Abstract
The antibody access to some conserved structures on the HIV-1 envelope glycoprotein (Env) is sterically restricted. We have hypothesized that the smallest independently folded antibody fragments (domains) could exhibit exceptionally potent and broadly cross-reactive neutralizing activity by targeting hidden conserved epitopes that are not accessible by larger antibodies. To test this hypothesis, we constructed a large (size 2.5 × 1010), highly diversified library of human antibody variable domains (domain antibodies) and used it for selection of binders to conserved Env structures by panning sequentially against Envs from different isolates. The highest affinity binder, m36, neutralized all tested HIV-1 isolates from clades A– D with an activity on average higher than that of C34, a peptide similar to the fusion inhibitor T20, which is in clinical use, and that of m9, which exhibits a neutralizing activity superior to known potent cross-reactive antibodies. Large-size fusion proteins of m36 exhibited diminished neutralizing activity but preincubation of virions with soluble CD4 restored it, suggesting that m36 epitope is sterically restricted and induced by CD4 (CD4i). M36 bound to gp120-CD4 complexes better than to gp120 alone and competed with CD4i antibodies. M36 is the only reported representative of a promising class of potent, broadly cross-reactive HIV-1 inhibitors based on human domain antibodies. It has potential for prevention and therapy and as an agent for exploration of the closely guarded conserved Env structures with implications for design of small molecule inhibitors and elucidation of mechanisms of virus entry and evasion of immune responses.
Keywords: neutralization, therapeutic, epitope, steric restriction
The HIV pandemic remains the most serious of infectious disease challenges to public health (1). According to estimates from the 2007 United Nations AIDS/World Health Organization (UNAIDS/WHO) AIDS Epidemic Update, >6,800 persons become infected and >5,700 persons die from AIDS every day; in 2007, 33.2 million (30.6–36.1 million) were living with HIV, 2.5 million (1.8–4.1 million) were infected, and 2.1 million (1.9–2.4 million) have died (http://data.unaids.org/pub/EPISlides/2007/2007_epiupdate_en.pdf) despite antiretroviral therapy, which reduced AIDS-related deaths among those who received it. The efficacy of HIV therapy is significantly compromised by resistance to antiretroviral drugs (2, 3). A significant percentage of patients with HIV-1 infection (>50% in the US) receiving antiretroviral therapy are infected with viruses that express resistance to at least one of the available antiretroviral drugs. In treatment-naïve patients, HIV can quickly develop resistance. Thus, the development of inhibitors and classes of inhibitors against HIV continues to be urgently needed.
Small molecules currently form the bulk of our weaponry against HIV and are highly effective especially in combinations. However, because of their small size, in most cases they are inherently not very specific and not very potent in inhibiting protein–protein interactions that are major targets for intervention. Antibodies are highly specific, safe, and potent inhibitors of such interactions. However, HIV-1 has acquired the ability to escape neutralization by antibodies generated by the immune system by using a variety of mechanisms including sterically restricted access to conserved epitopes (4). Thus, one can expect that the virus could quickly develop resistance to naturally occurring HIV-1-specific antibodies. However, engineered antibody fragments of smaller size could gain access to the highly guarded conserved structures on the HIV-1 envelope glycoprotein (Env). Such small fragments targeting sterically restricted regions on the Env could exhibit neutralization activity superior to larger antibodies as has been demonstrated for Fab and scFv X5, which are on average significantly more potent than the full-size antibody (IgG1 X5) (4). We have hypothesized that further decreasing the size of the antibody fragments to the smallest independently folded fragments, the antibody domains (∼10-fold smaller than an IgG), but maintaining high binding affinity could lead to exceptionally potent and broadly cross-reactive neutralizers.
Here, we describe the identification and characterization of an antibody heavy-chain variable domain (VH) (domain antibody, dAb), m36, targeting highly conserved but sterically restricted CD4-induced (CD4i) structures on the Env. M36 is the only reported representative of a class of potent and broadly cross-reactive HIV-1 inhibitors based on human dAbs. It has potential as a candidate therapeutic and a microbicide and as an agent for exploration of the highly protected conserved Env structures with implications for the design of small-molecule inhibitors and elucidation of the mechanisms of virus entry into cells and evasion of immune responses.
Results
Selection of m36 from a Newly Constructed Human Antibody VH Library.
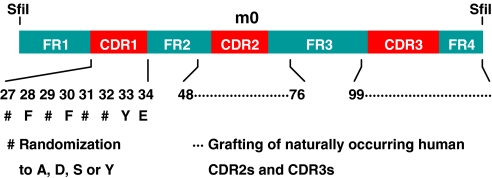
We have recently identified a phage-displayed heavy-chain-only antibody by panning of a large (size ≈1.5 × 1010) human naive Fab library against an Env (5). The VH of this Fab, designated as m0, was independently folded, stable, highly soluble, monomeric, and expressed at high levels in bacteria. M0 was used as a scaffold to construct a large (size ≈2.5 × 1010) highly diversified phage-displayed human VH library by grafting naturally occurring CDR2s and CDR3s of heavy chains from five human antibody Fab libraries and randomly mutating four putative solvent-accessible residues in CDR1 (Fig. 1). A VH, m36, was selected from this library as the highest affinity binder by using the sequential antigen panning (SAP) methodology (6) with HIV-1 Envs from clade B: A truncated Env lacking the transmembrane portion and the cytoplasmic tail from R2 (gp140R2) (7) or from JRFL (gp140JRFL) complexed with soluble CD4 (sCD4), and gp120 from Bal in complex with CD4 as a fusion protein (gp120Bal-CD4) (8). M36 is monomeric in PBS at pH 7.4 as determined by size exclusion chromatography, and runs on SDS/PAGE gels and size exclusion chromatography with an apparent molecular mass (MW) of 14–15 kDa, which is close to the calculated MW (14.972 kDa, including the His and FLAG tags) (data not shown). It is highly soluble, thermally stable, and is expressed at high levels in bacteria (≈30 mg/liter of culture) (data not shown). The m36 framework and CDR1 are closest to those encoded by the VH3–23 germ-line gene; the CDR2, to the VH4–34 [supporting information (SI) Fig. S1]. All m36 CDRs contain negatively charged and neutral but not basic residues. Its CDR3 is relatively short.
