multiple tumor cell colonies dispersed in Matrigel, which was vascularized and infiltrated by inflammatory cells (Fig. 6B). Introduction of WT-IKKα into TE13 cells resulted in tumor growth inhibition, as demonstrated by the reduced number and size of Matrigel tumor cell colonies, and this diminished the proliferation rate in comparison with control TE13 cells (Fig. 6 B and C). TE13 cells expressing WT-IKKα also displayed lower Myc expression and increased Mad1 expression (Fig. 6 B and C). Transduction of TE13 cells with the NLS-IKKα mutant had the opposite effect, as demonstrated by the increased size of tumor cell colonies, increased Ki-67 and Myc expression, and decreased Mad1 expression. These findings further support our hypothesis that IKKα needs a functional NLS to exert its tumor suppressor activity.
Fig. 6.
IKKα tumor suppressor activity in a mouse xenograft model correlates with Mad1 induction and Myc repression. (A) TE13 cells were infected with either empty (Ctr.) or WT-IKKα-expressing or NLS-IKKα-expressing retroviruses. After puromycine selection, polyclonal cell lines were lysed, and ectopic and endogenous IKKα expression was evaluated by immunoblotting. Relative IKKα expression levels were quantified using the ImageJ program. (B) Polyclonal cell lines (106 cells) were injected s.c. into SCID mice in the presence of 10 mg/ml Matrigel. At day 15, mice were killed and s.c. tumors were subjected to hematoxylin–eosin staining or IHC with specific antibodies to detect expression of Ki-67, Myc, and Mad1. Five mice were injected with each cell line. Representative fields are shown. (Magnification: 40×.) (C) Size of tumor cell colonies grown in Matrigel Matrix was evaluated by Image J software in 3 fields for each tumor. Histogram represents colony size relative to that of mice injected with Ctr.-infected tumor cells. For IHC quantization, positively stained cells were counted in 3 microscopic fields. Histograms indicate percentage of positive nuclei. Error bars represent 1 standard deviation. Asterisk (*) represents statistical significance (P < 0.05) with respect to control as determined by χ2 test.
Introduction of constitutively nuclear IKKα (Gal4-IKKα) led to the growth of tumor colonies negative for Gal4-IKKα, suggesting either that this molecule is toxic in vivo or that its strong tumor-suppressive activity only allows the clonal outgrowth of cells that lost Gal4-IKKα expression. Similar results were obtained when TE1 cells were used as the recipients for the different IKKα constructs (data not shown).
Discussion
TGF-β exerts dual effects in on a variety of biological events, such as cell proliferation, differentiation, angiogenesis, and inflammatory/immune responses (7, 26). For instance, TGF-β promotes proliferation of mesenchymal cells but acts as a potent inhibitor of epithelial, hematopoietic, and neural cell proliferation (6, 18, 19). Two major transcriptional responses are involved in TGF-β-mediated growth arrest—genes encoding CDK inhibitors and genes whose products restrain c-Myc expression and activity (7, 18). Mechanistically, these growth-inhibitory functions of TGF-β involve nuclear translocation of R-SMAD (19) and formation of specific transcriptionally active multiprotein complexes on target gene promoters. In human skin cancers, TGF-β is overexpressed throughout the tumor, including in basal cells (3). When overexpressed in cycling keratinocytes, TGF-β induces severe inflammation that is sufficient to override its growth-inhibitory effect, promote hyperproliferation, and thus favor the accumulation of UV-induced mutations and cell transformation (3). At later stages, TGF-β promotes epithelial–mesenchymal transition and progression to an invasive phenotype that culminates in metastasis (3–5).
Here, we have exploited the marked reduction in IKKα expression and failure to shuttle to the nucleus exhibited in poorly differentiated human SCCs, combined with gene profiling, to unveil an explanation for the previously documented tumor-suppressive function of IKKα (15, 16). We provide evidence for a role for IKKα in regulation of TGF-β-activated cytostatic responses in keratinocytes, and have translated these observations to human tumor cell models to explain how IKKα acts as a tumor suppressor in SCC derived from stratified epithelia.
