Abstract
Transcriptional control of metabolic circuits requires coordination between specific transcription factors and coregulators and is often deregulated in metabolic diseases. We characterized here the mechanisms through which the coactivator SRC-3 controls energy homeostasis. SRC-3 knock-out mice present a more favorable metabolic profile relative to their wild-type littermates. This metabolic improvement in SRC-3−/− mice is caused by an increase in mitochondrial function and in energy expenditure as a consequence of activation of PGC-1α. By controlling the expression of the only characterized PGC-1α acetyltransferase GCN5, SRC-3 induces PGC-1α acetylation and consequently inhibits its activity. Interestingly, SRC-3 expression is induced by caloric excess, resulting in the inhibition of PGC-1α activity and energy expenditure, whereas caloric restriction reduces SRC-3 levels leading to enhanced PGC-1α activity and energy expenditure. Collectively, these data suggest that SRC-3 is a critical link in a cofactor network that uses PGC-1α as an effector to control mitochondrial function and energy homeostasis.
Keywords: acetyltransferase, caloric restriction, cofactors, deacetylase, SIRT1
Transcriptional control is achieved through an interwoven and redundant molecular circuitry that involves individual transcription factors, the basal transcriptional machinery, and multiprotein coregulator complexes, which fine-tune metabolic homeostasis. Most of the coregulators interact directly with transcription factors and can either repress or enhance their transcriptional activities (1). Aberrant signaling by coregulators is known to generate abnormalities of cellular metabolism, and hence can contribute to abnormalities of systemic metabolic pathways and to the pathogenesis of several common disorders, such as obesity and type 2 diabetes (T2DM) (2). Steroid receptor coactivators (SRCs), also known as p160 proteins, which include SRC-1/NCoA-1, SRC-2/GRIP1/TIF2/NCoA2 and SRC-3/p/CIP/AIB1/ACTR/RAC3/TRAM-1, are transcriptional coactivators that interact with nuclear receptors and enhance their transactivation in a ligand-dependent manner (3–5). The main support for a role of the p160 proteins in the control of metabolism came from the characterization of various knock-out animal models. SRC-2−/− mice are protected against diet-induced obesity because of an enhanced energy expenditure (EE) in brown adipose tissue (BAT) (6) and in skeletal muscle (unpublished data). SRC-2−/− mice furthermore have an improved glucose tolerance and insulin sensitivity and a decreased adipocyte differentiation. In sharp contrast to SRC-2−/− mice, SRC-1−/− mice are prone to obesity due to reduced EE (6); SRC-3 is best known for its effects on cell proliferation and tumorigenesis (7, 8). The fact that SRC-3 decreases adipocyte differentiation (9, 10) and that the double SRC-1/3−/− mice exhibit an increased basal metabolic rate (10) suggested an impact of SRC-3 in metabolic control. The exact mechanisms through which SRC-3 affects metabolism, however, have not yet been fully characterized.
We here investigated the potential role of SRC-3 in energy homeostasis. As SRC-3−/− mice display clear signs of increased EE, subsequent to enhanced mitochondrial function in BAT and skeletal muscle, their body weight was lower and lipid and glucose homeostasis was improved. Mitochondrial activation in the absence of SRC-3 was caused by the deacetylation and increase in activity of the PPARγ coactivator-1α (PGC-1α), which coordinates mitochondrial function (11, 12). SRC-3 facilitates the acetylation and the consecutive inactivation of PGC-1α (13), through its effect on the expression of GCN5, the prime PGC-1α acetyltransferase (14). Furthermore, the expression of both SRC-3 and GCN5 is sensitive to fasting and high fat (HF) feeding, which subsequently, through its effect on PGC-1α-mediated EE, fine-tunes energy homeostasis. Taken together, these observations implicate SRC-3 as a critical component of a cofactor signaling network that converges on PGC-1α to control energy homeostasis.
Results
SRC-3−/− Mice Are Lean and Protected Against Diet-Induced Diabesity.