Fig. 1.
Construction of a human antibody VH library. A stable VH (m0) was used as a scaffold for grafting CDR2s and CDR3s from five human antibody Fab libraries. CDR1 residues 27, 29, 31, and 32 (ImMunoGeneTics numbering system) were randomized to A, D, S, or Y. The numbers denote the positions of the amino acid residues corresponding to the respective regions of the antibody VH gene where the CDRs were grafted; the # denotes the positions of the CDR1 randomization. The SfiI denotes the restriction enzyme sites used for cloning.
Potent Cross-Reactive Neutralization of Pseudotyped HIV-1 Isolates by m36.
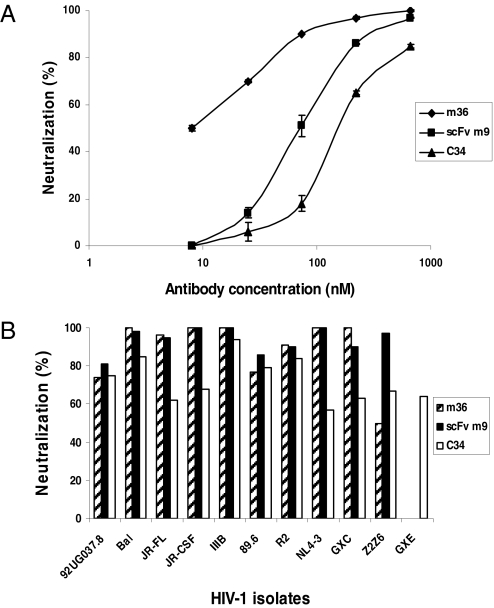
To determine the potency and breadth of HIV-1 neutralization by m36, viruses pseudotyped with Envs from HIV-1 isolates representing clades A– E were used. M36 neutralized six isolates from clade B, one isolate from clade C, and one isolate from clade A with potency on average twofold higher (twofold lower IC50s on molar basis) than that of the broadly cross-reactive neutralizing CD4i antibody scFv m9 (9) (Table 1); m9 is an in vitro matured derivative of X5 and exhibits superior neutralizing activity compared with known cross-reactive HIV-1 neutralizing antibodies (b12, 4E10, 2F5, 2G12, and X5) when tested against >100 isolates (M. Zhang, R. Ptak, V. Polonis, D. Montefiori, and D.S.D., unpublished work). M36 exhibited remarkable activity against the clade C isolate GXC-44 and the clade B isolate NL4–3 with very low IC90s (Table S1). It exhibited lower neutralization activity against the clade B isolate 89.6 and the clade D isolate Z2Z6 compared with scFv m9. M36 and scFv m9 did not neutralize the clade E isolate GXE at concentrations up to 667 nM. M36 was also on average more potent than the peptide C34 (Table 1); C34 (10) is a gp41-derived peptide that exhibits HIV-1 entry inhibitory activity comparable to or higher than that of the FDA approved peptide entry inhibitor T20 (DP178, brand name Fuzeon), which shares significant sequence homology with C34 although inhibits entry by a somewhat different mechanism involving binding to multiple sites (11). The inhibitory activity of m36 was dose dependent (Fig. 2A). Complete (100%) inhibition of four of seven clade B isolates and the isolate from clade C was achieved at 667 nM concentration; note that at the equivalent molar concentration, C34 did not completely inhibit any of the isolates tested, and m9 completely inhibited three of seven clade B isolates (Fig. 2B). These results suggest that m36 is a potent cross-reactive neutralizing antibody with potency and breadth of neutralization for this panel of isolates on average better than that of scFv m9 and C34. These three inhibitors exhibit differential neutralization profiles and could be used in combination.
Table 1.
Pseudotyped virus neutralization by m36 and its fusion proteins
| Virus | Clade | Antibodies |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| m36 | scFv m9 | C34 | m36SAbp | m36CH3 | m36b0Fc | m36h1Fc | m36c2Fc | m36h3Fc | ||
| 92UG037.8 | A | 147 ± 20* | 223 ± 7 | 297 ± 27 | 111 ± 44 | –† | – | – | – | – |
| Bal | B | 8 ± 0.7 | 73 ± 7 | 270 ± 27 | 13 ± 3 | – | – | – | – | – |
| JRFL | B | 25 ± 5 | 77 ± 17 | 270 ± 54 | 61 ± 6 | – | – | – | – | – |
| JR-CSF | B | 25 ± 11 | 73 ± 3 | 351 ± 81 | 24 ± 4 | – | – | – | – | – |
| IIIB | B | 8 ± 0.7 | 20 ± 2 | 119 ± 14 | 12 ± 3 | 167 ± 10 | – | 26 ± 0.9 | 26 ± 2 | 57 ± 2 |
| 89.6 | B | 173 ± 13 | 8 ± 0.7 | 12 ± 6 | 94 ± 22 | 102 ± 32 | >150 | 7 ± 0.5 | 5 ± 1 | 11 ± 6 |
| R2 | B | 21 ± 7 | 37 ± 3 | 12 ± 4 | 24 ± 8 | – | – | 364 ± 51 | >348 | >348 |
| NL4–3 | B | <8 | 29 ± 1 | 939 ± 135 | <8 | 75 ± 12 | >150 | <1 | <1 | <1 |
| GXC-44 | C | 16 ± 1 | 63 ± 10 | 351 ± 27 | 12 ± 4 | – | – | 25 ± 4 | 26 ± 10 | 28 ± 7 |
| Z2Z6 | D | 667 ± 60 | 22 ± 5 | 195 ± 35 | 667 ± 111 | – | – | – | – | – |
| GXE | E | – | – | 838 ± 27 | – | – | – | – | – | – |
*Antibody concentration (nM) resulting in 50% inhibition of virus infection (IC50).
