Abstract
Disrupted ERK1/2 (MAPK3/MAPK1) MAPK signaling has been associated with several developmental syndromes in humans; however, mutations in ERK1 or ERK2 have not been described. We demonstrate haplo-insufficient ERK2 expression in patients with a novel ≈1 Mb micro-deletion in distal 22q11.2, a region that includes ERK2. These patients exhibit conotruncal and craniofacial anomalies that arise from perturbation of neural crest development and exhibit defects comparable to the DiGeorge syndrome spectrum. Remarkably, these defects are replicated in mice by conditional inactivation of ERK2 in the developing neural crest. Inactivation of upstream elements of the ERK cascade (B-Raf and C-Raf, MEK1 and MEK2) or a downstream effector, the transcription factor serum response factor resulted in analogous developmental defects. Our findings demonstrate that mammalian neural crest development is critically dependent on a RAF/MEK/ERK/serum response factor signaling pathway and suggest that the craniofacial and cardiac outflow tract defects observed in patients with a distal 22q11.2 micro-deletion are explained by deficiencies in neural crest autonomous ERK2 signaling.
Keywords: 22q11 microdeletion, human syndromes, MAP kinase
The ERK1/2 intracellular signal transduction pathway represents one of the principal signaling cascades mediating the transmission of signals from cell surface receptors to cytoplasmic and nuclear effectors (1). ERK1/2 signaling is stimulated in response to various extracellular factors, often through the activation of receptor tyrosine kinases. The active receptors catalyze the assembly of complexes of multiple adapter proteins linking the receptors to a 3-tiered protein kinase cascade comprised of Raf kinases (A-Raf, B-Raf, C-Raf), MAP kinase kinases (MEK1/MEK2), and MAP kinases (ERK1/ERK2). Important effectors of the ERK1/2 cascade are transcription factors, including serum response factor (SRF), which directs the expression of a broad range of genes through interactions with the ternary complex factor family and myocardin-related transcription factor (MTRF) families of transcription factors (2, 3).
A number of clinical syndromes result from mutations within genes encoding proteins that are involved in the regulation of ERK1/2 signaling (4). These include Noonan (Shp2/Ptpn11, K-Ras, C-Raf, Sos1), LEOPARD (Shp2, K-Ras, C-Raf), Costello (H-Ras), and Cardio-facio-cutaneous (B-Raf, H-Ras, MEK1, MEK2) syndromes, which have been collectively termed neuro-cardio-facial-cutaneous syndromes (NCFCS) (5). There is also evidence from mouse models that suggests that ERK1/2 signaling may be disrupted in DiGeorge syndrome (DGS)/velocardiofacial syndrome (VCFS), the most common micro-deletion syndrome in humans (one in 4,000 live births) (6). The majority of patients with DGS/VCFS possess hemizygous deletions of proximal chromosome 22q11.2, most often encompassing a 3-Mb region that includes ≈35 genes (7, 8). Importantly, some of these genes (TBX1 and CRKL) may act within a common genetic pathway that regulates ERK1/2 signaling (9).
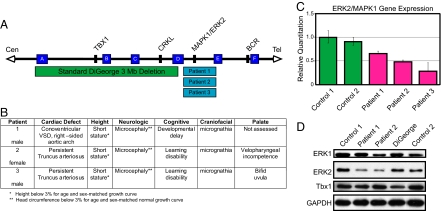
The ERK2 (MAPK1) gene also localizes to chromosome 22q11, but is positioned distal to and outside the 3-Mb DGS/VCFS region. We have identified patients with de novo micro-deletions in distal 22q11.2 (10) by FISH, and cloned the breakpoints of the deletion interval in patient 1 (11). The distal deletions include ERK2 as well as several other genes and transcripts, many of which are related to the Ig λ light chain (IGLL) gene cluster (Fig. 1A). Within this set of distal 22q11.2 genes, ERK2 represents an important candidate for the etiology of the cardiac and craniofacial defects found in these patients.
Fig. 1.
ERK2/MAPK1 protein and mRNA levels are decreased in patients with distal 22q11.2 deletions. (A) Deletions of chromosome 22q11.2 and their endpoints. Chromosome-specific low copy repeats are designated A through F (not to scale). The distal deletions seen in the patients in this study occur in the 1-Mb interval between low copy repeats (D and E). Relative locations of relevant genes are given. (B) Clinical findings in patients with distal micro-deletions of chromosome 22q11.2. (C) Gene expression plot from TaqMan assay of ERK2/MAPK1 is shown compared with 18s rRNA endogenous control. Error bars represent a composite of 2 separate experiments run in triplicate. (D) Western blotting of lymphoblastoid samples revealed decreased ERK2, but not TBX1, in 2 different patients with distal 22q11.2 micro-deletions compared with normal controls. Samples from a patient with DGS show decreased TBX1, but no change in ERK2.
Patients with either typical or distal 22q11.2 deletions exhibit defects in craniofacial and cardiac structures thought to be derived from neural crest (10, 12, 13), a pluripotent cell population that gives rise to and influences the development of a diverse array of tissues in the developing embryo. These include numerous craniofacial structures, the cardiac outflow tract, and endocrine glands (14). During development, neural crest cells are highly plastic and are known to be dependent on cues from the extracellular environment, such as sonic hedgehog, Wnts, bone morphogenetic proteins, and FGFs (14). Nonetheless, our understanding of the signal transduction pathways critical for neural crest development is incomplete. In particular, the effects of conditionally eliminating ERK1/2 signaling in developing neural crest has not previously been investigated.