To characterize the metabolic role of SRC-3, we monitored weight evolution of SRC-3+/+ and SRC-3−/− mice for three months. After weaning, SRC-3−/− mice weighed already significantly less. However, they gained weight at an rate equivalent to the SRC-3+/+ mice when fed chow (Fig. 1A). Strikingly, when fed an HF diet, SRC-3−/− mice gained less weight as compared to their WT littermates (Fig. 1A). These differences in body weight do not correlate with any change in caloric intake, spontaneous locomotor activity, or absorption [supporting information (SI) Fig. S1 A and B and data not shown] and are accounted for by a reduced fat mass accretion. Indeed, SRC-3−/− mice had a significantly lower body fat content than WT mice, as measured by dual energy x-ray absorptiometry (Fig. S1C) and as mirrored by the reduced weight of the epididymal white adipose tissue (WAT) and intrascapular BAT depots (Fig. S1D, ref. 10), whether they were fed chow or an HF diet.
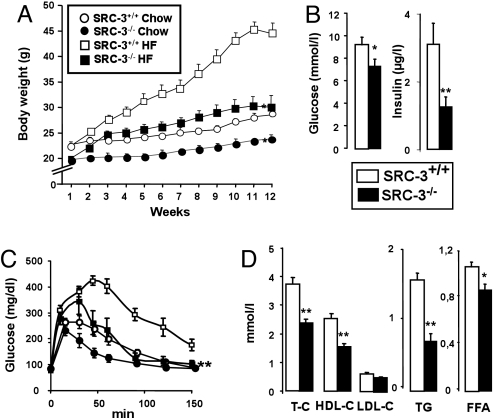
Fig. 1.
SRC-3−/− mice are lean with an increased insulin sensitivity and an improved lipid profile compared to SRC3+/+ mice (A) Body weight evolution of mice on either chow or HF diet (n = 8). (B) Serum glucose and insulin levels in mice fed an HF diet after a 6 h fasting (n = 8). (C) Serum glucose levels evolution during an i.p. glucose tolerance test performed after a glucose load (2g/kg) in chow or HF fed, overnight-fasted mice (n = 8). (D) Serum levels of total cholesterol (T-C), HDL-cholesterol (HDL-C), LDL-cholesterol (LDL-C), triglycerides (TG) and free fatty acids (FFA) in mice fed an HF diet (n = 8). *(P < 0.05) and **, P < 0.01 indicates significant differences as compared to SRC-3+/+ mice.
The reduction in body weight in SRC-3−/− mice is accompanied by an improved metabolic control as evidenced by the lower fasting plasma glucose and insulin levels (Fig. 1B). Furthermore, SRC-3−/− mice cleared glucose more effectively after i.p. glucose injection than SRC-3+/+ animals, in chow or HF fed conditions (Fig. 1C). The rate of glucose clearance upon i.p. insulin injection was also higher in SRC-3−/− mice, further suggesting insulin sensitization (Fig. S1E).
Another indication of an improved metabolic profile was provided by the lower fasting cholesterol, triglycerides, and free fatty acids levels in SRC-3−/− relative to WT mice (Fig. 1D). In addition, whereas the liver weight of SRC-3−/− and SRC-3+/+ mice under chow was indistinguishable, the livers of SRC-3−/− animals weighed less (Fig. S1D) and were smaller and less pale (Fig. S1F) when fed an HF diet, indicating an increased metabolic activity and a decreased lipid accumulation. Similar conclusions were derived from the analysis of hematoxylin and eosin (HE) stained liver sections of HF-fed animals and were confirmed by oil red O staining (Fig. S1F). Taken together these data suggest that the absence of SRC-3 improves both lipid and glucose homeostasis and induces a more favorable metabolic profile.
Energy Expenditure Is Enhanced in SRC-3−/− Mice.
Since changes in caloric intake or locomotor activity (Fig. S1 A and B) could not explain the weight differences, we investigated whether the protective effects against obesity observed in SRC-3−/− mice were mediated by an increase in basal metabolism. The higher O2 consumption and CO2 production evident in SRC-3−/− mice were consistent with an increased EE. The respiratory quotient was furthermore lower in SRC-3−/− mice, suggesting an increased fatty acid oxidation (FAO) (Fig. 2A).