†No significant neutralization at the highest antibody concentration (667, 667, 1000, 667, 167, 150, 364, 348, and 348 nM for m36, scFv m9, C34, m36SAbp, m36CH3, m36b0Fc, m36h1Fc, m36c2Fc, and m36h3Fc, respectively).
Fig. 2.
Potent m36-mediated neutralization of viruses pseodotyped with Envs of HIV-1 primary isolates. (A) Dose-dependent inhibition of Bal by m36, scFv m9, and C34, respectively. (B) Percentage inhibition of a panel of viruses by m36, scFv m9, and C34 at 667 nM, respectively.
M36 Binding to gp120 Is Enhanced by CD4 and Decreased by the CD4-Binding Site Antibody m14.
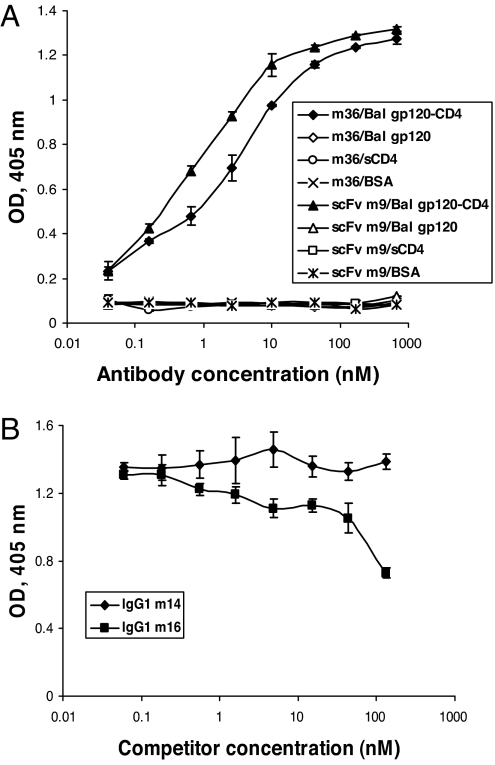
To approximately localize the m36 epitope and begin to elucidate the underlying mechanisms of neutralization, we measured binding of m36 to Envs from different isolates alone and in complex with CD4 and the m36 competition with well-characterized antibodies. M36 bound to gp120Bal-CD4 with high affinity (EC50 ≈ 2.5 nM) but not to gp120Bal or to sCD4 and BSA, as measured by an ELISA (Fig. 3A). It also bound to gp140 from a clade C isolate GXC-44 (gp140GXC-44) in the presence of sCD4 but notably it did bind, although weaker, to gp140GXC-44 alone too (data not shown), in contrast to gp120Bal (data not shown). Similarly, it also bound to another clade B Env, a tethered gp140 from 89.6, without complexation with CD4, but its binding was enhanced after gp140 bound to CD4 (data not shown). In both cases the binding as function on concentration deviated from a classical Langmuir-type isotherm likely because of a more complex multistage mechanism of antibody–antigen interactions. As expected for a CD4-induced (CD4i) antibody, m36 competed for binding to gp120Bal-CD4 with the CD4i antibody m16 (Fig. 3B) but not with the CD4 binding site (CD4bs) antibody m14, which was used as a negative control. Notably, it also competed for binding to gp140GXC-44 in the absence of CD4 with m14 (data not shown). These results suggest that m36 is a cross-reactive CD4i antibody that binds to an epitope localized close to the CD4 binding site.
Fig. 3.
Binding of m36 to gp120 is induced by the gp120 interaction with CD4. (A) Specific binding of m36 to gp120Bal-CD4 but not to gp120Bal alone. (B) Competition of m36 with a CD4i antibody (IgG m16) for binding to gp120Bal-CD4. The CD4bs antibody m14 was used as a negative control.
The m36 Epitope Is Sterically Restricted.
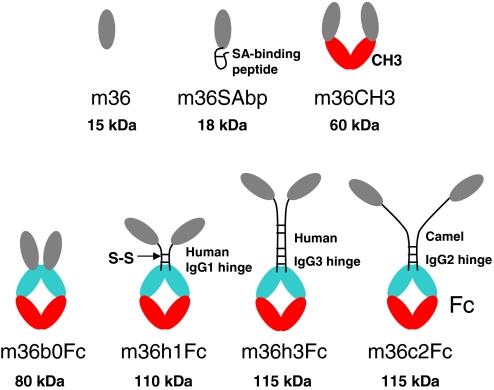
In an attempt to further elucidate possible mechanisms of neutralization we asked whether the access to the m36 epitope is sterically restricted. To answer this question we designed and generated several m36 fusion proteins with MWs ranging from 18–115 kDa (Fig. 4 and Fig. S2). We first measured their binding to Env complexes with CD4 to assure that the additional protein does not interfere with binding and then evaluated how the neutralizing activity was affected.
Fig. 4.
Design of m36 fusion proteins. Schematic representation of m36 fused with SAbp, human IgG1 CH3 domain, or Fc. The names of the constructs and their molecular weights are also shown. The sequences of the SAbp and the linkers used to join m36 with Fc by human IgG1 and IgG3 hinge, and camel IgG2 hinge are QRHPEDICLPRWGCLWGDDD, DKTHTCPPCP, EPKIPQPQPKPQPQPQPQPKPQPKPEPECTCPKCP, and ELKTPLGDTTHTCPRCPEPKSCDTPPPCPRCP, respectively.
