Abstract
Petrobactin, a virulence-associated siderophore produced by Bacillus anthracis, chelates ferric iron through the rare 3,4-isomer of dihydroxybenzoic acid (3,4-DHBA). Most catechol siderophores, including bacillibactin and enterobactin, use 2,3-DHBA as a biosynthetic subunit. Significantly, siderocalin, a factor involved in human innate immunity, sequesters ferric siderophores bearing the more typical 2,3-DHBA moiety, thereby impeding uptake of iron by the pathogenic bacterial cell. In contrast, the unusual 3,4-DHBA component of petrobactin renders the siderocalin system incapable of obstructing bacterial iron uptake. Although recent genetic and biochemical studies have revealed selected early steps in petrobactin biosynthesis, the origin of 3,4-DHBA as well as the function of the protein encoded by the final gene in the B. anthracis siderophore biosynthetic (asb) operon, asbF (BA1986), has remained unclear. In this study we demonstrate that 3,4-DHBA is produced through conversion of the common bacterial metabolite 3-dehydroshikimate (3-DHS) by AsbF—a 3-DHS dehydratase. Elucidation of the cocrystal structure of AsbF with 3,4-DHBA, in conjunction with a series of biochemical studies, supports a mechanism in which an enolate intermediate is formed through the action of this 3-DHS dehydratase metalloenzyme. Structural and functional parallels are evident between AsbF and other enzymes within the xylose isomerase TIM-barrel family. Overall, these data indicate that microbial species shown to possess homologs of AsbF may, like B. anthracis, also rely on production of the unique 3,4-DHBA metabolite to achieve full viability in the environment or virulence within the host.
Keywords: AsbF structure, dehydratase, siderophore, virulence factor
Siderophore production in pathogenic bacteria has gained considerable attention because of its crucial function in essential iron uptake by many microbes and the relevance of siderophore-associated proteins as molecular markers of various infectious agents (1). In Bacillus anthracis, the causative agent of anthrax, 2 siderophores, petrobactin and bacillibactin (Fig. 1A), play a significant role during iron acquisition (2–4), but only petrobactin is absolutely essential for full virulence within a mammalian host (5). This siderophore was initially discovered from the Gram-negative marine bacterium Marinobacter hydrocarbonoclasticus, whose genome bears a biosynthetic gene cluster homologous to the B. anthracis asb operon (2). Recent genetic and chemical analysis suggests that petrobactin biosynthesis may also be a prerequisite for virulence in related Bacillus species (6). These studies highlight the importance of elucidating the mechanisms of siderophore production in pathogenic microbes as a target for abrogating infection by organisms like B. anthracis, a rapidly virulent microbe with proven potential as a bioterrorism agent. Based on these factors, we have initiated studies to investigate key biosynthetic enzymes for petrobactin assembly in efforts to establish new antimicrobial targets to protect against anthrax. Eliminating petrobactin in pathogenic microbes is particularly important among catecholate siderophores, because the 3,4-dihydroxybenzoic acid (3,4-DHBA) moiety enables the molecule to escape sequestration by the mammalian immune protein siderocalin (7, 8). However, the genes responsible for mediating production of this metabolic precursor have remained undefined, as has the biochemical mechanism of its assembly.
Fig. 1.
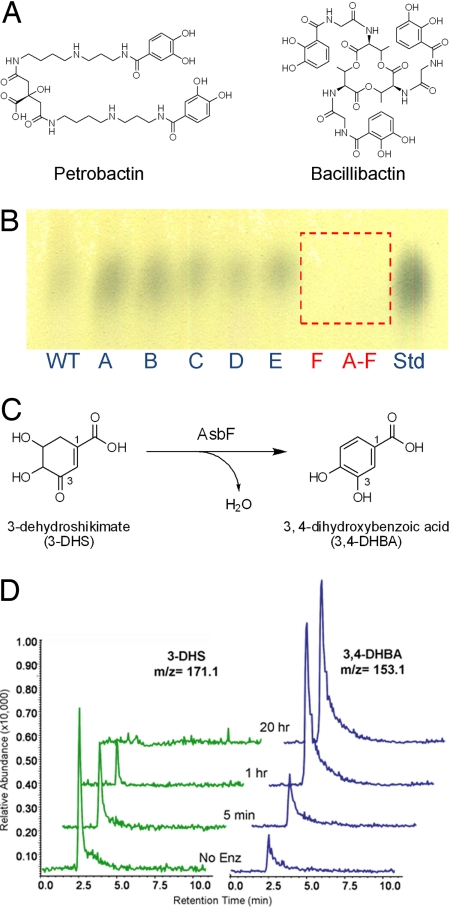
AsbF catalyzes dehydration of 3-DHS to 3,4-DHBA in B. anthracis. (A) Structures of petrobactin and bacillibactin, 2 siderophores of B. anthracis. (B) TLC analysis of culture supernatants from Δasb mutant strains stained with FeCl3. All strains generated a spot that comigrated with an authentic 3,4-DHBA standard except ΔasbABCDEF and ΔasbF. (C) Chemical transformation showing the dehydration of 3-DHS, a common bacterial metabolite, to generate the unique siderophore precursor 3,4-DHBA. (D) LC-MS traces of the AsbF reaction quenched at sequential time points to illustrate conversion of 3-DHS to 3,4-DHBA. No Enz, no enzyme control incubated for 20 h.