Here we show that patients with small ≈1 Mb, distal 22q11.2 micro-deletions exhibit haplo-insufficient ERK2 expression. To explore the cell-specific role of ERK2 in these phenotypes, we conditionally targeted components of the RAF/MEK/ERK pathway in mice using floxed alleles and a Cre driver line that mediates robust recombination in neural crest. We have identified ERK2 isoform–specific effects on mouse cardiac and craniofacial development that mimic defects observed in the haplo-insufficient patients, indicating a likely role as the key gene mediating the phenotype. Disruption of both ERK1 and ERK2 results in more severe cardiac and craniofacial abnormalities, as well as thymic and thyroid aplasia/hypoplasia. These effects are pheno-copied by the conditional inactivation of the upstream ERK1/2 regulators, MEK1/MEK2 and B-Raf/C-Raf, and remarkably by conditional inactivation of a single downstream transcription factor, SRF. Our results show that a Raf/MEK/ERK/SRF signaling pathway is necessary for mammalian neural crest development. These data also demonstrate a neural crest autonomous role for ERK2 signaling in developmental abnormalities associated with distal 22q11.2 micro-deletions.
Results
Patients with neural crest-related defects show haplo-insufficiency for MAPK1/ERK2.
We analyzed ERK2 expression in 3 patients with heterozygous 22q11.2 micro-deletions mediated by low copy repeats distal to the DGS/VCFS region. Although patients with larger deletions and similar phenotypes have been described, this cohort represents deletions of the smallest region of overlap exhibiting the full phenotype (10, 11, 13, 15) (Fig. 1A). The patients exhibit defects within the DGS/VCFS spectrum including conotruncal cardiac anomalies and craniofacial abnormalities, such as mandibular hypoplasia, bifid uvula, velopharyngeal incompetence, hypertelorism, smooth philtrum, and ear anomalies (Fig. 1B). The conotruncal cardiac defects in these patients are comparable to those seen in DGS/VCFS. Notably, the anatomic malformations in our patients are observed in neural crest-influenced or -derived structures.
The ERK2 gene is located within the distal 22q11.2 region and is a strong candidate to underlie the etiology of the malformations seen in these patients. Quantitative PCR and Western analysis of peripheral blood and lymphoblastoid cell lines derived from patients with distal 22q11.2 deletions demonstrate a significant reduction of ERK2 mRNA and protein (Fig. 1 C and D), consistent with ERK2 haplo-insufficiency in the patients. Importantly, the protein level of TBX1, a critical gene located within the 3-Mb DGS interval, was normal (Fig. 1D; n = 2 distal 22q11 patients; n = 4 normal controls). This suggests that the small deletion encompassing ERK2 is not producing a cis-acting position effect on the DGS region (Fig. 1D). Further, patients with a typical 3-Mb DGS 22q11.2 deletion and the same cardiac defect did not show decreased ERK2 protein levels, but did show decreased TBX1 expression consistent with its hemizygosity in the common DGS deletion. Together, these findings suggest that the related phenotypes result from the deletion of distinct regions containing unique sets of genes. It is possible the closely related ERK1 (MAPK3) isoform could contribute to these phenotypes; however, no consistent change was noted in the protein levels of ERK1 (MAPK3; Fig. 1D). Further, quantitative PCR did not reveal any changes in ERK1 mRNA levels between distal 22q11-deleted cases and controls (data not shown). This is consistent with the absence of ERK1 mutations in these patients following sequencing of all exons and exon–intron junctions (S.C.S., unpublished data). Thus, patients with distal 22q11 micro-deletions exhibit ERK2 haplo-insufficiency and defects in neural crest-derived structures.
Mouse Model of ERK2 Inactivation in Neural Crest.
To explore the role of ERK2 in the distal 22q11 micro-deletions and to further dissect the role of MAPK signaling in neural crest development, we used a mouse genetic approach. ERK2flox/flox mice were generated by inserting loxP sites surrounding exon 3 of ERK2 and mated with animals expressing a Wnt1:Cre transgene (16, 17), resulting in ERK2 inactivation in the developing neural crest. We also used ERK1−/− mice to evaluate contributions by this related isoform (18, 19).
Consistent with earlier reports, we found that Wnt1:Cre mediates robust recombination in neural crest cells by E9.5 within the pharyngeal arches, dorsal spinal cord, and dorsal root ganglion (20, 21) [supporting information (SI) Fig. S1A]. Analysis of later-stage embryos revealed extensive recombination in all neural crest derivatives including multiple craniofacial structures (Fig. S1B), the cardiac outflow tract (Fig. 3M), and the peripheral and enteric nervous system, in line with previous studies (16, 21). The dorsal root ganglia, a neural crest-derived structure, in E12.5 ERK1−/− ERK2fl/fl Wnt1:Cre mice, exhibited significantly reduced expression of ERK2 and the complete absence of ERK1 (Fig. S1C). These data demonstrate that the ERK1−/− ERK2fl/fl Wnt1:Cre mouse provides a robust animal model for studying the conditional deletion of ERK1/2 signaling during neural crest development in vivo.