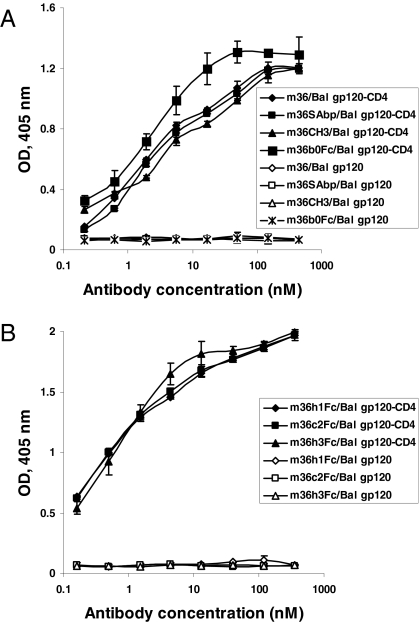
The m36 was fused to a serum albumin-binding peptide (SAbp), human IgG1 CH3 domain and Fc without a peptide linker; these fusion proteins, m36SAbp (MW ≈ 18 kDa), m36CH3 (MW ≈ 60 kDa), and m36b0Fc (MW ≈ 80 kDa), respectively (Fig. 4), were expressed in Escherichia coli HB2151 and purified. In the other three fusion proteins m36 was joined with human IgG1 Fc by a human IgG1 hinge (m36h1Fc, MW ≈ 110 kDa), a camel IgG2 hinge (m36c2Fc, MW ≈ 115 kDa), or a human IgG3 hinge (m36h3Fc, MW ≈ 115 kDa), respectively (Fig. 4), and expressed in mammalian 293 suspension cells. All fusion proteins, except m36SAbp, were dimeric in PBS, pH 7.4 as shown on nonreducing SDS/PAGE gels (Fig. S3). They all exhibited comparable to or higher than m36 binding to gp120Bal-CD4 as measured by ELISA. M36SAbp and m36CH3 showed binding comparable to that of m36 (Fig. 5A); m36b0Fc bound slightly better. The three fusion proteins expressed in mammalian cells (m36h1Fc, m36c2Fc, and m36h3Fc) exhibited the highest binding strengths with EC50 ≈ 0.5 nM (Fig. 5B). All antibodies at concentrations up to 870 nM did not show significant binding to gp120Bal in the absence of CD4.
Fig. 5.
Comparable binding activity of m36 and its fusion proteins. (A) Binding of m36, m36SAbp, m36CH3, and m36b0Fc to gp120Bal-CD4 and gp120Bal, respectively. (B) Binding of m36h1Fc, m36c2Fc, and m36h3Fc to gp120Bal-CD4 and gp120Bal, respectively.
Despite the preserved or even higher affinity (avidity), all fusion proteins, except m36SAbp, exhibited significantly weaker neutralizing activity when compared with m36 side by side in the same experiment (Table 1 and Table S1). The increased size of m36CH3 resulted in loss of neutralization against 7 of 10 isolates compared with m36, and for those neutralized (IIIB, 89.6, and NL4–3) the IC50s were significantly higher than the corresponding ones for m36. With an additional increase in molecular size, m36b0Fc further lost neutralization against the T cell line adapted (TCLA) isolate, IIIB, and had a decreased inhibitory activity against 89.6 and NL4–3. Notably, the three fusion proteins with long flexible linkers neutralized HIV-1 significantly better than the bacterially expressed m36b0Fc, which does not have a linker and m36CH3, which has much smaller molecular size (Table 1 and Table S1). They neutralized five isolates, one of them (89.6) even with potency higher than that of m36, likely because of their bivalency leading to avidity and other effects. These isolates were neutralized equally well by m36h1Fc, m36c2Fc, and m36h3Fc indicating that further increase in the length of the linker may not effect the neutralization activity. Fusion of m36 with the relatively much smaller SAbp resulted only in a slight but not significant decrease of the neutralizing activity (Table 1).
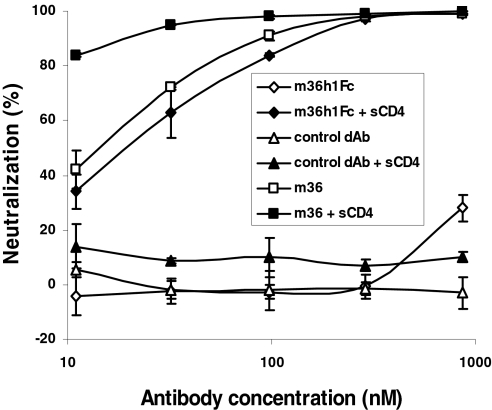
To find out whether the access to the m36 epitope on intact virions before entry into cells can be enhanced by CD4, we preincubated pseudovirus with m36 and its fusion proteins in the presence of low concentration of sCD4. The m36h1Fc fusion protein alone at up to 870 nM and sCD4 alone at 8 nM or combined with a control domain antibody-Fc fusion protein exhibited low neutralizing activity of ∼25 and 10%, respectively (Fig. 6 and data not shown). Preincubation of the Bal pseudovirus with both m36h1Fc and sCD4 resulted in a dramatic increase in neutralization—up to 100% (Fig. 6); a similar increase was also observed with JRFL pseudovirus (data not shown). Taken together these results suggest that the m36 epitope is sterically obstructed and fully accessible during virus entry only by relatively small-size molecules.
Fig. 6.