Biosynthesis of petrobactin in B. anthracis requires citrate, spermidine, and 3,4-DHBA as biosynthetic units (3, 9). Whereas citrate and spermidine are readily available primary metabolites in bacteria, the presence of a 3,4-DHBA in the precursor pool is unusual. Because 3,4-DHBA is critical for petrobactin to function as a virulence factor, we hypothesized that the asb operon would likely code for its synthesis. Indeed, it was indicated by earlier reports (10), by precursor incorporation studies (11), and through our own analysis that supernatants of a B. anthracis Sterne culture contain 3,4-DHBA. Specifically, we demonstrated that supernatants of an asbABCDEF mutant fail to accumulate this key petrobactin subunit, indicating that 1 or more asb-encoded polypeptides are involved in its synthesis. Further examination of supernatants isolated from cultures of individual asb deletion mutants (3) revealed that each strain is capable of producing 3,4-DHBA except ΔasbF. To substantiate its functional role, we sought to determine the biochemical function of recombinant AsbF exogenously expressed and purified from Escherichia coli. X-ray crystallographic analysis of AsbF provides structural details of this unique polypeptide and has enabled new insights into AsbF enzymatic activity and a proposed mechanism in formation of the 3,4-DHBA moiety essential to petrobactin biosynthesis. Moreover, this study identifies AsbF as a potential new target in efforts to identify inhibitors for development of effective therapeutics against B. anthracis and other pathogens where petrobactin operates as a stealth virulence factor.
Results and Discussion
The Requirement for AsbF Is Revealed Through Analysis of B. anthracis Mutant Strains.
The role of AsbF was initially assessed by determining the level of 3,4-DHBA production from various B. anthracis strains, including ΔasbA, ΔasbB, ΔasbC, ΔasbD, ΔasbE, ΔasbF, and ΔasbABCDEF (3), as well as a wild-type Sterne 34F2, that were grown in iron-depleted medium (IDM) (3). The presence of 3,4-DHBA was assayed by cellulose plate TLC on organic solvent extracts obtained from filtrates of the IDM cultures of each strain. TLC analysis revealed a single spot corresponding to the iron-chelating compound 3,4-DHBA evident in the extracts of B. anthracis Sterne 34F2 and the mutants ΔasbA, ΔasbB, ΔasbC, ΔasbD, and ΔasbE at Rf = 0.91–0.93 (Fig. 1B). In contrast, spots corresponding to the subunit were absent in the ΔasbF and ΔasbABCDEF strains. The ferri-DHBA complex is a strong chromophore, forming blue-colored complexes when sprayed with 1% FeCl3 after TLC development. Extract samples were further analyzed with HPLC by using a C18 reverse-phase semipreparative column. The retention time and UV spectrum of the corresponding 3,4-DHBA peaks in each sample were coincident with an authentic standard (data not shown). Peak areas were determined, quantified, and compared based on a standard curve constructed by injecting a series of 6 concentrations (ranging from 0 to 500 μg/mL) of authentic 3,4-DHBA [supporting information (SI) Table S1]. Analysis of these B. anthracis mutants provided compelling evidence for the function of the BA1986 gene product (AsbF) as the 3,4-DHBA synthase necessary for petrobactin production. Moreover, in a mouse model of inhalation anthrax (12), the ΔasbF mutant strain was found to be completely avirulent at day 10, whereas the parental Sterne strain (34F2) resulted in 50% fatality over the same period (Fig. S1).
Bioinformatic analysis revealed that the asb cluster is conserved in a variety of bacterial genomes, including both Gram-positive and Gram-negative organisms (Fig. S2A). Interestingly, these microbes do not share a common niche; they are found in diverse environments with marine species dominating. The significance of this trend is unclear because it is difficult to assess how widespread petrobactin synthesis is within marine bacterial genomes. However, the relative phylogenetic diversity of bacteria bearing an asb-type operon (Fig. S2B), together with evidence that these gene clusters appear to have been acquired long ago (they have diverged considerably in sequence, and each has assumed the global sequence characteristics of its host genomes), suggests that petrobactin may confer a competitive advantage in iron acquisition within the ocean environment as well. We expect that as the number of sequenced bacterial genomes increases, the evolutionary history of the asb cluster will become clearer, and clues to the significance of petrobactin in various habitats will emerge.
AsbF Is Responsible for Conversion of 3-Dehydroshikimate (3-DHS) to 3,4-DHBA.
To guide our initial studies of AsbF catalytic activity, bioinformatic analyses were conducted to identify potential functional homologs and to suggest reaction substrates. However, these efforts revealed only sequence similarity to conserved proteins of unknown function or irrelevant catalytic properties (data not shown). Alternatively, a search of the biochemical pathway databases revealed several anabolic processes leading to assembly of 3,4-DHBA, in addition to the many well studied catabolic pathways. In some bacteria (exemplified by Pseudomonas putida PobA), 3,4-DHBA is generated by hydroxylation of 4-hydroxybenzoic acid (4-HBA) to use the substrate as a carbon source (13). However, reaction of purified AsbF with 4-HBA and NAD(P)H failed to yield 3,4-DHBA (data not shown).