Down-regulation of IKKα expression in normal keratinocytes interferes with induction of several TGF-β-regulated genes whose products inhibit CDKs as well as c-Myc. Specifically, IKKα is required for expression of 2 negative regulators of c-Myc transcription and activity, Mad1 and Ovol1, both of which regulate cell cycle exit keratinocyte differentiation (22, 27). Mad1 is a direct binding partner of the Max protein, thereby disrupting Myc:Max oligomers and contributing to down-regulation of c-Myc transcriptional activity, whereas Ovol1 represses c-Myc expression through association with its promoter (22, 28). As suggested by ChIP experiments, IKKα may serve as a direct coactivator of Ovol1 and Mad1 transcription as it is recruited to their promoters upon TGF-β stimulation.
The results obtained in normal keratinocytes are supported by observations made in cancerous counterparts. Indeed, IKKα down-regulation in SCCs from skin, esophagus, oral cavity, and lung is accompanied by progressive reduction in its nuclear localization and reduced expression of its target Mad1. This is most evident in the area where the tumor becomes invasive, suggesting that IKKα down-regulation may be involved in the invasion step of tumor progression. This conclusion conforms to the finding that IKKα down-regulation occurs after loss of p53 function by mutations in a model of chemical skin carcinogenesis in mice (15).
We suggest that IKKα is actively shuttled into nuclei of keratinocytes undergoing terminal differentiation or following TGF-β stimulation (Fig. 4C and Fig. S6B), to activate an antiproliferative program. IKKα nuclear translocation is dependent on TGF-β-induced SMAD2/3 activation, because inhibition of SMAD2/3 expression by siRNA or phoshorylation using TGF-βRI/II kinase inhibitors also inhibits IKKα accumulation in the nucleus. The SCC cell lines from esophageal cancer examined herein fail to mount this response and show very low levels of TGF-β-induced SMAD2 phosphorylation. In these cells, Mad1 and Ovol1 are not efficiently up-regulated by TGF-β, and c-Myc transcription is only partially repressed in TE1 cells and increased in TE13 cells after TGF-β stimulation. This correlates with the inability of TGF-β to induce IKKα nuclear translocation in these cells. Forced expression of nuclear IKKα restores the ability of these cells to undergo cell cycle arrest upon TGF-β stimulation, suggesting that the suppressive activity of nuclear IKKα is mediated in part through activation of the TGF-β antiproliferative response. This notion is supported by the strong tumor-suppressive activity of IKKα in the mouse xenograft model, which correlates with decreased Myc and increased Mad1 expression (Fig. 6C). Based on these observations, we speculate that activation of oncogenic signaling pathways responsible for progression to an invasive tumor stage may cause IKKα down-regulation and cytoplasmic sequestration. This may occur through interference with TGF-β signaling. In this context, it is noteworthy that besides resulting in deregulated keratinocyte proliferation through interference with Myc down-regulation, IKKα down-modulation entails up-regulation of the TGF-β pathway inhibitor SMAD7 and reduced E-cadherin transcription, as well as defective keratinocyte differentiation. These coordinate events may render the malignant cell progressively less responsive to the antiproliferative activity of TGF-β, and may promote its invasive phenotype.
Materials and Methods
Tumor Samples and Immunohistochemistry.
Paraffin sections were purchased from SuperBioChip (cod. CX1, CR1, CH2, VA2, VB2, and VC2), whereas normal skin and SCC sections were taken from healthy volunteers and from patients with SCC referring to our Dermatology Clinic. Sections were stained with an avidin-biotin-peroxidase technique by using 3-amino-9-ethylcarbazole as a substrate (DAKO). Details about the IHC and semiquantitative digital image analysis to determine the intensity of IKKα expression are provided in SI Materials and Methods.
Cell Culture.