Pretriggering (sensitization) of virus by sCD4 dramatically increases neutralization by large molecules fused with m36. Viruses were preinoculated with different concentrations of antibodies and/or sCD4 at 8 nM for 1 h at 37°C, and then the mixture was added to 1.5 × 104 HOS-CD4-CCR5 cells grown in each well of 96-well plates. Luminesence was measured 48-h postinfection and percentage inhibition was calculated as described in Materials and Methods.
Discussion
A major advancement described in this article is the identification and characterization of an HIV-1 inhibitor conceptually different from the existing ones—a human antibody domain. Isolated stable human antibody variable domains (called also dAbs) have been described (12) and are promising candidate therapeutics against cancer and autoimmune diseases. Recently, one dAb (against TNFα) was successfully evaluated in a phase I clinical trial and very recently a phase II trial has commenced (http://www.arana.com); another one from camelid origin (called nanobody) was also safe (http://www.ablynx.com). The possibility that dAbs could efficiently access cryptic conserved epitopes has been discussed in the past but no evidence has been presented that dAbs targeting such epitopes exist and could be useful for development of potential therapeutics. Here, we describe evidence for our hypothesis that dAbs could bind to conserved structures that are inaccessible or partially accessible during virus entry for molecules of larger size comparable to that of full-size antibodies generated by the human immune system. Such antibodies could be potentially useful not only as candidate therapeutics against viruses, including HIV-1, which can protect highly conserved structures that are vital for virus replication, but can also help identify those conserved structures with implications for the development of small-molecule inhibitors and elucidation of mechanisms of entry and evasion of immune responses.
We have hypothesized in the past that binding of the Env to receptor and coreceptor molecules may result in the exposure of conserved structures that could be used as antigens for selection of cross-reactive neutralizing antibodies (13, 14). The identification and characterization of the potent broadly cross-reactive human Fab X5 (14) provided evidence confirming this hypothesis. The crystal structure of its complex with gp120-CD4 allowed the precise localization to be determined of its highly conserved epitope overlapping the putative coreceptor binding site very close to the CD4 binding site (15). However, unexpectedly, IgG1 X5 on average exhibited lower potency than Fab and scFv, likely because of its larger size (4). Because the crystal structure of Fab X5 complexed with gp120-CD4 suggested that only its heavy-chain contacts gp120 (15), we have hypothesized that decreasing the size to a single VH could further increase the potency of X5. However, our efforts to isolate stable VH X5 or VH X5-like dAbs by rational design, mutagenesis, and screening have failed (M. Zhang, Y. Wu, and D.S.D., unpublished work). Similarly, we have not been successful in developing a stable highly soluble VH dAb based on an HIV-1 gp120-specific heavy-chain-only antibody (M. Zhang and D.S.D., unpublished work), likely because of a certain extent of hydrophobicity that is important for its structural stability. Accidentally, while panning a large naïve Fab library against the Env, we identified another heavy-chain-only antibody, which as an isolated VH, m0, exhibited high stability and solubility. Thus, we decided to change the strategy and construct a large highly diversified library by using as a scaffold m0. The high diversification was achieved by two strategies—grafting highly diverse CDR2s and CDR3s from five of our other libraries including one from HIV-1-infected individuals and randomly mutating four residues in the CDR1 to residues frequently found in antibody CDRs. This library was used for selection of m36 and could also be useful for isolation of dAbs against other antigens.
Access of full-size antibodies to CD4i epitopes can be restricted during virus entry into cells (4, 16). The crystal structures of two CD4i antibodies, X5 and 17b, in complex with gp120 and sCD4, indicate that access to their epitopes requires long protruding heavy-chain CDR3s (15, 17). Most of the known CD4i antibodies have long CDR3s that could play an important role in accessing sterically restricted areas. Their CDR3s are highly acidic (Fig. S1) and the closest germ-line VH gene for this group of antibodies is VH1–69 (18). A smaller group of CD4i antibodies has relatively short CDR3s, acidic CDR2s, and VH1–24 gene usage (18). M36 appears to be the only representative of a third group characterized by short CDR3, acidic CDR1, and VH3–23 gene usage (Fig. S1). Because of its small size it may not need a long CDR3 for access to sterically restricted structures.
M36 exhibited on average higher neutralizing activity than scFv m9, which has been recently shown to be superior to the best characterized cross-reactive HIV-1 neutralizing antibodies b12, 2G12, 2F5, and 4E10 (M. Zhang, R. Ptak, V. Polonis, D. Montefiori, and D.S.D., unpublished work), and than C34, a peptide similar to the fusion inhibitor T20 (Fuzeon), which is in clinical use. We hypothesized that a dimer of m36 could have even higher potency because of avidity effects and used CH3 as a dimerization domain. However, m36CH3 was significantly weaker than m36 indicating that the m36 epitope is fully accessible during virus entry only by antibody domains or smaller molecules (Table 1). A larger fusion protein, m36b0Fc, was a poorer inhibitor most likely because of the increased molecular size. However, a fusion protein with a human IgG1 hinge region as a linker between m36 and Fc, m36h1Fc, neutralized several isolates better than m36CH3 and m36b0Fc despite an increase in size (Table 1). For some isolates (89.6, NL4–3, and GXC-44) its potency was as high as that of m36. We hypothesized that the long linker may provide a flexibility needed for the m36 to reach its epitope combined with an increased binding because of avidity effects resulting from the m36h1Fc bivalency. However, fusion proteins with even longer hinge regions (from camel antibodies, m36cFc, or from human IgG3, m36h3Fc) did not exhibit higher potency than m36h1Fc (Table 1) possibly because of compensation of the flexibility effect by an increase in the effective hydrodynamic size of the molecules leading to a decrease in the accessibility of the m36 epitope.