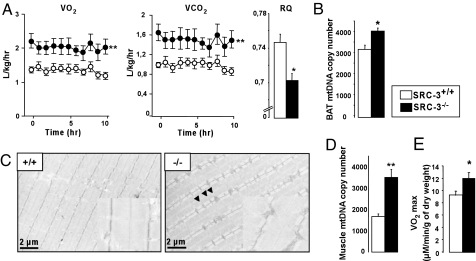
Fig. 2.
Enhanced energy expenditure and endurance in SRC-3−/− mice relative to SRC-3+/+ mice (A) O2 consumption (VO2) and CO2 production (VCO2) in mice fed regular chow. The respiratory quotient (RQ = VCO2/VO2) was calculated between 2 and 3 a.m. (n = 8). (B) Mitochondrial DNA (mtDNA) copy number in brown adipose tissue from mice fed an HF diet. (C) Representative pictures of gastrocnemius muscle sections from mice fed an HF diet, analyzed by electron microscopy. The inset shows an enlarged image of the muscle. Mitochondria are indicated by arrowheads. (D) Mitochondrial DNA copy number in the gastrocnemius muscle from SRC-3+/+ and SRC-3−/− mice fed an HF diet. (E) Maximum O2 consumption measured ex vivo in isolated gastrocnemius fibers of SRC-3+/+ and SRC-3−/− mice fed a chow in the presence of increasing ADP concentration (n = 5). *, P < 0.05 and **, P < 0.01 indicates significant differences as compared to SRC-3+/+ mice.
To identify the site(s) responsible for the increased EE, we performed a detailed histological analysis of key metabolic tissues. Transmission electron microscopy analysis of BAT sections showed significantly smaller lipid vacuoles and more mitochondria in SRC-3−/− mice (Fig. S2A). This amplification of the mitochondria was reflected by the increase in BAT mitochondrial DNA (mtDNA) content (Fig. 2B). Furthermore, the brown adipocytes size was reduced in SRC-3−/− mice, as assessed by an HE staining (Fig. S2B). Finally, consistent with the general increase in EE and with the BAT morphology, rectal temperature of SRC-3−/− mice remained significantly higher upon cold exposure, demonstrating an enhanced adaptive thermogenesis (Fig. S2C).
The number and the size of mitochondria were also increased in SRC-3−/− mice muscle (Fig. 2C). This was confirmed by immunohistochemical studies using an antibody directed against Optic Atrophy 1, a protein of the inner mitochondrial membrane (15), which showed a more intense staining of the SRC-3−/− mice muscle sections (Fig. S2D), as well as by the robust increase in muscle mtDNA content (Fig. 2D). In line with the morphological aspect of the muscle, the distance run to exhaustion on a treadmill was significantly increased in SRC-3−/− mice (Fig. S2E). Moreover, the maximum O2 consumption monitored in isolated muscle fibers was significantly higher in SRC-3−/− mice, testifying to an increased oxidative capacity (Fig. 2E). These results together indicate a major contribution of BAT and muscle to the enhanced EE observed in SRC-3−/− mice.
SRC-3 Controls PGC-1α Activity by Modifying Its Acetylation Status.
To further understand the molecular role of SRC-3 in the regulation of EE, we analyzed, in BAT and muscle, the expression of a set of representative genes known to be involved in mitochondrial function. With few exceptions, the expression of all of the transcription factors and genes that control FAO and oxidative phosphorylation (OXPHOS) that we analyzed was up-regulated in the BAT and muscle of SRC-3−/− compared to WT mice, confirming our phenotypic observations (Fig. 3A). Most notable was the change in the expression of PGC-1α, the master regulator of mitochondrial activity (11).
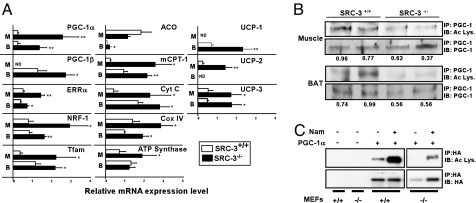
Fig. 3.