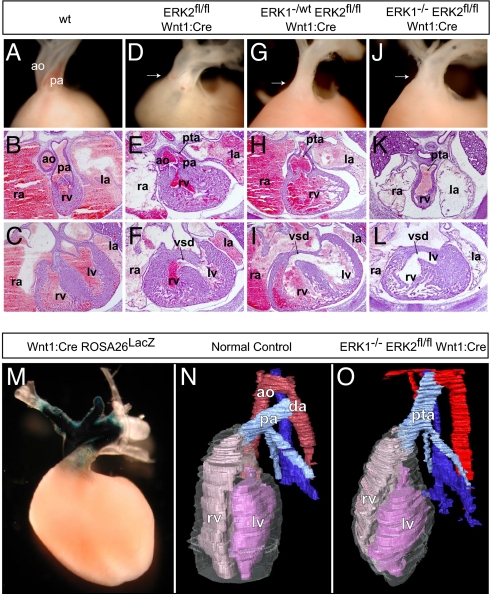
Fig. 3.
Disruption of ERK1/2 signaling results in cardiac outflow tract defects. Compared with controls (A–C), ERK2fl/fl Wnt1:Cre embryos displayed variable penetrance of cardiac outflow defects, including double-outlet right ventricle (D), PTA (E), and VSDs (F). E16.5 cross-sections and dissected E17.5 embryos (atria dissected away) from ERK1−/wt ERK2fl/fl Wnt1:Cre (G–I) and ERK1−/− ERK2fl/fl Wnt1:Cre (J–L) embryos consistently exhibited PTA and VSDs (I and L). Whole-mount LacZ staining of Wnt1:Cre Rosa26LacZ hearts reveals the distribution of neural crest derivatives in the embryonic conotruncus (M). Three-dimensional reconstructions (N) of cross-sections from E16.5 control embryos show a normal heart with 2 separate vessels, the aorta (red) and pulmonary artery (light blue), connected distally by the ductus arteriosus. Cross-sectional reconstructions of ERK1−/− ERK2fl/fl Wnt1:Cre hearts further illustrate PTA in these embryos (O). (ao = aorta, pa = pulmonary artery, rv = right ventricle, lv = left ventricle, la = left atrium, ra = right atrium, da = ductus arteriosus.)
ERK2 Signaling Is Necessary for the Development of Neural Crest-Derived Craniofacial Structures In Vivo.
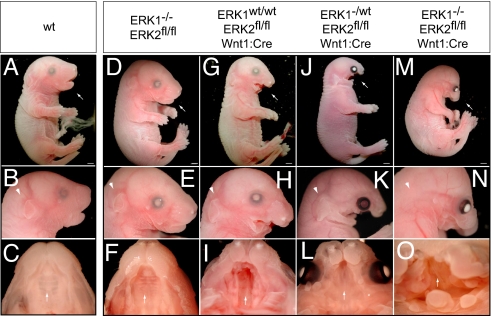
We have reported previously that the ERK1−/− mice possess deletions in exon 1 through 6 and express no detectable ERK1, but are viable and develop normally (Fig. 2 A–F), presumably as a result of compensation by ERK2 (18, 19). Heterozygous conditional inactivation of ERK2 in neural crest has no detectable effect on viability, and mice are born at expected Mendelian frequencies (e.g., 22 of 76). However, in contrast to the situation with ERK1, homozygous conditional inactivation of ERK2 in neural crest resulted in embryonic lethality with no viable neonates detected (0 of 102). Although the expected ratio of ERK2fl/fl Wnt1:Cre embryos was present in late gestation (8 of 28), embryos exhibited a spectrum of major craniofacial defects including a shortened maxilla, mandibular hypoplasia, and cleft palate (Fig. 2 G-I; n = 8). Many of these defects and the resulting cardiac defects (as described later) resemble abnormalities noted in patients with distal 22q11 micro-deletions.
Fig. 2.
The development of craniofacial structures in mice is critically dependent on ERK2 expression in neural crest. In comparison to E17.5 WT embryos (A–C) ERK1−/− embryos exhibit no significant defects (D–F). E17.5 ERK2fl/fl Wnt1:Cre embryos exhibit deficits in maxilla formation and mandibular hypoplasia (G and H), cleft palate (I, arrow), and absence of the tongue. Interestingly, ERK1−/wt ERK2fl/fl Wnt1:Cre show more severe defects in craniofacial development (J–L). ERK1−/− ERK2fl/fl Wnt1:Cre embryos exhibited the most significant defects, including truncation of the maxilla (M and N), mandibular aplasia (M and N), absence of an external ear (arrowhead in N), overall decrease in crown–rump length, eye placement anomalies, and palatal defects (O, arrow). Scale bars, 2 mm.
If ERK1 alleles were also deleted, craniofacial defects were significantly exacerbated. ERK1−/wt ERK2fl/fl Wnt1:Cre embryos demonstrate more severe mandibular hypoplasia, maxillary truncation, cleft palate, absence of the tongue, and eye placement anomalies (Fig. 2 J-L; n = 8 of 8). Homozygous inactivation of both ERK1/2 genes (ERK1−/− ERK2fl/fl Wnt1:Cre) resulted in more robust truncation of the maxilla, mandibular aplasia, eye placement anomalies, and absent tongue (Fig. 2 M-O; n = 18 of 18). In addition, these animals exhibited shortened crown–rump length and absence of an external ear. We further analyzed the early effects of ERK1/2 inactivation on the development of the pharyngeal arches. In E10.5 ERK1−/− ERK2fl/fl Wnt1:Cre embryos, but not ERK2fl/fl Wnt1:Cre embryos, pharyngeal arch morphogenesis was significantly hypoplastic compared with WT controls, suggesting a defect in either neural crest migration or survival (Fig. S2 A–E). These data demonstrate that the severity of the perturbations in craniofacial development is regulated in a gene dose-dependent manner by the expression of ERK1/2.