The neutralizing activity of the m36 fusion proteins was dramatically increased for viruses sensitized (pretriggered) by sCD4 to expose the m36 epitope (Fig. 6). These data not only provide evidence for the restricted nature of the m36 epitope during entry but also suggest the possibility of developing m36-based potential therapeutics, e.g., fusion proteins of m36 with sCD4 or small-molecule mimics of CD4. These molecules could neutralize the virus before it binds to cell surface-associated CD4 whereas m36 is likely to exert its major neutralizing activity after the virus binds to the cell surface-associated CD4, which triggers the conformational changes in gp120 leading to exposure of its epitope.
To our knowledge m36 is the only reported HIV-1 neutralizing human dAb. It could have potential as a therapeutic adding a new target to the growing family of entry inhibitors (19). Although its half-life in vivo is likely to be very short, our findings that a fusion protein with an SAbp retains approximately the same neutralizing activity as m36 indicates the possibility of improving its pharmacokinetics. We found that the m36SAbp binds to serum albumins from human (HSA), bovine (BSA), and mouse (MSA) (data not shown) indicating that such a possibility is realistic, although only experiments in animals and humans can definitely prove it. The epitope of m36 is sterically restricted and may not be directly used to develop potential vaccine immunogens. However, it is highly conserved and therefore could be useful as a tool to explore mechanisms of entry and to understand how HIV-1 guards its conserved structures and evades neutralizing immune responses.
Materials and Methods
Cells, Viruses, Plasmids, gp120, gp140, and Antibodies.
We purchased the 293T cells from ATCC. Other cell lines and plasmids used for expression of various HIV-1 Envs were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (ARRRP). Recombinant gp140s were kindly provided by C. Broder (Uniformed Services University of the Health Sciences, Bethesda, MD). Gp120Bal and the single-chain fusion protein gp120Bal-CD4 (8) were gifts from T. Fouts (Institute of Human Virology, Baltimore; currently at Profectus, Baltimore, MD). Horseradish peroxidase (HRP)-conjugated anti-FLAG tag antibody and HRP-conjugated anti-human IgG (Fc-specific) antibody were purchased from Sigma-Aldrich (St. Louis).
Library Construction and Selection of VHs Against HIV-1 Antigens.
A large phage-displayed human VH library (≈2.5 × 1010 individuals) was constructed by grafting of naturally occurring heavy-chain CDR2s and CDR3s from other five-template libraries to a VH framework scaffold, m0 (5) and randomly mutating four putative solvent-accessible residues in its CDR1 (Fig. 1). These template libraries include: (a) a naive human Fab library (5 × 109 members) from peripheral blood B cells of 10 healthy donors (20); (b) a naïve human Fab library (1.5 × 1010 members) from peripheral blood B cells of 22 healthy donors, spleens of 3 donors, and lymph nodes of 34 healthy donors (W.C., Z.Z., and D.S.D., unpublished work); (c) two naïve human Fab libraries (6 × 108 and 7.2 × 108 members, respectively) from cord blood of two healthy babies, respectively (W.C. and D.S.D., unpublished work); and (d) an immune human Fab library from the bone marrow of three long-term nonprogressors whose sera exhibited the broadest and most potent HIV-1 neutralization among 37 HIV-infected individuals (provided by T. Evans, currently at Novartis, Boston) (6). This dAb library was used for selection of VHs against HIV-1 antigens conjugated to magnetic beads (Dynabeads M-270 epoxy; DYNAL Inc., New Hyde Park, NY) as described previously (20). For sequential panning, 10 and 5 μg of gp120Bal-CD4 was used in the first round and third round, respectively; antigens were alternated with 5 μg of gp140R2 or gp140JRFL during the second round and fourth round. Clones that bound to HIV-1 antigens were identified from the third and fourth round of biopanning by using monoclonal phage ELISA as described (20).
Construction and Cloning of m36 Fusion Proteins.
The following primers were used: m36F, 5′-TGGTTTCGCTACCGTGGCCCAGCCGGCCCAGGTGCAGCTGGTG-3′ (sense); m36F1, 5′-TGGTTTCGCTACCGTGGCCCAGGCGGCCCAGGTGCA G C TGGTG-3′ (sense); m36R1, 5′-GTGAGTTTTGTCGGGCCCTGAGGAGACGGTGAC-3′ (antisense); m36R2, 5′-TGGTTGTGGTTGGGGTATCTTGGGTTCTGAGGAG A C GGTGAC-3′ (antisense); m36R3, 5′-GTCACCAAGTGGGGTTTTGAGCTCTGAGGAGACGGTGAC-3′ (antisense); m36R4, 5′-TTCTCGGGGCTGCCCTGAGGAGACGGTGAC-3′ (antisense); m36R5, 5′-CAGGAGTTCAGGTGCTGAGGAGACGGTGAC-3′ (antisense); CH3F, 5′-GGGCAGCCCCGAGAACCA-3′ (sense); CH3R, 5′-GTGGTGGTGGTGGTGGCCGGCCTGGCCTTTACCCGGAGACAG-3′ (antisense); FcF1, 5′-ACGTGTCCCAAATGTCCAGCACCTGAACTCCTGGGG-3′ (sense); FcF2, 5′-CCGTGCCCACGGTGCCCAGCACCTGAACTCCTGGGG-3′ (sense); FcF3, 5′-GCACCTGAACTCCTGGGG-3′ (sense); FcR, 5′-GTCGAGGCTGATCAGCGG-3′ (antisense); FcR1, 5′-CTCCTATGCGGCCGCTTTACCCGGAGACAG-3′ (antisense); HcF, 5′-CCCCAACCACAACCAAAACCACAACCACAACCACAACCACAACCAAAACCAC-3′ (sense); HcR, 5′-TGGACATTTGGGACACGTGCATTCTGGTTCAGGTTTTGGTTGT G G T T T TGGTTGTGG-3′ (antisense); HhF, 5′-ACCCCACTTGGTGACACAACTCACACATGCCCACGGTGCCCAGAGCCCAAA-3′ (sense); HhR, 5′-TGGGCACCGTG G G C ACGGGGGAGGTGTGTCACAAGATTTGGGCTCTGGGCA-3′ (antisense); HSAPR2, 5′-CTCCTATGCGGCCGCATCATCGTCGCCCCACAAACACCCCCAGCGTGGCAA-3′ (antisense); HSAPR3, 5′-CCCCCAGCGTGGCAAACATATATCTTCTGGGTGG C G C TGGCCCTTATCGTCATC-3′ (antisense).