We considered an alternative metabolic process found in Neurospora crassa (qa-4), Aspergillus nidulans (QutC), and Klebsiella pneumoniae (aroZ) where 3,4-DHBA is generated by dehydration of 3-dehydroshikimate (3-DHS), a shikimate pathway intermediate, to use aromatic compounds as a carbon source (Fig. 1C) (14–16). A reaction mixture including AsbF with 3-DHS (purified from cultures of recombinant E. coli) (17) resulted in a product bearing a new absorbance maximum at 290 nm that corresponded to a DHBA chromophore (Fig. S3A). Upon analysis by LC-MS in negative SIM mode, the same reaction mixture resulted in disappearance of 3-DHS (m/z = 171) and generation of a product consistent with the structure of 3,4-DHBA (m/z = 153) (Fig. 1D). Both starting material and product eluted from a C18 reverse-phase column with retention times identical to authentic standards (data not shown).
Enzymatic Characterization.
With a clear catalytic role for AsbF identified as a 3-DHS dehydratase, we next sought to determine the efficiency of the AsbF-catalyzed 3-DHS dehydration (3,4-DHBA production) reaction. The reaction rate at room temperature increased monotonically with increasing pH (from 6.0 to 9.2) with a plateau between 7.2 and 8.0 (Fig. S3B). Earlier studies of a fungal 3-DHS dehydratase suggested a requirement for divalent cations (15, 16). Significantly, the AsbF-catalyzed dehydration reaction was inhibited at EDTA concentrations >0.1 mM. Supplementation of this experimental reaction mixture with 5 mM MgCl2 restored activity. After this observation, a suite of metals was titrated into reactions containing AsbF inactivated by EDTA. While a series of divalent cations including cobalt, calcium, and magnesium reactivated the enzyme, manganese was found to maximize catalytic efficiency whereas equal concentrations of zinc completely abrogated it. In conjunction with these biochemical findings, a fluorescent scan of crystallized recombinant AsbF showed manganese to be the predominant enzyme-bound metal (data not shown, see below). Similar dependencies, in which manganese is the preferred metal and zinc is inhibitory, are found among a structural class of sugar isomerases (18).
Kinetic parameters for AsbF at pH 7.5 were determined spectrophotometrically. Recorded initial velocities were fit to a Michaelis–Menten curve demonstrating the enzyme to have a Km of ≈290 μM and a kcat of ≈80 min−1. The Km value is on an order of magnitude similar to those reported by groups describing fungal dehydroshikimate dehydratases from N. crassa (590 μM) (15) and A. nidulans (530 μM) (16).
Structural Characterization of AsbF Bound to Its Product, 3,4-DHBA.
Because of the lack of sequence similarity to known 3-DHS dehydratases, we sought the crystal structure of AsbF and to explore a potential reaction mechanism based on these insights. The structure of AsbF adopts a (β/α)8-barrel (TIM barrel) configuration (Fig. 2A). As expected from a prior TEV protease cleavage experiment (Materials and Methods), the N terminus of the structure is partially buried at the bottom part of the barrel whereas the C terminus is completely exposed to the solvent channel. With the His6 tag in place, the C termini could be involved in lattice interactions with neighboring protein molecules, which might have led to twinning.
Fig. 2.
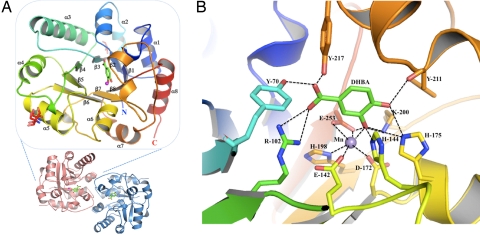
Structural analysis of AsbF. (A) Crystal structure form of AsbF. An overall view of the AsbF monomer is enlarged and rotated 90° relative to the dimeric structure below. Monomer is depicted with a color ramp of blue to red (N to C terminus). The manganese atom is shown as a purple sphere in the middle of the TIM barrel adjacent to the ligand molecule 3,4-DHBA (green). Numbering of main TIM barrel secondary structures is shown as α1–8 and β1–8. (B) Active site of liganded AsbF showing 3,4-DHBA coordinated to manganese interacting with side chains. Manganese is coordinated with 4 carboxyl group oxygen atoms from 2 glutamate residues and an aspartate as well as an amine of His ranging in distance between 2.08 and 2.38 Å. All hydrogen bonds are indicated by dashed lines.
AsbF belongs to AP endonuclease 2 TIM barrel proteins (19), with the closest homologues including xylose isomerase and the myoinositol catabolism protein IolI (20). The internal section of AsbF is well packed by relatively large side chains with a narrow bottom part of the barrel relative to the top. The barrel opens up at the top to form a pocket with a number of hydrophilic residues including His, Glu, Thr, Lys, Tyr, and Gln to contain a specific ligand. Unexpectedly, the 3,4-DHBA reaction product was found in the active/binding site of AsbF, surrounded by several aromatic amino acid residues including F255, F211, F104, H144, and Y217. The 3,4-DHBA molecule makes direct contacts with hydroxyl groups of Y70 and Y217, a guanidinium of R102, an amine of K200, an imine of H175, a carboxyl of E253, and the main-chain carbonyl of F211 using mainly hydrogen bonding interactions. Moreover, 3,4-DHBA is capped tightly under the helical loop (residues 206–224) located between the seventh β-strand (β7) and the seventh α-helix (α7) and is directly coordinated with a Mn2+ ion that is also found in the structure (Fig. 2A). Both 3,4-DHBA and a metal ion were evident from the electron density map calculation using experimental phases with the weighted coefficient (Fw). Metal identity was determined from the specific emission peak during the fluorescence spectrum scan. Inspection of the AsbF structure reveals that Mn2+ is coordinated with the carboxylic acid side chains of E142, D172, E253 (both oxygen atoms), the deprotonated imine of H198, and the 3-hydroxy group of 3,4-DHBA, with the distance range of 2.08–2.38 Å that is typical of Mn2+ ions (Fig. 2B) (21). Although there is only 1 AsbF protein molecule in the asymmetric unit, they apparently form a dimer (Fig. 2A) that excludes 2,460 Å of surface area according to PISA (22). Analysis by static light scattering and size-exclusion chromatography also supports a dimeric state of the enzyme in solution (data not shown).