Elimination of ERK2 Signaling in Developing Neural Crest Leads to Conotruncal Cardiac Defects.
Conotruncal defects are typical in DGS/VCFS, and are seen in patients with distal 22q11 micro-deletions (10, 13, 15). Cardiac neural crest cells play an important role in septation of the outflow tract and formation and remodeling of the great arteries (16, 22). Crosses with the ROSA26 reporter line further illustrate these contributions in the Wnt1:Cre mouse heart (Fig. 3M) (20). A disruption in septa formation between the aorta and pulmonary artery, valves, and outflow regions results in persistent truncus arteriosus (PTA) and ventricular septal defects (VSDs). ERK2fl/fl Wnt1:Cre embryos displayed variable penetrance of these cardiac defects, with 2 of 5 embryos exhibiting VSDs at E16.5, one of which also had PTA. One of 4 E17.5 animals showed double-outlet right ventricle on gross dissection (Fig. 3 D–F). Thus, conditional elimination of ERK2 in developing neural crest resulted in variable penetrance in abnormalities similar to those observed in humans with distal 22q11 micro-deletions.
As with the craniofacial features, elimination of ERK1 alleles exacerbated these effects. We consistently detect PTA and VSDs in cross-sections of E16.5 ERK1−/wt ERK2fl/fl Wnt1:Cre (n = 3 of 3) embryos, ERK1−/− ERK2fl/fl Wnt1:Cre (n = 5 of 5) embryos, and dissected E17.5 hearts (Fig. 3 G–L). Three-dimensional reconstructions derived from the E16.5 cross-sections further illustrate PTA in the ERK1/2-deficient animals (Fig. 3 N and O).
Thymus and Thyroid Abnormalities in the Absence of Neural Crest ERK1/2 Signaling.
Thymus and thyroid abnormalities are an important element of the DGS/VCFS phenotypic spectrum, so these tissues were examined. Compared with WT controls (Fig. S3 A–C), the thymus and thyroid appeared morphologically normal in 4 of 5 E16.5 ERK2fl/fl Wnt1:Cre embryos evaluated by histology in cross-sections, even though craniofacial defects were present (Fig. S3 D–F). This is consistent with the patient phenotypes, as those with the recurrent small deletion that contains ERK2 have not been shown to have significant glandular anomalies. In contrast, ERK1−/wt ERK2fl/fl Wnt1:Cre embryos possess a single-lobed, fused thymus that is misplaced (Fig. S3G; n = 6), whereas the thymus failed to develop in ERK1−/− ERK2fl/fl Wnt1:Cre embryos (Fig. S3J; n = 6). Additionally, the thyroid gland is misplaced and does not develop normally in ERK1−/wt ERK2fl/fl Wnt1:Cre and ERK1−/− ERK2fl/fl Wnt1:Cre embryos. Instead of the normal bilateral lobules, these embryos develop a medially localized, single-lobed thyroid, identified by thyroglobulin expression (Fig. S3 H–L; n = 3 of each). These results indicate that thyroid and thymus development is dependent on ERK1/2 signaling in neural crest.
Inactivation of Upstream ERK1/2 Regulators Revealed a Conserved Pathway.
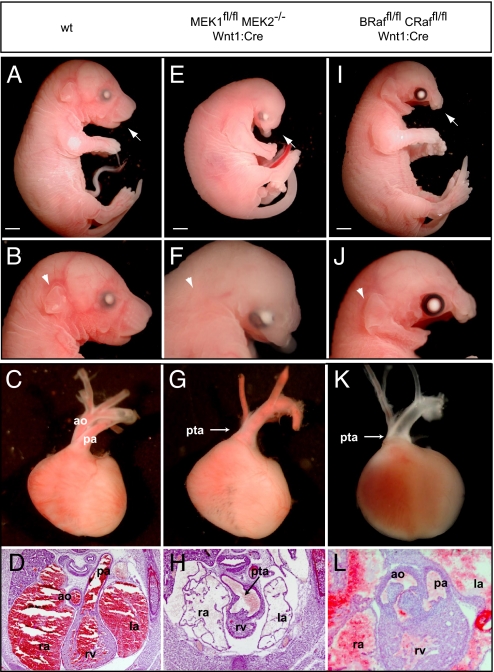
We sought to establish whether the phenotypic effects arising from inactivation of ERK1/2 would be recapitulated upon inactivation of upstream elements of the ERK1/2 cascade, including those linked to NCFCS. Embryos in which both MEK1 and MEK2 were inactivated (MEK1fl/fl MEK2−/− Wnt1:Cre) provided a nearly exact phenocopy of ERK1−/− ERK2fl/fl Wnt1:Cre embryos, exhibiting analogous craniofacial, heart, thymus, and thyroid defects (Fig. 4; n = 3 of 3). These animals exhibited late embryonic lethality with mandibular hypoplasia and maxilla truncation, decreased crown–rump length, and absence of the tongue and external ear (Fig. 4 E and F). Cardiac defects were present, including fully penetrant PTA and VSDs (Fig. 4 G and H), as well as thymus aplasia and thyroid malpositioning (Fig. S3 M–O). No isoform specific effects for MEK1 or MEK2 were identified, and defects were only detected in embryos following homozygous inactivation of both isoforms.