SRC-3 affects PGC-1α acetylation and activity. (A) mRNA levels of PGC-1α, PGC-1β, ERRα, NRF-1, Tfam, mCPT-1, ACO, Cyt C, Cox IV, ATP synthase, UCP-1, UCP-2, UCP-3, SRC-1 and SRC-2 in gastrocnemius muscle (Muscle) and brown adipose tissue (BAT) of SRC-3+/+ and SRC-3−/− mice determined by quantitative PCR (n = 8). (B) Acetylation level of PGC-1 in the gastrocnemius muscle and brown adipose tissue (BAT) of 2 representative SRC-3+/+ and SRC-3−/− mice fed a high fat diet. PGC-1 was first immunoprecipitated (IP). Total and acetylated PGC-1 were then detected through an immunoblot (IB) using anti-PGC-1 and anti-acetylated lysine (Ac Lys.) antibodies respectively. The values under the blots represent the ratio of acetylated over total PGC-1. (C) Acetylation level of PGC-1α in SRC-3+/+ and −/− MEFs. Where indicated, MEFs were infected with an adenovirus expressing PGC-1α (PGC-1α) and incubated with 20 mM nicotinamide (Nam). PGC-1α was first immunoprecipitated. Total and acetylated PGC-1α were detected through an immunoblot using anti-HA and anti-acetylated lysine antibodies respectively. *, P < 0.05 and **, P < 0.01 indicate significant differences as compared to SRC-3+/+ mice.
We also evaluated whether the genetic absence of SRC-3 could alter the acetylation levels of PGC-1α, one of the prime ways PGC-1α activity is regulated (13). Interestingly, the ratio of acetylated over total PGC-1α protein was significantly decreased in both muscle and BAT isolated from SRC-3−/− compared to SRC-3+/+ mice (Fig. 3B). To further substantiate that the absence of SRC-3 decreases the acetylation of PGC-1α, we compared PGC-1α acetylation in SRC-3+/+ and SRC-3−/− mouse embryonic fibroblasts (MEFs) that were infected with an adenovirus encoding for PGC-1α. Relative to SRC-3+/+ MEFs, the acetylation level of PGC-1α in SRC-3−/− MEFs is significantly decreased (Fig. 3C). Even in the presence of nicotinamide, an inhibitor of the PGC-1α deacetylase SIRT1, PGC-1α acetylation was significantly lower in the SRC-3−/− MEFs (Fig. 3C).
To consolidate our hypothesis, we evaluated whether reduction of SRC-3 expression in C2C12 cells that were infected with an adenovirus expressing PGC-1α—and hence express comparable levels of PGC-1α—would decrease PGC-1α acetylation levels and subsequently induce the expression of PGC-1α target genes. As predicted, the siRNA-mediated knock-down of SRC-3 in these C2C12 myotubes decreased PGC-1α acetylation significantly (Fig. S3A). Furthermore the expression levels of mRNAs encoding for a set of genes involved in mitochondrial function were all induced concomitantly with the reduction of SRC-3 expression in these C2C12 cells (Fig. S3B). In combination, these in vivo and in vitro data prove that SRC-3 facilitates the acetylation of PGC-1α and controls its activity.
SRC-3 Facilitates PGC-1α Acetylation Through Promoting GCN5 Expression.
As PGC-1α was recently reported to be acetylated by GCN5, resulting in its inactivation (14), and as SRC-3 is an important coactivator for gene expression, we investigated the possibility that SRC-3 could control GCN5 expression and hence affect PGC-1α acetylation. Interestingly, GCN5 gene expression was significantly decreased in both muscle and BAT of SRC-3−/− mice (Fig. 4A). The cell autonomous effects of SRC-3 on GCN5 expression were then investigated by both gain and loss of function approaches. In a cellular context, SRC-3 overexpression enhanced GCN5 mRNA expression (Fig. 4B); conversely, the reduction of its expression by siRNA-mediated knock-down reduced GCN5 mRNA levels (Fig. 4C), demonstrating that SRC-3 controls the expression of GCN5. This transcriptional regulation of GCN5 expression is the consequence of a direct and specific binding of SRC-3 on the promoter of GCN5 as indicated by the results of a ChIP assay (Fig. 4D). Finally, we could recapitulate the transcriptional effect of SRC-3 on the expression of a luciferase reporter construct under the control of a specific region of the GCN5 promoter that was identified by ChIP to bind the coactivator (Fig. 4E). All together, these data suggest that GCN5 expression is transcriptionally controlled by the coactivator SRC-3.