Structural comparisons revealed that AsbF can be reasonably well superimposed with the 3-dehydroquinate dehydratase (DHQ) from Salmonella typhimurium (1qfe) (Fig. S4A), an enzyme that functions as part of the shikimate pathway for biosynthesis of aromatic compounds (19). Indeed the AsbF structure is generally homologous to this type of dehydratase enzyme comprised of a TIM barrel fold with a few variations, including the lid loop located at the top of the pocket. For AsbF, this lid loop extends from residue 202 to residue 227 as the partly α-helical loop between strands β7 and α7. In contrast, for the enzyme DHQ it comes from the loop between β3 and α3 spanning residues 82–93 covering approximately the opposite side of the pocket mouth. Another important difference in DHQ is an additional small β-ribbon at the C terminus that closes the bottom of the β-barrel. Each ligand in its corresponding binding pocket occupies a different location and orientation (Fig. S4A). One of the key distinctions between TIM barrel enzymes is the location of metal binding sites and the type of metal involved. The AsbF manganese ion is bound to 6 atoms, including 1 from 3,4-DHBA and 5 amino acid residues that may also contribute to catalysis by stabilizing an intermediate ligand and/or a product ligand differing from other dehydratases, particularly DHQ (such as 1qfe) that does not contain a metal cofactor (19).
Proposed Mechanism for AsbF Dehydratase.
With the crystal structure as a guide, we used site-directed mutagenesis to probe further the key amino acid residues involved in catalysis of AsbF. This analysis revealed that a K200R mutant form of the protein was partially active, confirming the lack of a covalent imine intermediate shown to occur in structurally similar DHQs (19). Characterization of additional AsbF site-directed mutants demonstrated (Table 1) that most inactive forms of the enzyme lacked residues believed to be crucial for metal or substrate coordination ascertained through inspection of the 3,4-DHBA orientation within the AsbF crystal structure (Fig. 2B).
Table 1.
Kinetic parameters for AsbF mutants
| Mutant | Km, μM | kcat, min−1 | % activity |
|---|---|---|---|
| WT | 288.72 ± 38.93 | 79.84 ± 0.96 | 100 |
| K200E | — | — | Inactive |
| K200A | — | — | Inactive |
| K200R | 30.25 ± 8.48 | 2.56 ± 0.05 | 3.2 |
| H144A | — | — | Inactive |
| H175A | — | <1 | Trace |
| R102A | — | — | Inactive |
| Y70A,Y217A | ≈2,511 | 48.35 ± 3.82 | 60.6 |
Site-directed mutagenesis was performed on BA 1986 (asbF) to probe for residues involved in catalysis. The Km of WT AsbF is comparable to 2 eukaryotic 3-DHS dehydratases. Inactive mutants are presumably disrupted in coordination of the metal or substrate. Replacement of Y70 and Y217 is likely to expose the active site to solvent and reduce substrate specificity while loss of the proposed catalytic H144 renders the enzyme inactive.
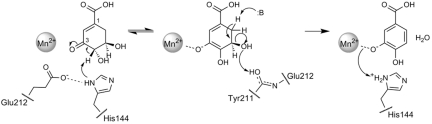
Based on the crystal structure and mutant analysis, the surrounding amino acid residues comprising the putative AsbF active site suggest an E1CB (elimination unimolecular via conjugate base) mechanism that requires a base to abstract the axial proton from the adjacent aliphatic carbon atom (C4) of 3-DHS (Fig. 3). In the AsbF structure, H144 is orthogonal to the catechol ring of the 3,4-DHBA product, an ideal position for removal of the C4 axial proton (the hydroxyl is equatorial) of the substrate. H144 and E142 are both well conserved among members of the xylose isomerase family (Fig. S5). The nearby E142 could elevate basicity of H144 by forming a hydrogen bond to NE2 of H144, improving its proton abstraction ability. Histidine functions as a base for initial proton removal during many enzymatic reactions, including some type I 3-dehydroquinate dehydratases (23, 24). Furthermore, although the activity of a H175A mutant is attenuated, the adjacent AsbF H144A mutant is completely inactive. Proton removal from the C4 position of 3-DHS leads to formation of a noncovalently bound enolate intermediate with the resulting negative charge stabilized by the divalent Mn2+ cation.
Fig. 3.
Proposed mechanism for AsbF-catalyzed conversion of 3-DHS to 3,4-DHBA. Mn2+ stabilizes an enolate intermediate after proton removal at C4 by H144. This is followed by dehydration/aromatization of the molecule to generate the key petrobactin subunit 3,4-DHBA.
To assess enolate formation as a first step in conversion of 3-DHS to 3,4-DHBA, we conducted a deuterium labeling experiment using the AsbF reaction in D2O buffer. Rapid and reversible exchange of deuterium from the conversion of C3 keto to enol (Fig. 3) was expected to result in accumulation of the substrate bearing a 1 mass unit increase detectable by Fourier-transform ion cyclotron resonance MS. This rapid exchange was readily detectable (Fig. S6), supporting the proposed mechanistic hypothesis regarding initial formation of the 3-DHS enolate upon substrate binding to AsbF.