Fig. 4.
Inactivation of upstream elements of the MAPK cascade results in analogous defects in neural crest development. Compared with WT controls (A–D), mandibular hypoplasia and maxillary truncation is observed in MEK1fl/fl MEK2−/− Wnt1:Cre E17.5 embryos (E and F). Disruption of MEK1/2 signaling also resulted in decreased crown–rump length, eye placement anomalies, absence of the tongue, and a disruption of external ear development (E and F). PTA was consistently detected in E16.5 cross-sections and dissected E17.5 MEK1fl/fl MEK2−/− Wnt1:Cre embryos (G and H). The conditional inactivation of B-Raf/C-Raf did not alter embryonic size or external ear development, but mandibular and maxilla hypoplasia (I and J) was observed. Analysis of B-Raffl/fl C-Raffl/fl Wnt1:Cre embryos revealed partial penetrance of cardiac defects. An E17.5 embryo is displayed (K) that clearly demonstrates PTA whereas cross-sections from the E16.5 embryo (L) show mild conotruncal defects, namely double-outlet right ventricle. Scale bars, 2 mm.
We also examined B-Raffl/fl C-Raffl/fl Wnt1:Cre embryos and found that they exhibit mandibular and maxilla hypoplasia and death in late gestation (Fig. 4 I and J). These defects are similar to those seen in ERK1−/wt ERK2fl/fl Wnt1:Cre or ERK2fl/fl Wnt1:Cre embryos, in which the crown–rump length and external ear are unaffected. This may be a result of an incomplete reduction in ERK1/2 signaling resulting from compensation by the third Raf isoform, A-Raf, which is also expressed in neural crest (23). Outflow tract defects were of variable penetrance, and of the 6 mice examined, 4 exhibited PTA and VSDs; one exhibited VSDs with a DORV, but not overt PTA; and one was grossly normal (Fig. 4 K and L). Thymus hypoplasia was observed in 5 of 8 E16.5/17.5 embryos whereas the thyroid was hypoplastic or malpositioned in 2 of 3 E16.5 embryos (Fig. S3 P and R).
SRF May Mediate the Effects of ERK1/2 Signaling on Neural Crest.
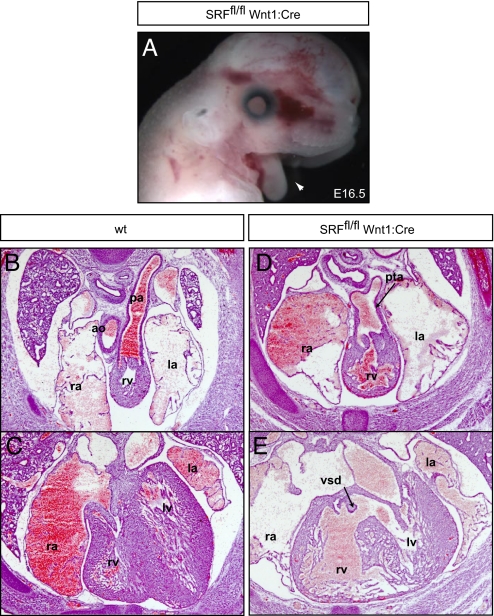
The SRF mediates some of the downstream effects of ERK1/2 on gene expression through interactions with the ternary complex factor and MTRF families. Remarkably, genetic inactivation of SRF in the neural crest resulted in embryos that were phenotypically similar to those in which ERK1/2 and its upstream regulators were inactivated. SRFfl/fl Wnt1:Cre embryos died at late embryonic stages and displayed fully penetrant mandibular hypoplasia (Fig. 5A), PTA and VSDs (Fig. 5 D and E), and thymic and thyroid aplasia (Fig. S3 S–U; n = 3). Maxillary hypoplasia was less severe and the external ear and tongue were present, indicating that these developmental aspects may be controlled by other downstream ERK1/2 effectors (Fig. 5A). Thus, neural crest specific inactivation of this single transcription factor results in craniofacial and cardiac defects analogous to those seen in embryos in which ERK1/2 or its upstream regulators were inactivated.
Fig. 5.
Neural crest–specific inactivation of SRF disrupts mouse development. E16.5 SRFfl/fl Wnt1:Cre embryos exhibit fully penetrant mandibular hypoplasia (A, arrowhead). These embryos possess cardiac defects, including PTA (D) with VSDs (E). Scale bars, 2 mm.
Discussion
Craniofacial and cardiac defects thought to arise principally from perturbation of neural crest development are prominent features of DGS/VCFS, which is caused by a recurrent deletion in chromosome 22q11 (12, 24). Significantly, deletions distal to the typical DGS/VCFS region present with a similar spectrum of defects, including cardiac outflow tract abnormalities, although different genes are deleted (10, 12, 13). These distal hemizygous deletions include the ERK2/MAPK1 gene (10), and we have shown here that deleted patients show haplo-insufficiency for ERK2. We further demonstrate that loss of ERK2 in mouse neural crest recapitulates many of the malformations seen in our patients, particularly craniofacial and conotruncal defects. Our model organism analysis provides support for a critical role for ERK2 in the pathogenesis of distal 22q11 micro-deletions, likely through the cell-autonomous disruption of neural crest development.