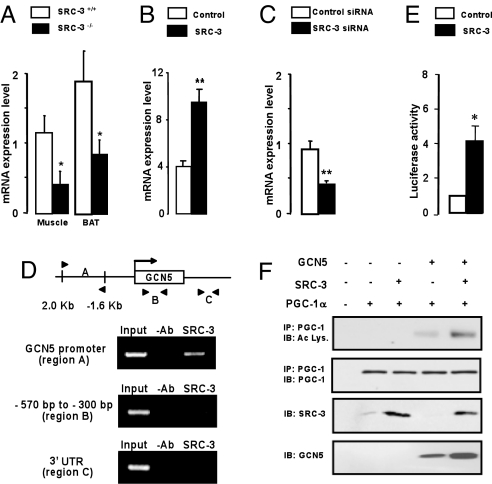
Fig. 4.
SRC-3 promotes GCN5 expression (A) GCN5 mRNA level in gastrocnemius muscle and brown adipose tissue (BAT) of SRC-3+/+ and SRC-3−/− mice fed a high fat diet (n = 8). (B) GCN5 mRNA level in C2C12 cells infected with an adenovirus expressing SRC-3 (C) GCN5 mRNA level in C2C12 cells transfected with either a control siRNA or a SRC-3 siRNA (n = 6). (D) ChIP assay showing the specific recruitment of SRC-3 on the promoter but not on the coding or 3′UTR of the mGCN5 gene. (E) Luciferase activity detected in extracts of C2C12 cells transfected with a GCN5 promoter luciferase reporter construct and cotransfected or not with SRC-3 (n = 6). (F) Acetylation level of PGC-1α determined from total extracts of C2C12 cells infected with a combination of adenoviruses, expressing PGC-1α, GCN5 and/or SRC-3. Western-blot analysis of SRC-3 and GCN5 are shown in the bottom panels, whereas the levels of acetylated (Ac-Lys) and total PGC-1α are depicted in the top panels. *, P < 0.05 and **, P < 0.005 are indicative of significant differences.
To validate the effect of SRC-3 on GCN5-mediated PGC-1α acetylation, C2C12 cells were infected with adenoviruses expressing PGC-1α, SRC-3 or GCN5 respectively (Fig. 4F). As expected, GCN5 induced PGC-1α acetylation in C2C12 cells, whereas SRC-3 by itself did not enhance PGC-1α acetylation. Interestingly, the acetylation level of PGC-1α was further enhanced in cells coinfected with SRC-3 and GCN5-expressing adenoviruses (Fig. 4F). In a similar experiment in HEK293 cells, SRC-3 also failed to acetylate PGC-1α directly, but again facilitated GCN5-mediated PGC-1α acetylation (Fig. S4). It is of note that in both cellular models, SRC-3 overexpression is also correlated with an increased GCN5 protein level, which fits with our previous observations showing that SRC-3 stimulates GCN5 expression (Fig. 4F, Lane 5).
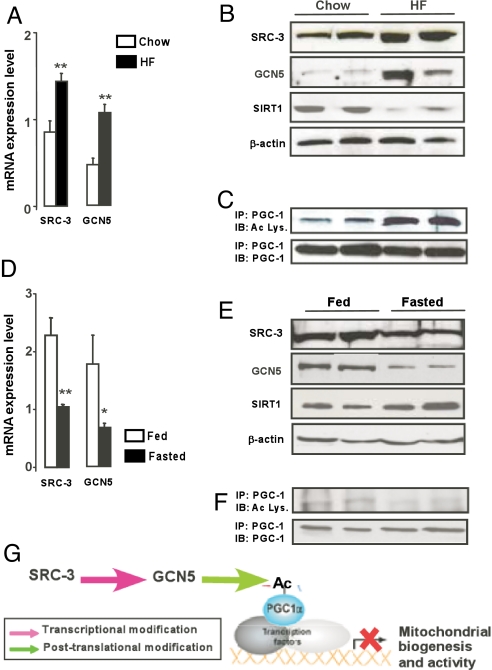
SRC-3 and GCN5 Levels Are Reciprocally Regulated by HF Diet and Fasting.