In a second, more rapid step of the reaction, dehydration presumably occurs through a general acid-catalyzed elimination of the C5 hydroxyl and product formation after aromatization (Fig. 3). The most likely candidate for the acid-catalyzed removal of the C5 hydroxyl is the main-chain carbonyl of Y211 located at the C terminus of the short α-helix as a part of the helical loop between β7 and α7 capping the binding pocket. A main-chain carbonyl is generally not a good acid, but the main-chain carbonyl of Y211 is the closest to C5, and its acidity is likely to be elevated by positioning at the C terminus of the α-helix (with partial negative charge from the helix dipole) that includes conserved amino acid residues among the family (Fig. 2B).
To understand further the role of specific active-site residues in catalysis, we modeled 3-DHS in place of the 3,4-DHBA in AsbF (Fig. S4B). This motivated analysis of Y217 and Y70, which were initially believed to be necessary for excluding solvent from the active site. Both were found to be inconsequential for normal enzymatic function when mutated individually and reduced activity by only 40% when the corresponding double Tyr mutation was constructed (Table 1). Unexpectedly, an R102A mutation completely inactivates AsbF indicating its role in maintaining a proper active-site conformation, whereas substitution of K200 for arginine significantly reduces substrate binding and overall catalytic efficiency.
Elucidation of the AsbF Structure and Function Is Important to Understanding Siderophore Biosynthesis of B. anthracis.
Recently, other pathogenic Bacillus sp. were found to produce petrobactin (6), whereas the ability for uptake and utilization of exogenous petrobactin is even more widespread among prokaryotes (25). Moreover, some evidence suggests that the siderophore or its analogs play additional biological roles beyond iron sequestration (26–28). Although we have now characterized the enzymatic activity of AsbF as well as its role in iron acquisition, the complete nature of the enzyme within bacteria has yet to be fully understood. Indeed research on other members of the asb gene cluster suggests that a pool of 3,4-DHBA, and thus functioning AsbF, is necessary for the initial steps of petrobactin biosynthesis (3, 9).
The entire asb gene cluster exemplifies the coevolution of a pathogen with its host. AsbF itself, with its metal coordination structure reminiscent of other proteins, most likely evolved from enzymatic precursors with a purpose very different from siderophore biosynthesis. The presence of the unusual 3,4 isomer of DHBA in petrobactin is vital to its ability to acquire iron for the bacterium while avoiding detection by innate host defenses. Without petrobactin, B. anthracis would be limited to the ability of bacillibactin to compete for free iron, a process that is highly compromised because of siderocalin sequestration (7). Because the generation of 3,4-DHBA is a key step for petrobactin biosynthesis (9), AsbF represents an attractive target for small-molecule antibiotic therapy. In addition to the prevention of petrobactin assembly, disrupting AsbF function would thwart uptake of any iron bound by 3,4-DHBA subunits that may be exported out of the cell (9). This approach is supported by the fact that B. anthracis cells carrying an asbF gene deletion grow significantly more poorly in iron-depleted media (3) and display a highly attenuated lethality to mice (Fig. S1). The search for AsbF inhibitors with possible therapeutic application is already underway. Questions regarding how AsbF is regulated and the precise timing and order of 3,4-DHBA incorporation into petrobactin remain to be addressed. This research will serve to further articulate how even insidious microbes are fundamentally reliant on finding a method to surpass innate immune defenses in the acquisition of nutrients and will likely uncover innovative strategies for combating life-threatening infections.
Materials and Methods
Analysis of Sterne Strain Mutants.
B. anthracis asb mutant strains ΔasbA, ΔasbB, ΔasbC, ΔasbD, ΔasbE, ΔasbF, and ΔasbABCDEF (3) as well as a wild-type Sterne 34F2 were generated as described previously and grown in IDM (3). Filtrates (100 mL) of the growth medium were prepared as described previously (3). Sample preparation for TLC analysis and quantification of 3,4-DHBA were performed by extracting acidified (pH 2.0) IDM culture supernatants (10 mL) with an equal volume of ethyl acetate 3 times. The organic layers were pooled, and the solvents were concentrated to dryness in vacuo. The dried pellets were dissolved in 50 μL of methanol and used for analysis. TLC was performed with the extracts on a cellulose plate (Merck). TLC plates were developed with a solvent system of butanol-acetic acid-water (12:3:5, vol/vol/vol), and 3,4-DHBA was detected by spraying with 1% (wt/vol) ferric chloride.
For quantification of 3,4-DHBA (Table S1), samples were analyzed by HPLC. Analytical results and detailed methods are available online in the Table S1 legend.
Intratracheal Inoculation of DBA/2 Mice.
Mouse infection was performed as described previously (12). All mouse experiments were performed by using protocols approved by the University of Michigan Committee on the Use and Care of Animals.
Cloning and DNA Manipulation.
The asbF gene of the B. anthracis Sterne strain was cloned into the expression vector pMCSG7 (29) by using ligation-independent cloning (30). Detailed cloning and mutagenesis methods are available in SI Methods.
Bioinformatic Analysis.
Bioinformatic analyses of the B. anthracis asb cluster and homologs were performed with tools implemented within the Integrated Microbial Genomes web site (http://img.jgi.doe.gov) and the EMBL-EBI web site (www.ebi.ac.uk).
Protein Purification for Enzymatic Analysis.