For construction of m36CH3, the m36 gene was amplified by PCR (primer: m36F1 and m36R4) with m36-encoding plasmid pCom36 as a template. The CH3 gene of human IgG1 was PCR (primer: CH3F and CH3R) amplified from plasmid pSecTagB-Fc, which encoded the human IgG1 Fc portion (Fig. S2). M36 fragment was joined to CH3 by overlapping PCR performed in a volume of 100-μl by using both templates (in the same molarities) for 7 cycles in the absence of primers and 15 additional cycles in the presence of primers (500 pM m36F1 and CH3R, respectively). The m36CH3 products appended with SfiI restriction sites on both sides were digested and cloned into pComb3X (a gift from D. Burton, The Scripps Research Institute, La Jolla, CA) (Fig. S2). To generate m36b0Fc, m36 gene was amplified by PCR by using primers m36F1 and m36R5. Human IgG1 Fc gene was obtained by PCR amplification by using pSecTagB-Fc as the template and primers FcF3 and FcR1. M36 fragment was joined to Fc as described above. The products were digested with SfiI and NotI, and cloned into pZYD-N1, which was developed in our laboratory and has a NotI site after the FLAG tag. The vector pSecTagB-Fc was used for construction of m36h1Fc, m36c2Fc, and m36h3Fc. The m36 fragment was PCR amplified by using primer m36F and m36R1, digested with SfiI and ApaI, and cloned into pSecTagB-Fc to generate m36h1Fc. M36c2Fc was cloned by amplifying m36 fragment (primers: m36F and m36R2), human IgG1 Fc (primers: FcF1 and FcR), and camel IgG2 hinge (primers: HcF and HcR). The m36 fragment was fused to camel IgG2 hinge and the product was subsequently joined to Fc by overlapping PCR. The resultant full-length m36c2Fc product was digested with SfiI and PmeI, and cloned into pSecTagB-Fc vector. In the same way m36h3Fc was constructed except for the use of primer m36F and m36R3 for m36 amplification, primers FcF2 and FcR for human IgG1 Fc amplification, and primer HhF and HhR for human IgG3 hinge amplification. To generate m36SAbp, the m36 fragment was amplified by using primer m36F1 and HSAPR3, purified, and further extended by PCR using primers m36F1 and HSAPR2. The products were digested with SfiI and NotI and cloned into pZYD-N1.
Expression and Purification of m36 and Its Fusion Proteins.
M36, m36SAbp, m36CH3, and m36b0Fc were expressed in E. coli HB2151, as described previously (20). The bacterial pellet was collected after centrifugation at 5,000 × g for 10 min and resuspended in PBS (pH 7.4) containing 0.5 million-unit polymixin B (Sigma-Aldrich). After 30 min incubation with rotation at 50 rpm at room temperature, it was centrifuged at 25,000 × g for 25 min at 4 °C. The supernatant was used for purification of m36, m36SAbp, and m36CH3 by immobilized metal ion affinity chromatography (IMAC) by using Ni-NTA resin (Qiagen, Valencia, CA) according to manufacturer's protocols. For purification of m36b0Fc, nProtein A Sepharose 4 Fast Flow (GE Healthcarezcomx, Piscataway, NJ) was used. M36h1Fc, m36c2Fc, and m36h3Fc were expressed in 293 free style cells. CellFectin (Invitrogen, Carlsbad, CA) was used to transfect 293 free style cells according to the instructions of the manufacturer. Three days post-transfection, the culture supernatant was harvested and used for purification of m36h1Fc, m36c2Fc, and m36h3Fc by using nProtein A Sepharose 4 Fast Flow.
Binding ELISA.
Antigens were coated on Corning EIA/RIA high-binding 96-well plates (Corning Inc., Corning, NY) at 50-ng per well overnight at 4 °C and blocked with 3% nonfat milk in PBS. Threefold serially diluted antibody was added in the absence or presence of sCD4 at 2 μg/ml final concentration and incubated at room temperature for 2 h. The plates were washed with PBS containing 0.05% Tween 20. Bound m36, m36SAbp, or m36CH3 was detected by HRP-conjugated anti-FLAG tag antibody (Sigma-Aldrich). The m36 fusion proteins with human IgG1 Fc were detected by HRP-conjugated anti-human IgG (Fc-specific) antibody (Sigma-Aldrich). The assay was developed at 37 °C with ABST substrate (Roche, Indianapolis, IN) and monitored at 405 nm. The half-maximal binding (EC50) was calculated by fitting the data to the Langmuir adsorption isotherm.
Competition ELISA.
Antigens were coated and blocked as described above. M36 at a concentration leading to 90% maximum binding was premixed with threefold serially diluted competitors without or with sCD4 at 2 μg/ml final concentration. Mixtures were subsequently added to each well and incubated. Bound m36 was detected and the assay was developed as described above.
Measurement of m36 Oligomerization.