Since it was clear from the data that SRC-3, together with GCN5, is a critical component of the regulatory machinery that converges on PGC-1α to control energy homeostasis, we tested whether such a regulation has a physiological impact. We evaluated the expression of SRC-3 and GCN5 in C57BL/6J mice that were exposed either to a caloric excess, by feeding them an HF diet, or to a caloric deficit, by fasting them. Expression levels of both SRC-3 and GCN5 mRNA were robustly induced in the muscle after HF feeding (Fig. 5A), as were the protein levels (Fig. 5B, Top). Conversely, after a 6 h fast, SRC-3 and GCN5 mRNA and protein levels were decreased (Fig. 5 D and E). Interestingly, the coordinate regulation of SRC-3 and GCN5 expression upon HF feeding was associated with an increase in the ratio of acetylated over total PGC-1α protein (Fig. 5C), whereas the changes in the expression levels of both cofactors upon fasting reduced this ratio in the muscle (Fig. 5F). Strikingly, the expression of SIRT1, which is known to be regulated by caloric restriction (13) and to deacetylate and activate PGC-1α, showed almost a mirror image of that of SRC-3 and GCN5 (Fig. 5 B and E). This converse regulation of SIRT1 expression could clearly reinforce the changes in PGC-1α acetylation and activity, induced by the alterations in SRC-3 and GCN5 expression. Altogether, the changes in cofactor expression induced by caloric excess or restriction hence control PGC-1α acetylation and activity and govern the dynamic adaptation of energy homeostasis as summarized in Fig. 5G.
Fig. 5.
Modulation of SRC-3 in diet-induced obese mice and under fasting condition. (A) SRC-3 and GCN5 mRNA levels in gastrocnemius muscle of C57BL/6J male mice fed either a chow or a high fat (HF) diet for 4 months (n = 6). (B) SRC-3 and GCN5 protein levels in animals treated as described in A. β-actin levels are shown as loading controls. (C) PGC-1 acetylation level determined from gastrocnemius muscle nuclear extracts of C57BL/6J male mice fed either regular chow or HF diet for 4 months. PGC-1 was immunoprecipitated (IP). Then total and acetylated PGC-1 were detected through an immunoblot (IB) using anti-PGC-1 and anti-acetylated lysine (Ac Lys.) antibodies respectively. (D) SRC-3 and GCN5 mRNA levels in gastrocnemius muscle of C57BL/6J male mice under fed or fasted (6 h) conditions (n = 6). (E) SRC-3 and GCN5 protein levels in animals treated as described in D. β-actin is used as loading control. (F) PGC-1 acetylation level determined from gastrocnemius muscle nuclear extracts of 2 C57BL/6J male mice under fed or fasted conditions. PGC-1 acetylation level is detected as described in C. (G) Schematic illustration of the role of SRC-3 on the expression of GCN5, on PGC-1α acetylation and subsequent activity. *, P < 0.05 and **, P < 0.01 indicate significant differences as compared to the control mice.
Discussion
Mitochondrial dysfunction in general and reduced PGC-1α activity in particular, has recently been linked with the pathogenesis of metabolic diseases as it increases metabolic and cardiovascular risk and precedes the development of T2DM (16–18). Conversely, an increase in physical activity and dietary restriction, the cornerstone of the therapy of obesity and the metabolic syndrome, are both inducing mitochondrial function (19). It is also clear in our study that enhanced PGC-1α activity contributes in a major fashion to the mitochondrial activation observed in BAT and muscle of SRC-3−/− mice. PGC-1α activity is predominantly regulated by various posttranslational modifications, including changes in its acetylation status (for review see ref. 12). Consistent with this type of regulation, we demonstrate here that SRC-3 facilitates PGC-1α acetylation in muscle and BAT, through enhancing the expression of the PGC-1α acetyltransferase, GCN5 (14). How SRC-3 exactly controls GCN5 expression remain to be investigated. We hypothesize that SRC-3 could act through transcription factors such as C/EBPδ, for which response elements are present within the GCN5 promoter in the region where SRC-3 is binding. In addition, our data do not permit us to exclude the possibility that SRC-3 could also control GCN5 levels via posttranslational processes such as protein stabilization, rendering the interplay between both acetyltransferases even more complex.