All pMCSG7-asbF constructs were subsequently used to transform BL21 expression cells (Novagen). The enzyme was purified by using standard His-tag Ni-affinity chromatography. Detailed protein purification methods are available in SI Methods.
Enzymatic Assays.
Initial substrate determination assays were performed with 75 mM Hepes (pH 7.5), 100 mM substrate (3-DHS), and 1 μM enzyme. Fifty-microliter reactions were tracked spectrophotometrically by observing ΔA290 using a SpectraMax M5 plate reader (Molecular Devices). Standard serial dilutions of synthesized 3,4-DHBA (Fluka) in similar reaction conditions were used to quantify product yield. LC-MS analysis of substrate to product conversion was performed by addition of 9 reaction volumes of methanol followed by evaporation of the resulting supernatant and loading onto a 3.5-μm C18 reverse-phase column (Waters) in line with a Shimadzu LCMS-2010A for detection. Elution was performed with a stepwise 5–70% gradient of MeCN in 20 mM aqueous NH4HCO3. Changes to reaction conditions were performed as noted in the text. Characterization of kinetic parameters was performed with 75 mM Hepes (pH 7.5), 250 nM enzyme, and various concentrations (0–2 mM) of 3-DHS in 80-μL reactions. Both substrate and enzyme were equilibrated in reaction buffer at ambient temperature for 15 min before the reaction was initiated by mixing. 3,4-DHBA production was tracked by observing ΔA290 using a Flexstation III plate reader (Molecular Devices). Reactions were run in quadruplicate, and nonlinear regression using Kaleidagraph software (Synergy) was performed on a plot of V0 vs. [3-DHS] for determination of Km and Vmax.
Gene Cloning, Expression, and Protein Purification for Structure Determination.
After failing to obtain diffraction-quality crystals from the initial pMCSG7-asbF construct, asbF was cloned in pMCSG26 vectors with 2 variations, including an uncleavable C-terminal His6 tag and a C-terminal His6 tag with a TEV protease cleavage site. Recombinant E. coli cells for AsbF overexpression were grown by using selenomethionine (Se-Met)-containing enriched M9 medium (pink medium) under conditions known to inhibit methionine biosynthesis (31, 32). Protein was purified by 2-step Ni-affinity chromatography following the standard protocol described previously (33). Detailed cloning, overexpression, and purification methods are available in SI Methods.
Protein Crystallization.
Crystallization screening was executed for all AsbF protein variants, including the one bearing an N-terminal His6 tag (expressed from pMCSG7) and the ones from pMCSG26 vectors with and without the C-terminal His6 tag cleavage. They were screened for crystallization conditions by using a Mosquito robot (TTP Labtech) using sitting drops in 96-well plates (Greiner), with drops of 0.4 μL of protein at 60 mg/mL mixed with 0.4 μL of crystallization screen solutions and equilibrated over 135 μL of crystallization screen solutions in the well at 18 °C. A number of commercially available screens including Index (Hampton Research) and ANL-1 and ANL-2 (Qiagen) were used for crystallization screening. AsbF with N-terminal His6 tag failed to yield a diffraction-quality crystal. The best crystals of AsbF without the TEV cleavage site appeared after 1–2 days from Index #25 containing 3.5 M sodium formate (pH 7.0). The same condition and a few others including ANL-2 #88 and #89 gave diffraction-quality crystals for the AsbF without the C-terminal His6 tag. Before data collection crystals were flash-frozen in liquid nitrogen in the presence of a number of different cryoprotectants. The chunky plate shape of a 0.15-mm side crystal of the AsbF with the C-terminal tag diffracted to 2.0 Å. However, the data were twinned, and the phasing stalled. One of the rod-shape crystals of AsbF lacking a His6 tag from condition ANL-2 #89 (0.1 M Tris, pH 8.5/3.2 M sodium chloride) with the saturated sucrose cryoprotectant diffracted to 2.12 Å and was used for data collection.
Data Collection, Structure Determination, Refinement, and Deposition.
The single-wavelength anomalous dispersion (SAD) data near the Se absorption edge (0.9793Å) up to 2.12 Å were collected from a single Se-Met-labeled rod-shape protein (30 × 30 × 300 μm) crystal at 100 K at the 19ID beam line of the Structural Biology Center at the Advanced Photon Source, Argonne National Laboratory. The crystal was exposed for 5 s per 1.0° rotation of ω with the crystal to detector distance of 320 mm. The data were recorded on an ADSC Q315 detector by the scanning of 200°. The space group was P6522 with cell dimension of a = b = 134.48 Å and c = 72.81 Å. All data were processed and scaled with HKL3000 (34) (Table 2).
Table 2.
AsbF data collection, phasing and refinement
| Data Collection (anomalous peak) | |
|---|---|
| Wavelength, Å | 0.9793 |
| Resolution (last shell), Å | 49.4 to 2.11 (2.19 to 2.11) |
| Reflections measured/unique | 22,726/2,210 |
| Multiplicity | 13.9 (12.3) |
| Completeness (last shell), % | 99.9 (99.4) |
| R sym (last shell), %* | 14.4 (53.4) |
| I/σ (last shell) | 9.1 (7.1) |
| Phasing | |
| Resolution, Å | 49.4 to 2.12 |
| Phasing power | 1.06 |
| Figure of merit | 0.112 |
| After density modification | 0.801 |
| Refinement | |
| Resolution, Å | 49.4 to 2.12 |
| R work/R free† | 14.8/17.8 |
| rms deviation bond lengths, Å | 0.017 |
| rms deviation bond angles, Å | 1.47 |
| PDB code | 3DX5 |
*Rsym = ΣΣj Ij − <I> /ΣIj, where Ij is the intensity measurement for reflection j, and <I> is the mean intensity for multiply recorded reflections.