The distal 22q11/2 deletion contains other known genes and transcripts including HIC2, YPEL1, PPIL2, UBE2L3, SDF2L1, and PPM1F. Immediately distal to this group of genes is the IGLL gene cluster. Several genes including SUHW1 and 2, VPREB, TOP3B, and PRAME are located within the IGLL locus. None of these genes has thus far been associated with human disorders. We have sequenced the coding regions of HIC2, YPEL, PPIL2, UBE2L3, PRAME, and MAPK1 in 25 patients with features of DGS/VCFS who do not show evidence of a deletion in chromosome 22q, and also in another 20 patients with isolated PTA. No potential pathogenic mutations were noted in these cohorts (S.C.S., unpublished data).
Recent genetic analyses have shown that a number of syndromes are associated with mutations in upstream elements of the ERK1/2 cascade (Fig. S4). In humans, these defects are typically recognized as syndromes in the NCFCS spectrum. Mutations associated with the NCFCS usually result in gain of function with respect to signaling through ERK1/2, with the exception of certain LEOPARD syndrome mutations in SHP2/PTPN11, which are associated with loss of function (4, 25–30). Apparently, human cardiac and craniofacial development is exquisitely sensitive to alterations in the magnitude and dynamics of signaling through the ERK1/2 cascade. Our data indicate a potential mechanistic link between the NCFCS and DGS spectrum, via regulation of ERK1/2 activity in the developing neural crest.
The development of the neural crest is known to be reliant upon extracellular signals that may act through ERK1/2 signaling, particularly FGF8 (14). Mice in which the expression of FGF8 has been reduced or eliminated exhibit defects similar to those noted in ERK-inactivated mice, including cardiac defects involving PTA and outflow tract anomalies, as well as mandibular hypoplasia and cleft palate (31–34). Embryonic studies have shown a consistent correlation between the spatial location of activated ERK1/2 and known sites of FGF stimulation (35). FGF8 binding to FGF receptors stimulates the ERK1/2 cascade by promoting the association of the adapter proteins SHP2 and CRKL with activators of members of the RAS family of small G-proteins. The mutation or inactivation of SHP2 results in animals with severe cardiac defects and craniofacial abnormalities that are associated with dysregulation of ERK1/2 activation (36–38). Inactivation of Crkl results in a very similar phenotype (39). CRKL is positioned within the classical recurrent DGS deletion, as is TBX1, which has been shown to regulate the expression of FGF8 (40). Tbx1 mutant mice exhibit more subtle craniofacial defects than those observed in the ERK1/2 inactivated mice, but the conotruncal defects are comparable (41). Importantly, Fgf8 and Crkl interact within a common genetic pathway critical for craniofacial and outflow tract morphogenesis (6). Further, ERK1/2 activation is attenuated in the pharyngeal epithelia in response to decreased Fgf8 and Crkl gene dosage (6). These findings imply that the common developmental abnormalities observed in individuals with the distal 22q11 deletion or with the classical 3-Mb DGS deletion arise from perturbation of the same pathway.
Our data show that ERK2 is required for normal neural crest development. Whether the requirement for ERK2 represents an isoform-specific effect or an effect on overall ERK1/2 activity is unclear (42). Indeed, elimination of ERK1 alleles exacerbated neural crest phenotypic defects. Of course, we cannot exclude complex compensatory interactions between these 2 isoforms. Importantly, conditional inactivation of MEK1/2 or B-Raf/C-Raf, upstream canonical components of the ERK1/2 pathway, resulted in near-identical developmental abnormalities in craniofacial and conotruncal structures. Functions for RAF, MEK, or ERK outside of the established signaling module have been identified; however, the effects we have detected are consistent with their actions within the ERK1/2 signaling cascade (43).
One of the remarkable outcomes of these studies is the discovery that deletion of SRF in the developing neural crest results in embryos that exhibit the same range of cardiac, craniofacial, and glandular phenotypes as upstream elements of the pathway. SRF forms dimeric complexes with the Ets family of ternary complex factors, most prominently Elk1. Phosphorylation of Elk1 by ERK1/2 results in the induction of expression of a number of genes that regulate cell cycle and growth (44). Members of the MTRF family also directly associate with SRF and act to regulate the transcription of a large number of genes that participate in regulation of the actin cytoskeleton. The most widely expressed member of this family, MKL1, is phosphorylated by ERK1/2, which induces gene expression (45, 46). Thus, SRF target genes are functionally linked to the ability of cells to proliferate and migrate, consistent with the phenotypes we have observed in embryos in which SRF was conditionally deleted from neural crest. Our findings suggest that SRF plays a dominant role in the regulation of ERK1/2-dependent gene expression that governs neural crest development.
Conclusion
We have shown here that patients with distal 22q11 micro-deletions are haplo-insufficient for human ERK2 expression. Elimination of ERK2 specifically in mouse neural crest leads to craniofacial and cardiac defects analogous to those seen in patients with ERK2 haplo-insufficiency, suggesting a neural crest autonomous role for ERK2 signaling in the etiology of distal 22q11 micro-deletions. Further genetic analysis demonstrated the importance of a Raf/MEK/ERK/SRF pathway for mammalian neural crest development. These findings suggest that many aspects of the pathogenesis of DGS/VCFS, as well as the broad spectrum of NCFCS, may be related to perturbation of a common genetic pathway necessary for the activation of ERK1/2 (Fig. S4).