Acetylation of PGC-1α is an important way to reduce PGC-1α activity and EE. Conversely, the type III histone deacetylase SIRT1 (20) was shown to promote EE, through deacetylation and activation of PGC-1α (13, 21). PGC-1α acetylation and activity seems to be controlled by a delicate balance between cellular acetyltransferases and deacetylases. For its deacetylase activity, SIRT1 is strictly dependent on cellular NAD+ levels, which reflect cellular energy status. Also the activity of acetyltransferases, such GCN5, that use acetyl-CoA as substrate, is coupled to the metabolic state of the cells (22). Like changes in cellular NAD+ levels, that affect SIRT1 deacetylase activity (20), variations in acetyl-CoA substrates for acetyltransferases (22), can therefore lead to changes in histone and protein acetylation, that impact on transcription (23). The relative activity of acetyltransferases and deacetylases therefore informs PGC-1α about the cellular energy status, which then adapts cellular energy production through its commanding role on mitochondrial biogenesis and function. Such additional cofactors act along with PGC-1α to influence energy balance (reviewed in ref. 2) and the p160 family of cofactors is a good case in point, as all of its member have been shown to impact on energy homeostasis. In fact, mice lacking the SRC-2 protein are protected against obesity due to enhanced adaptative thermogenesis, whereas SRC-1 deficient animals are prone to obesity because of reduced EE (6). Double SRC-1/3 germline mutant mice have also increased basal metabolic rate, although it was difficult to assign the effects to either SRC-1, SRC-3, or an epistatic interaction between the two proteins (10).
Interestingly, we show here that the expression of several acetyltransferases and deacetylases is highly regulated by the energy status. In fact, SRC-3 and GCN5 expression is induced during HF feeding, whereas their expression is reduced upon fasting. When energy is in excess, such as encountered during the consumption of an HF diet, the increased expression of SRC-3 and GCN5 promotes GCN5-mediated acetylation of PGC-1α, leading thereby to its inactivation, attenuating as such mitochondrial function and FAO. Under such conditions, EE cannot compensate for the higher energy intake, thus promoting the storage of excess energy in WAT. Our data are fully in line with a recent report indicating that an HF diet attenuates gene pathways necessary for OXPHOS and mitochondrial biogenesis and provides a mechanistic explanation for the observed phenotype in that study (24). Conversely, in situations of caloric restriction, mimicked by fasting in the current study, SRC-3 and GCN5 expression decreases, reducing the acetylation but enhancing the activity of PGC-1α, which facilitates the use of fatty acids stored in adipose tissues as a source of energy. Interestingly, as previously reported and confirmed in the present study, the expression levels of SIRT1, the major PGC-1α deacetylase, mirror those of SRC-3 and GCN5, resulting in a convergent regulation of PGC-1α acetylation by both acetyltransferases and deacetylases. From an evolutionary perspective, the regulation of PGC-1α acetylation by energy levels would favor the storage of energy during periods of feast, which could later be spend during times of famine. In our current industrialized societies, where food and energy are abundant, such a thrifty response is maladapted and could contribute to development of obesity and insulin resistance. It is therefore tempting to speculate that this converging cofactor network can be exploited to design new preventive and therapeutic strategies to combat obesity and associated metabolic disorders as T2DM.