†Rwork/Rfree = Σ∥Fobs − Fcalc∥/Fobs, where the working and free R factors are calculated by using the working and free reflections sets. The free reflections (5% of the total) were held aside throughout refinement.
The structure was determined by SAD phasing using the anomalous signal from Se atoms with HKL3000 (34), SHELX (35), SOLVE/RESOLVE (36), MLPHARE (37), DM (38), CCP4 (39), and arp/Warp (40) using the peak data to 2.12 Å. The model from HKL3000 after the high-resolution model building routine by arp/Warp contained most of the residues with side chains except 4 N-terminal residues, 7 C-terminal residues, and all artificial residues from cloning. The extra electron density for the 3,4-DHBA was evident from the electron density map calculated from the experimental phases. The subsequent refinement was performed iteratively by REFMAC 5.2 (41) in CCP4 and manual adjustment using Coot (42) until it converged to the R factor of 0.148 and the free R of 0.178 with the rms bond distances of 0.017 and the rms bond angles of 1.47°. The final model included residues 1–274 of 1 chain of AsbF, 1 molecule of 3,4-DHBA, a Mn ion, a disordered Tris, a glycerol molecule, and 320 ordered water molecules. The C terminus residues including those introduced during cloning could not be resolved. The stereochemistry of the structure was checked with PROCHECK (43) and the Ramachandran plot. Atomic coordinates and experimental structure factors of AsbF have been deposited in the Protein Data Bank under ID code 3DX5.
Supporting Information.
For full experimental details, see SI Methods, Table S1, and Figs. S4–S6.
Supplementary Material
Acknowledgments.
We are grateful to Prof. John Frost for providing 3-DHS for assays; Prof. Carol Fierke for discussions; William Eschenfeldt and Lucy Stols for creating the vectors for this project; and Prof. Jason Gestwicki, Liangcai Gu, Nicole B. Lopanik, Tom McQuade, Zach A. Beck, Doug A. Burr, Ellen Swenson, and Robin Guo for their contributions. We thank members of the Structural Biology Center at Argonne National Laboratory for their help with data collection at the 19ID beamline. This work was supported by a development grant from the Great Lakes Regional Center of Excellence for Bio-defense and Emerging Infectious Diseases Research (Grant U54AI57153), by the Hans W. Vahlteich Professorship (to D.H.S.), and by National Institutes of Health Grant HHSN266200400059C/N01-AI-40059 (to P.C.H.). B.F.P. and J.B.S. are recipients of a Great Lakes Regional Center of Excellence postdoctoral training fellowship. Y.K. and N.M. were supported by National Institutes of Health Grant GM074942 and the U. S. Department of Energy, Office of Biological and Environmental Research, under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3DX5).
This article contains supporting information online at www.pnas.org/cgi/content/full/0808118105/DCSupplemental.
References
- 1.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.Koppisch AT, et al. Petrobactin is the primary siderophore synthesized by Bacillus anthracis str. Sterne under conditions of iron starvation. Biometals. 2005;18:577–585. doi: 10.1007/s10534-005-1782-6. [DOI] [PubMed] [Google Scholar]
- 3.Lee JY, et al. Biosynthetic analysis of the petrobactin siderophore pathway from Bacillus anthracis. J Bacteriol. 2007;189:1698–1710. doi: 10.1128/JB.01526-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson MK, Abergel RJ, Raymond KN, Arceneaux JE, Byers BR. Siderophores of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Biochem Biophys Res Commun. 2006;348:320–325. doi: 10.1016/j.bbrc.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 5.Cendrowski S, MacArthur W, Hanna P. Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol Microbiol. 2004;51:407–417. doi: 10.1046/j.1365-2958.2003.03861.x. [DOI] [PubMed] [Google Scholar]
- 6.Koppisch AT, et al. Petrobactin is produced by both pathogenic and non-pathogenic isolates of the Bacillus cereus group of bacteria. Biometals. 2008;21:581–589. doi: 10.1007/s10534-008-9144-9. [DOI] [PubMed] [Google Scholar]
- 7.Abergel RJ, et al. Anthrax pathogen evades the mammalian immune system through stealth siderophore production. Proc Natl Acad Sci USA. 2006;103:18499–18503. doi: 10.1073/pnas.0607055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischbach MA, Lin H, Liu DR, Walsh CT. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol. 2006;2:132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 9.Pfleger BF, et al. Characterization and analysis of early enzymes for petrobactin biosynthesis in Bacillus anthracis. Biochemistry. 2007;46:4147–4157. doi: 10.1021/bi6023995. [DOI] [PubMed] [Google Scholar]
- 10.Garner BL, Arceneaux JE, Byers BR. Temperature control of a 3,4-dihydroxybenzoate (protocatechuate)-based siderophore in Bacillus anthracis. Curr Microbiol. 2004;49:89–94. doi: 10.1007/s00284-004-4286-7. [DOI] [PubMed] [Google Scholar]
- 11.Koppisch AT, et al. Biosynthesis of the 3,4-dihydroxybenzoate moieties of petrobactin by Bacillus anthracis. J Org Chem. 2008;73:5759–5765. doi: 10.1021/jo800427f. [DOI] [PubMed] [Google Scholar]