Cell lines used in this study were: HaCaT cells, TE13 (p53 null), TE1 (harboring a temperature-sensitive mutated p53; gift from Pierre Hainaut, IARC, Lyon, France), and SCC25 cells (harboring a mutated p53 allele). Cells were grown using standard culture techniques as described previously (14, 29). Unless otherwise indicated, treatment of cells with TGF-β1 (Calbiochem) was at 5 ng/ml. Details of cell culture and clonogenic assays are provided in SI Materials and Methods.
GeneChip and Gene Expression Analysis.
A total of 10 μg TRIzol-extracted total RNA from siCtr- or siIKKα-transfected cells was purified by RNeasy MicroKit (Qiagen) and reverse transcribed using a T7-Oligo(dT) promoter primer and MuLV reverse transcriptase (Applied Biosystems). Microarray profiling was conducted as described previously (30). Details of statistical comparison of gene expression are provided in SI Materials and Methods.
TE13 SCC Xenograft Model in SCID Mice.
All animal procedures were approved by the Institutional Animal Care and Use Committee at The University of Rome “Tor Vergata.” Standard rodent sterile techniques were used. Beige SCID mice 6 to 10 weeks of age from Charles River Laboratories were injected s.c. on both flanks with 106 TE13 cells expressing WT-IKKα, NLS-IKKα, or Gal4-IKKα dispersed in 0.4 ml of 10 mg/ml Matrigel (BD Biosciences). At 15 days after injection, mice were killed, and tumors grown in Matrigel were formalin-fixed, paraffin-embedded, and processed as for human tissues.
Supplementary Material
Acknowledgments.
We thank B. Amati, A. Rossi, L. Guerrini, and G. Natoli for critically reading the manuscript and for helpful suggestions. This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (to A.C. and S.A.); Fondo Italiano Ricerca di Base (FiRB)-IDEAS Call, Ministero dell'Istruzione, dell'Università e della Ricerca (PRIN 2005), and the European Commission (EC-FP6 “Active p53 Consortium”) (to A.C.); and National Institutes of Health Grant AI043477 (to M.K.). M.K. is an American Cancer Society Research Professor. B.M. and P.D. were supported by Italian Foundation for Cancer Research (FIRC) and a Human Frontier Science Program fellowship, respectively.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809288105/DCSupplemental.
References
- 1.Akhurst RJ, Balmain A. Genetic events and the role of TGFβ in epithelial tumor progression. J Pathol. 1999;187:82–90. doi: 10.1002/(SICI)1096-9896(199901)187:1<82::AID-PATH248>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 2.Mangan PR, et al. Transforming growth factor-β induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 3.Han G, et al. Distinct mechanisms of TGFβ1-mediated epithelial-to-mesenchymal transition and metastasis during skin carcinogenesis. J Clin Invest. 2005;115:1714–1723. doi: 10.1172/JCI24399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oft M, Heider KH, Beug H. TGFβ signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol. 1998;8:1243–1252. doi: 10.1016/s0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- 5.Oft M, et al. TGFβ1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 1996;10:2462–2477. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 6.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGFβ in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 7.Li AG, Lu SL, Han G, Hoot KE, Wang XJ. Role of TGFβ in skin inflammation and carcinogenesis. Mol Carcinog. 2006;45:389–396. doi: 10.1002/mc.20229. [DOI] [PubMed] [Google Scholar]
- 8.Gomis RR, et al. A FoxO-Smad synexpression group in human keratinocytes. Proc Natl Acad Sci USA. 2006;103:12747–12752. doi: 10.1073/pnas.0605333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomis RR, et al. C/EBPb at the core of the TGFβ cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 10.Häcker H, Karin M. Regulation and function of IKK and IKK related kinases. Sci STKE. 2006;357:re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- 11.Hu Y, et al. IKKα controls formation of the epidermis independently of NF-kappaB. Nature. 2001;410:710–714. doi: 10.1038/35070605. [DOI] [PubMed] [Google Scholar]