Superdex75 column was calibrated with protein molecular mass standard of 14 (ribonuclease A), 25 (chymotrypsin), 44 (ovalbumin), 67 (albumin), 158 (aldolase), 232 (catalase), 440 (ferritin), and 669 (thyroglobulin) kDa. Purified m36 in PBS were loaded onto the column that had been preequilibrated. The proteins were eluted with PBS at 0.5 ml/min.
Pseudovirus Neutralization Assay.
Viruses pseudotyped with HIV-1 Envs were prepared by cotransfection of 70–80% confluent 293T cells with pNL4–3.luc.E-R- and pSV7d constructs encoding HIV-1 Envs (a gift from G. Quinnan, USUHS, Bethesda, MD) by using the PolyFect transfection reagent (Qiagen) according to manufacturer's instruction. Pseudotyped viruses were obtained after 24 h by centrifugation and filtration of cell culture through 0.45-μm filters. For neutralization, viruses were mixed with different concentrations of antibodies and/or sCD4 at 8 nM for 1 h at 37 °C, and then the mixture was added to ≈1.5 × 104 HOS-CD4-CCR5 (used for all R5 and dual tropic viruses) or HOS-CD4-CXCR4 cells grown in each well of 96-well plates. Luminesence was measured after 48 h by using the Bright-Glo Luciferase Assay System (Promega, Madison, WI) and a LumiCount microplate luminometer (Turner Designs). Mean relative light units (RLU) for duplicate wells were determined. Percentage inhibition was calculated by the following formula: (1 − average RLU of antibody-containing wells/average RLU of virus-only wells) × 100. IC50 and IC90 of neutralization were assigned for the antibody concentration at, which 50 and 90% neutralization were observed, respectively.
Supplementary Material
Acknowledgments.
We thank Christopher Broder, Gerald Quinnan, Dennis Burton, and Tim Fouts for providing reagents and the members of our group John Owens, Meiyun Zhang, and Xiadong Xiao for help. This project was supported by the Intramural AIDS Targeted Antiviral Program of the National Institutes of Health (NIH), by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, by federal funds from the National Cancer Institute, NIH, under contract N01-CO-12400, and by the Gates Foundation (D.S.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805297105/DCSupplemental.
References
- 1.Fauci AS. 25 years of HIV. Nature. 2008;453:289–290. doi: 10.1038/453289a. [DOI] [PubMed] [Google Scholar]
- 2.Perno CF, et al. Overcoming resistance to existing therapies in HIV-infected patients: The role of new antiretroviral drugs. J Med Virol. 2008;80:565–576. doi: 10.1002/jmv.21034. [DOI] [PubMed] [Google Scholar]
- 3.Prabakaran P, Dimitrov AS, Fouts TR, Dimitrov DS. Structure and function of the HIV envelope glycoprotein as entry mediator, vaccine immunogen, and target for inhibitors. Adv Pharmacol. 2007;55:33–97. doi: 10.1016/S1054-3589(07)55002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Labrijn AF, et al. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol. 2003;77:10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen W, Zhu Z, Feng Y, Xiao X, Dimitrov DS. Construction of a large phage-displayed human antibody domain library with a scaffold based on a newly identified highly soluble, stable heavy chain variable domain. J Mol Biol. 2008;382:779–789. doi: 10.1016/j.jmb.2008.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang MY, et al. Broadly cross-reactive HIV neutralizing human monoclonal antibody Fab selected by sequential antigen panning of a phage display library. J Immunol Methods. 2003;283:17–25. doi: 10.1016/j.jim.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Quinnan GV, Zhang PF, Fu DW, Dong M, Alter HJ. Expression and characterization of HIV type 1 envelope protein associated with a broadly reactive neutralizing antibody response. AIDS Res Hum Retroviruses. 1999;15:561–570. doi: 10.1089/088922299311088. [DOI] [PubMed] [Google Scholar]
- 8.Fouts TR, et al. Expression and characterization of a single-chain polypeptide analogue of the human immunodeficiency virus type 1 gp120-CD4 receptor complex. J Virol. 2000;74:11427–11436. doi: 10.1128/jvi.74.24.11427-11436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang MY, et al. Improved breadth and potency of an HIV-1-neutralizing human single-chain antibody by random mutagenesis and sequential antigen panning. J Mol Biol. 2004;335:209–219. doi: 10.1016/j.jmb.2003.09.055. [DOI] [PubMed] [Google Scholar]
- 10.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 11.Liu S, et al. Different from the HIV fusion inhibitor C34, the anti-HIV drug Fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. J Biol Chem. 2005;280:11259–11273. doi: 10.1074/jbc.M411141200. [DOI] [PubMed] [Google Scholar]
- 12.Holt LJ, Herring C, Jespers LS, Woolven BP, Tomlinson IM. Domain antibodies: Proteins for therapy. Trends Biotechnol. 2003;21:484–490. doi: 10.1016/j.tibtech.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Dimitrov DS. Fusin: A place for HIV-1 and T4 cells to meet. Identifying the coreceptor mediating HIV-1 entry raises new hopes in the treatment of AIDS. Nat Med. 1996;2:640–641. doi: 10.1038/nm0696-640. [DOI] [PubMed] [Google Scholar]
- 14.Moulard M, et al. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc Natl Acad Sci USA. 2002;99:6913–6918. doi: 10.1073/pnas.102562599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang CC, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Decker JM, et al. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med. 2005;201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang CC, et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci USA. 2004;101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ray N, Doms RW. HIV-1 coreceptors and their inhibitors. Curr Top Microbiol Immunol. 2006;303:97–120. doi: 10.1007/978-3-540-33397-5_5. [DOI] [PubMed] [Google Scholar]
- 20.Zhu Z, et al. Potent neutralization of Hendra and Nipah viruses by human monoclonal antibodies. J Virol. 2006;80:891–899. doi: 10.1128/JVI.80.2.891-899.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.