Materials and Methods
Animal experiments: The generation of the SRC-3−/− mice has been described (23). All mice were maintained on a pure C57BL/6J background. Only male, aged-matched mice were used for phenotyping. Animals were maintained in standard conditions described in the supplemental method section. The methods used to assess the metabolic phenotype and for histological analysis were performed as outlined in the standard operating procedures linked to the EMPReSS website http://empress.har.mrc.ac.uk and as described (6, 21). Glucose, insulin, and lipids serum levels were measured as described (21).
ex Vivo O2 Consumption Measurements.
The O2 consumption was measured in isolated gastrocnemius muscle fibers as described (21).
RNA and DNA Analysis.
RNA was extracted from tissues or cultured cells and quantified by quantitative RT-PCR as described in the SI Methods. MtDNA content was measured exactly as described (21).
Immunoblotting and Immunoprecipitations.
Tissue homogenization and preparation of cytosolic and nuclear fractions were performed as described (25). Protein extracts were separated by SDS/PAGE. The list of the antibodies used is available in the SI Methods. PGC-1α acetylation was analyzed by immunoprecipitation of PGC-1 from nuclear lysates as described in SI Methods (26).
ChIP analyses in C2C12 cells were performed using an anti-SRC-3 antibody (BD Biosciences). The sets of primers used to amplify the different regions of the GCN5 mouse promoter and PCR conditions are available in SI Methods.
Cell Culture, Transient Transfections and Adenoviral Infections.
these protocols can be found in the SI Methods section online.
Statistics.
Statistical analyses were performed with the Student's test for independent samples (nonparametric), and data are expressed as means ± SEM. P value < 0.05 was considered as statistically significant.
Supplementary Material
Acknowledgments.
The authors thank Aurélie Auburtin, Adeline Jauffre, Nadia Messaddeq, Marie-France Champy, Jérôme Feige, Carles Canto, and Charles Foulds for helpful discussions and technical assistance. This work was supported by grants from CNRS, INSERM, Hôpitaux Universitaires de Strasbourg, EU (QLRT-2001–00930 and LSHM-CT-2004–512013), and NIH (DK59820 and HD07857). ML was supported by Nestlé.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808207105/DCSupplemental.
References
- 1.Lonard DM, O'Malley BW. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–414. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 2.Feige JN, Auwerx J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007;17:292–301. doi: 10.1016/j.tcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Smith CL, O'Malley BW. Coregulator function: A key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25(1):45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 4.Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 5.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 6.Picard F, et al. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- 7.Torres-Arzayus MI, et al. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell. 2004;6:263–274. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 8.Coste A, et al. Absence of the steroid receptor coactivator-3 induces B-cell lymphoma. EMBO J. 2006;25:2453–2464. doi: 10.1038/sj.emboj.7601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louet J-F, et al. Oncogenic steroid receptor coactivator-3 is a key regulator of the white adipogenic program. Proc Natl Acad Sci USA. 2006;103:17868–17873. doi: 10.1073/pnas.0608711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, et al. Critical roles of the p160 transcriptional coactivators p/CIP and SRC-1 in energy balance. Cell Metab. 2006;3:111–122. doi: 10.1016/j.cmet.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 12.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 14.Lerin C, et al. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 15.Cipolat S, et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 16.Petersen KF, et al. Mitochondrial dysfunction in the elderly: Possible role in insulin resistance. Science. 2003;300:1140–1142. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 18.Patti ME, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci USA. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auwerx J. Improving metabolism by increasing energy expenditure. Nat Med. 2006;12:44–45. doi: 10.1038/nm0106-44. [DOI] [PubMed] [Google Scholar]
- 20.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 21.Lagouge M, et al. Resveratrol Improves Mitochondrial Function and Protects against Metabolic Disease by Activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 22.Ladurner AG. Rheostat control of gene expression by metabolites. Mol Cell. 2006;24:1–11. doi: 10.1016/j.molcel.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 24.Sparks LM, et al. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes. 2005;54:1926–1933. doi: 10.2337/diabetes.54.7.1926. [DOI] [PubMed] [Google Scholar]
- 25.Schoonjans K, et al. PPARalpha and PPARgamma activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 1996;15:5336–5348. [PMC free article] [PubMed] [Google Scholar]
- 26.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.