Experimental Procedures
Real-Time Expression Studies.
Patients were ascertained as described previously (10). Peripheral blood samples were collected from each patient after informed consent, under an institutional review board-approved research protocol of the Children's Hospital of Philadelphia, and used to extract protein, DNA, and RNA, and establish lymphoblastoid cell lines. RNA was isolated from cells and cDNA was synthesized using a high-capacity cDNA reverse transcription kit (Applied Biosystems). Taqman (Hs_00177066_m1; Applied Biosystems) was performed in triplicate for each sample toward a 127-bp amplicon spanning the boundary of exons 1 and 2 of MAPK1, and the endogenous control, 18s rRNA, by means of a relative quantitation assay. Amplification reactions were performed as per the manufacturer's instructions. All data were analyzed using the Applied Biosystems SDS-7500 machine and SDS software version 1.4.
Western Blotting.
Human lymphoblasts were collected in lysis buffer and equal amounts of protein were separated in SDS/PAGE gels, and transferred to Immobilon-P PVDF (Millipore). Membranes were blocked for 2 h at room temperature, then incubated overnight at 4 °C with primary antibodies including mouse anti-Erk2 (Becton Dickinson, 1:3,000), anti-Tbx1 (Lifespan Biosciences, 1:250), and anti-G3PDH (Trevigen, 1:5,000). After rinsing, membranes were incubated with secondary antibodies—HRP-conjugated goat anti-mouse or anti-rabbit antibody (1:4,000; GE Healthcare)—in 5% milk in buffered saline solution–Tween 20 for 2 h at room temperature. Blots were washed with buffered saline solution–Tween 20 and detection was performed using chemiluminescence reagents.
Transgenic Animals.
All animal experiments were performed in accordance with established protocols approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill and mice were handled in accordance with National Institutes of Health guidelines for the use and care of laboratory animals. Mouse genotyping was performed by PCR using genomic DNA isolated from tail samples collected at various stages. Primers used for gene amplification and details regarding other transgenic mice in this work are described in the SI Methods.
Histology and Immunohistochemistry.
Embryos were fixed in 4% paraformaldehyde. For cryoprotection, embryos were incubated in 30% sucrose/PBS solution at 4 °C for 2 to 3 days before embedding in Tissue-Tek OCT compound (Sakura Finetek). Serial, transverse sections were collected on Superfrost/Plus slides (Fisher Scientific). For general histological analysis, hematoxylin and eosin staining was performed. For 3D reconstruction, images were collected, segmented into lumens, and reconstructed with Amira. A surface smoothing algorithm was used and the resulting 3D model was rendered as a surface. For immunofluorescence microscopy, cryostat sections were incubated for 10 min in 0.1 M citrate buffer at 95 °C for antigen retrieval, incubated in polyclonal rabbit anti-human Thyroglobulin (DakoCytomation), and diluted in blocking solution (PBS solution, 10% normal goal serum, 0.5% Triton X-100) overnight at 4 °C. Following rinsing, slides were incubated in 1:1,000 Alexa Fluor 488 goat anti-rabbit antibody (Invitrogen) in blocking solution for 2 h at room temperature and mounted.
Supplementary Material
Acknowledgments.
We thank Dr. Beverly Emanuel and Dr. Betsy Goldmuntz for assistance with patient materials; Levi Goins, Anne Catherwood, and the animal facility staff for technical assistance; Dr. David L. Wilson for advice during the 3D reconstruction; and Drs. Jeffrey Vergales, Jonathan Rassi, and Ke Yang for assistance in phenotyping. This work is supported by National Institutes of Health Grants NS031768 (to W.D.S.), HL080637 and HL074731 (to S.C.S.), and NS34814 (to D.D.G); National Science Foundation Grant IBN97–23147 (to G.E.L.); the University of North Carolina Developmental Biology Training Program (J.M.N.); the National Institutes of Health Medical Scientist Training Program (S.R.W.); and Kirschstein NRSA award F31-MH074241 (I.S.). D.D.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805239105/DCSupplemental.