- 12.Hu Y, et al. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKα subunit of IkappaB kinase. Science. 1999;284:316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- 13.Sil AK, Maeda S, Sano Y, Roop DR, Karin M. IkappaB kinase-α acts in the epidermis to control skeletal and craniofacial morphogenesis. Nature. 2004;428:660–664. doi: 10.1038/nature02421. [DOI] [PubMed] [Google Scholar]
- 14.Descargues P, et al. IKKα is a critical coregulator of a Smad4-independent TGFβ-Smad2/3 signaling pathway that controls keratinocyte differentiation. Proc Natl Acad Sci USA. 2008;105:2487–2492. doi: 10.1073/pnas.0712044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B, et al. A critical role for I kappaB kinase a in the development of human and mouse squamous cell carcinomas. Proc Natl Acad Sci USA. 2006;103:17202–17207. doi: 10.1073/pnas.0604481103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda G, Chiba T, Kawashiri S, Satoh T, Imai K. Epigenetic inactivation of IkappaB Kinase-α in oral carcinomas and tumor progression. Clin Cancer Res. 2007;13:5041–5047. doi: 10.1158/1078-0432.CCR-07-0463. [DOI] [PubMed] [Google Scholar]
- 17.Van Waes C, Yu M, Nottingham L, Karin M. Inhibitor-kappaB kinase in tumor promotion and suppression during progression of squamous cell carcinoma. Clin Cancer Res. 2007;13:4956–4959. doi: 10.1158/1078-0432.CCR-07-1287. [DOI] [PubMed] [Google Scholar]
- 18.Massague J. How cells read TGF-β signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 19.Massague J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 20.Cordenonsi M, et al. Links between tumor suppressors: p53 is required for TGF-β gene responses by cooperating with Smads. Cell. 2003;113:301–314. doi: 10.1016/s0092-8674(03)00308-8. [DOI] [PubMed] [Google Scholar]
- 21.Dai X, et al. The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev. 1998;12:3452–3463. doi: 10.1101/gad.12.21.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nair M, et al. Ovol1 regulates the growth arrest of embryonic epidermal progenitor cells and represses c-myc transcription. J Cell Biol. 2006;173:253–264. doi: 10.1083/jcb.200508196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satterwhite DJ, White RL, Aakre ME, Moses HL. TGF-β1 regulates the expression of multiple max-interacting transcription factors in Balb/MK cells: Implications for understanding the mechanism of action of TGF-beta1. Pediatr Res. 2001;50:67–75. doi: 10.1203/00006450-200107000-00014. [DOI] [PubMed] [Google Scholar]
- 24.Hurlin PJ, et al. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1996;15:2030–2041. [PMC free article] [PubMed] [Google Scholar]
- 25.Osawa H, Nakajima M, Kato H, Fukuchi M, Kuwano H. Prognostic value of the expression of Smad6 and Smad7, as inhibitory Smads of the TGF-β superfamily, in esophageal squamous cell carcinoma. Anticancer Res. 2004;24:3703–3709. [PubMed] [Google Scholar]
- 26.Feng XH, Derynck R. Specificity and versatility in TGFβ signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 27.Werner S, Beer HD, Mauch C, Luscher B, Werner S. The Mad1 transcription factor is a novel target of activin and TGF-β action in keratinocytes: Possible role of Mad1 in wound repair and psoriasis. Oncogene. 2001;20:7494–7504. doi: 10.1038/sj.onc.1204937. [DOI] [PubMed] [Google Scholar]
- 28.Rottmann S, Luscher B. The Mad side of the Max network: Antagonizing the function of Myc and more. Curr Top Microbiol Immunol. 2006;302:63–122. doi: 10.1007/3-540-32952-8_4. [DOI] [PubMed] [Google Scholar]
- 29.Barnas C, et al. Inactivation of the p53 protein in cell lines derived from human esophageal cancers. Int J Cancer. 1997;71:79–87. doi: 10.1002/(sici)1097-0215(19970328)71:1<79::aid-ijc14>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 30.Koster MI, et al. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci USA. 2007;104:3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.