- 12.Fisher N, et al. The dltABCD operon of Bacillus anthracis Sterne is required for virulence and resistance to peptide, enzymatic, and cellular mediators of innate immunity. J Bacteriol. 2006;188:1301–1309. doi: 10.1128/JB.188.4.1301-1309.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosokawa K, Stanier RY. Crystallization and properties of p-hydroxybenzoate hydroxylase from Pseudomonas putida. J Biol Chem. 1966;241:2453–2460. [PubMed] [Google Scholar]
- 14.Frost JW, Draths KM. Biocatalytic syntheses of aromatics from D-glucose: Renewable microbial sources of aromatic compounds. Annu Rev Microbiol. 1995;49:557–579. doi: 10.1146/annurev.mi.49.100195.003013. [DOI] [PubMed] [Google Scholar]
- 15.Stroman P, Reinert WR, Giles NH. Purification and characterization of 3-dehydroshikimate dehydratase, an enzyme in the inducible quinic acid catabolic pathway of Neurospora crassa. J Biol Chem. 1978;253:4593–4598. [PubMed] [Google Scholar]
- 16.Wheeler KA, Lamb HK, Hawkins AR. Control of metabolic flux through the quinate pathway in Aspergillus nidulans. Biochem J. 1996;315:195–205. doi: 10.1042/bj3150195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi J, Li K, Draths KM, Frost JW. Modulation of phosphoenolpyruvate synthase expression increases shikimate pathway product yields in E. coli. Biotechnol Prog. 2002;18:1141–1148. doi: 10.1021/bp020101w. [DOI] [PubMed] [Google Scholar]
- 18.Fuxreiter M, Farkas O, Naray-Szabo G. Molecular modelling of xylose isomerase catalysis: The role of electrostatics and charge transfer to metals. Protein Eng. 1995;8:925–933. doi: 10.1093/protein/8.9.925. [DOI] [PubMed] [Google Scholar]
- 19.Gourley DG, et al. The two types of 3-dehydroquinase have distinct structures but catalyze the same overall reaction. Nat Struct Biol. 1999;6:521–525. doi: 10.1038/9287. [DOI] [PubMed] [Google Scholar]
- 20.Zhang RG, et al. Crystal structure of Bacillus subtilis IolI shows endonuclase IV fold with altered Zn binding. Proteins. 2002;48:423–426. doi: 10.1002/prot.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng H, Chruszcz M, Lasota P, Lebioda L, Minor W. Data mining of metal ion environments present in protein structures. J Inorg Biochem. 2008;102:1765–1776. doi: 10.1016/j.jinorgbio.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Deka RK, Kleanthous C, Coggins JR. Identification of the essential histidine residue at the active site of Escherichia coli dehydroquinase. J Biol Chem. 1992;267:22237–22242. [PubMed] [Google Scholar]
- 24.Leech AP, et al. Re-evaluating the role of His-143 in the mechanism of type I dehydroquinase from Escherichia coli using two-dimensional 1H,13C NMR. J Biol Chem. 1998;273:9602–9607. doi: 10.1074/jbc.273.16.9602. [DOI] [PubMed] [Google Scholar]
- 25.Abergel RJ, Zawadzka AM, Raymond KN. Petrobactin-mediated iron transport in pathogenic bacteria: Coordination chemistry of an unusual 3,4-catecholate/citrate siderophore. J Am Chem Soc. 2008;130:2124–2125. doi: 10.1021/ja077202g. [DOI] [PubMed] [Google Scholar]
- 26.Gardner RA, Kinkade R, Wang C, Phanstiel O. Total synthesis of petrobactin and its homologues as potential growth stimuli for Marinobacter hydrocarbonoclasticus, an oil-degrading bacteria. J Org Chem. 2004;69:3530–3537. doi: 10.1021/jo049803l. [DOI] [PubMed] [Google Scholar]
- 27.Harris WR, Amin SA, Kupper FC, Green DH, Carrano CJ. Borate binding to siderophores: Structure and stability. J Am Chem Soc. 2007;129:12263–12271. doi: 10.1021/ja073788v. [DOI] [PubMed] [Google Scholar]
- 28.Hickford SJ, et al. Petrobactin sulfonate, a new siderophore produced by the marine bacterium Marinobacter hydrocarbonoclasticus. J Nat Prod. 2004;67:1897–1899. doi: 10.1021/np049823i. [DOI] [PubMed] [Google Scholar]
- 29.Stols L, et al. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expression Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 30.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 32.Walsh MA, Dementieva I, Evans G, Sanishvili R, Joachimiak A. Taking MAD to the extreme: Ultrafast protein structure determination. Acta Crystallogr D. 1999;55:1168–1173. doi: 10.1107/s0907444999003698. [DOI] [PubMed] [Google Scholar]
- 33.Kim Y, et al. Automation of protein purification for structural genomics. J Struct Funct Genomics. 2004;5:111–118. doi: 10.1023/B:JSFG.0000029206.07778.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: The integration of data reduction and structure solution—From diffraction images to an initial model in minutes. Acta Crystallogr D. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 35.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 36.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otwinowski Z. In: Wolf W, Evans PR, Leshe AGW, editors. Daresbury Study Weekend Proceedings; Warrington, UK: SERC Daresbury Laboratory; 1991. pp. 80–85. [Google Scholar]
- 38.Cowtan KKC. An automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newslett Protein Crystallogr. 1994;31:34–38. [Google Scholar]
- 39.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 40.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 41.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.