References
- 1.Pearson G, et al. Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 2.Posern G, Treisman R. Actin' together: Serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Shaw PE, Saxton J. Ternary complex factors: Prime nuclear targets for mitogen-activated protein kinases. Int J Biochem Cell Biol. 2003;35:1210–1226. doi: 10.1016/s1357-2725(03)00031-1. [DOI] [PubMed] [Google Scholar]
- 4.Schubbert S, Bollag G, Shannon K. Deregulated Ras signaling in developmental disorders: New tricks for an old dog. Curr Opin Genet Dev. 2007;17:15–22. doi: 10.1016/j.gde.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Bentires-Alj M, Kontaridis MI, Neel BG. Stops along the RAS pathway in human genetic disease. Nat Med. 2006;12:283–285. doi: 10.1038/nm0306-283. [DOI] [PubMed] [Google Scholar]
- 6.Moon AM, et al. Crkl deficiency disrupts Fgf8 signaling in a mouse model of 22q11 deletion syndromes. Dev Cell. 2006;10:71–80. doi: 10.1016/j.devcel.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindsay EA, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 8.Scambler PJ. The 22q11 deletion syndromes. Hum Mol Genet. 2000;9:2421–2426. doi: 10.1093/hmg/9.16.2421. [DOI] [PubMed] [Google Scholar]
- 9.Moon AM. Mouse models for investigating the developmental basis of human birth defects. Pediatr Res. 2006;59:749–755. doi: 10.1203/01.pdr.0000218420.00525.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saitta SC, et al. A 22q11.2 deletion that excludes UFD1L and CDC45L in a patient with conotruncal and craniofacial defects. Am J Hum Genet. 1999;65:562–566. doi: 10.1086/302514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaikh TH, et al. Low copy repeats mediate distal chromosome 22q11.2 deletions: Sequence analysis predicts breakpoint mechanisms. Genome Res. 2007;17:482–491. doi: 10.1101/gr.5986507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobrynski LJ, Sullivan KE. Velocardiofacial syndrome, DiGeorge syndrome: The chromosome 22q11.2 deletion syndromes. Lancet. 2007;370:1443–1452. doi: 10.1016/S0140-6736(07)61601-8. [DOI] [PubMed] [Google Scholar]
- 13.Ben-Shachar S, et al. 22q11.2 distal deletion: A recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet. 2008;82:214–221. doi: 10.1016/j.ajhg.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trainor PA. Specification of neural crest cell formation and migration in mouse embryos. Semin Cell Dev Biol. 2005;16:683–693. doi: 10.1016/j.semcdb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Rauch A, et al. A novel 22q11.2 microdeletion in DiGeorge syndrome. Am J Hum Genet. 1999;64:659–666. doi: 10.1086/302235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang X, et al. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 17.Samuels IS, et al. Deletion of ERK2 mitogen-activated protein kinase identifies its key roles in cortical neurogenesis and cognitive function. J Neurosci. 2008;28:6983–6995. doi: 10.1523/JNEUROSCI.0679-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selcher JC, et al. Mice lacking the ERK1 isoform of MAP kinase are unimpaired in emotional learning. Learn Mem. 2001;8:11–19. doi: 10.1101/lm.37001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nekrasova T, et al. ERK1-deficient mice show normal T cell effector function and are highly susceptible to experimental autoimmune encephalomyelitis. J Immunol. 2005;175:2374–2380. doi: 10.4049/jimmunol.175.4.2374. [DOI] [PubMed] [Google Scholar]
- 20.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 21.Danielian PS, et al. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 22.Kirby ML, Waldo KL. Neural crest and cardiovascular patterning. Circ Res. 1995;77:211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- 23.Luckett JCA, et al. Expression of the A-raf proto-oncogene in the normal adult and embryonic mouse. Cell Growth Differ. 2000;11:163–171. [PubMed] [Google Scholar]
- 24.Emanuel BS, Saitta SC. From microscopes to microarrays: Dissecting recurrent chromosomal rearrangements. Nat Rev Genet. 2007;8:869–883. doi: 10.1038/nrg2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Razzaque MA, et al. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat Genet. 2007;39:1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- 26.Pandit B, et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 27.Tartaglia M., et al. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 28.Roberts AE, et al. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet. 2007;39:70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- 29.Hanna N, et al. Reduced phosphatase activity of SHP-2 in LEOPARD syndrome: Consequences for PI3K binding on Gab1. FEBS Lett. 2006;580:2477–2482. doi: 10.1016/j.febslet.2006.03.088. [DOI] [PubMed] [Google Scholar]
- 30.Kontaridis MI, et al. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J Biol Chem. 2006;281:6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 31.Abu-Issa R, et al. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129:4613–4625. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- 32.Frank DU, et al. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trumpp A, et al. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park EJ, et al. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corson LB, Yamanaka Y, Lai KM, Rossant J. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–4537. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
- 36.Jopling C, van Geemen D, den Hertog J. Shp2 knockdown and Noonan/LEOPARD mutant Shp2-induced gastrulation defects. PLoS Genet. 2007;3:e225. doi: 10.1371/journal.pgen.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Araki T, et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura T, et al. Mediating ERK1/2 signaling rescues congenital heart defects in a mouse model of Noonan syndrome. J Clin Invest. 2007;117:2123–2132. doi: 10.1172/JCI30756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guris DL, et al. Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat Genet. 2001;27:293–298. doi: 10.1038/85855. [DOI] [PubMed] [Google Scholar]
- 40.Kochilas L, et al. The role of neural crest during cardiac development in a mouse model of DiGeorge syndrome. Dev Biol. 2002;251:157–166. doi: 10.1006/dbio.2002.0819. [DOI] [PubMed] [Google Scholar]
- 41.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 42.Lefloch R, Pouyssegur J, Lenormand P. Single and combined silencing of ERK1 and ERK2 reveals their positive contribution to growth signaling depending on their expression levels. Mol Cell Biol. 2008;28:511–527. doi: 10.1128/MCB.00800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hindley A, Kolch W. Extracellular signal regulated kinase (ERK)/mitogen activated protein kinase (MAPK)-independent functions of Raf kinases. J Cell Sci. 2002;115:1575–1581. doi: 10.1242/jcs.115.8.1575. [DOI] [PubMed] [Google Scholar]
- 44.Miano JM, Long X, Fujiwara K. Serum response factor: Master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 45.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 46.Kalita K, Kharebava G, Zheng JJ, Hetman M. Role of megakaryoblastic acute leukemia-1 in ERK1/2-dependent stimulation of serum response factor-driven transcription by BDNF or increased synaptic activity. J Neurosci. 2006;26:10020–10032. doi: 10.1523/JNEUROSCI.2644-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.