Abstract
We previously have shown that DNA demethylation by chicken embryo 5-methylcytosine DNA glycosylase (5-MCDG) needs both RNA and proteins. One of these proteins is a RNA helicase. Further peptides were sequenced, and three of them are identical to the mammalian G/T mismatch DNA glycosylase. A 3,233-bp cDNA coding for the chicken homologue of human G/T mismatch DNA glycosylase was isolated and sequenced. The derived amino acid sequence (408 aa) shows 80% identity with the human G/T mismatch DNA glycosylase, and both the C and N-terminal parts have about 50% identity. As for the highly purified chicken embryo DNA demethylation complex the recombinant protein expressed in Escherichia coli has both G/T mismatch and 5-MCDG activities. The recombinant protein has the same substrate specificity as the chicken embryo 5-MCDG where hemimethylated DNA is a better substrate than symmetrically methylated CpGs. The activity ratio of G/T mismatch and 5-MCDG is about 30:1 for the recombinant protein expressed in E. coli and 3:1 for the purified enzyme from chicken embryos. The incubation of a recombinant CpG-rich RNA isolated from the purified DNA demethylation complex with the recombinant enzyme strongly inhibits G/T mismatch glycosylase while slightly stimulating the activity of 5-MCDG. Deletion mutations indicate that G/T mismatch and 5-MCDG activities share the same areas of the N- and C-terminal parts of the protein. In reconstitution experiments RNA helicase in the presence of recombinant RNA and ATP potentiates the activity of 5-MCDG.
Keywords: recombinant protein, hemimethylated DNA substrate
The generation and maintenance of specific DNA methylation patterns in vertebrates requires a complex interplay of DNA methyltransferases, demethylation reactions combined with cis and trans regulatory elements (for reviews see refs. 1 and 2). For the demethylation of DNA there are basically two possible reactions: the passive and the active demethylation. The passive DNA demethylation occurs by the inhibition of the maintenance DNA methyltransferase throughout cycles of replication, whereas active DNA demethylation requires specific enzymatic reactions. Among them, there are the replacement of 5-methylcytosine (5-Me) by cytosine (3–6) or the direct removal of the methyl group from 5-Me (7). Demethylation also can be obtained by a combination of both a passive and an active mechanism. In this case the product of the passive reaction is the formation of a hemimethylated DNA, which becomes the substrate of 5-MeC-DNA glycosylase (5-MCDG) (8). The presence of such an enzymatic activity has been detected in developing chicken embryos (9), mouse myoblasts (10), and mouse embryos and embryonic stem cells (J.-P.J., unpublished results). Recently we have shown that the demethylation of hemimethylated DNA by purified 5-MCDG requires both proteins and RNA (11–13). Peptides derived from the highly purified DNA demethylation complex have been characterized by mass spectrometry. One of the proteins present in the demethylation complex is a RNA helicase closely related to the human P68 DEAD box protein (14). Here we show that other peptides from the same preparation of the enzyme complex are identical to human G/T mismatch DNA glycosylase. We also show that the chicken recombinant protein expressed in Escherichia coli has both the G/T mismatch DNA glycosylase and the 5-MCDG activities.
Materials and Methods
Purification of 5-MCDG from 12-Day-Old Chicken Embryos and Microsequencing of Peptides.
The purification and sequencing of peptides derived from 100,000-fold purified 5-MCDG were carried out as reported (5, 14).
Construction and Screening of a cDNA Library.
As in our previously published work (14) we used the same cDNA library for screening and the same expression vectors for the production of recombinant proteins in E. coli. The following antisense oligonucleotides were designed for the screening of the cDNA library: for the N-terminal part of G/T mismatch DNA glycosylase we used 5′-CCACTGGTTGTTTTGGTTCTGTTGTTCTGGGTTTTCTTTTTCTTCCTTTTGG-3′ and 5′-GGTTTTGCAGACTTGCCAGATTTTTTTGACTCAACAGGTTTTTTGGG-3′, and for the C-terminal 5′-GCACCTCTTGAACGTCCATATTTCGTTCAATGCCTTTCAACTGATCTCTTA-3′ and 5′-CAGCCATCTTCTTTGCATCCTCTTGGGCAAGCTGTAGGTCAAATGTATATT-3′. Positive clones were characterized by restriction mapping and DNA sequencing. DNA sequencing was carried out with an AB1 Prism 377 DNA sequencer from Perkin–Elmer.
Construction of Deletion Mutants and Production of Recombinant Protein.
The full-length recombinant protein construct named pRE408a was generated by the ligation of a 1,282-bp BamHI/SacI fragment from the positive λ clone and the BamHI/SacI-digested pQE30 vector (Qiagen, Chatsworth, CA). In this construct, in addition to the 408 aa of the chicken G/T mismatch DNA glycosylase there is a stretch of 27 aa including six histidines. To produce the C-terminal truncated protein Δ C63 to Δ C152, plasmid pRE408a was cut at the restriction sites PvuI, PvuII, AviII, or KpnI. The cohesive ends obtained by PvuI or KpnI digestion were blunt-ended with T4 DNA polymerase. Then the samples were digested with BamHI. The 1,086-, 938-, 889-, and 818-bp fragments were recovered from 0.7% agarose gel and inserted into the BamHI/SmaI sites of pQE30 vector. The recombinant plasmids were named pREC345 (Δ C63), pREC296 (Δ C112), pREC280 (Δ C128), and pREC256 (Δ C152), respectively. For the expression of N-terminal deletion mutants ΔN42-ΔN139, plasmid pRE408a was cut at the restriction sites XbaI, DraI, or NciI. The cohesive ends obtained by XbaI digestion were blunt-ended with T4 DNA polymerase. All of the samples then were digested with SacI. The 1,107-bp XbaI/SacI fragment and 933-bp DraI/SacI fragment were ligated into the BamHI (blunt-ended)/SacI sites of pQE32 vector. The 827-bp NciI/SacI fragment was ligated into the BamHI (partially filled up, leaving a “G” overhang)/SacI sites of pQE32 vector. The recombinant plasmids were named pREN43 (ΔN42), pREN101 (ΔN100), and pREN140 (ΔN139), respectively. All of the deletion mutants were fused in-frame with the His tag of the vectors.
The full-length recombinant protein construct and the deletion mutants were transformed into E. coli strain XL1-Blue. For each recombinant protein, 1 liter of liquid culture derived from a single colony was grown at 37°C to A600 = 0.6. The culture then was treated with 1 mM isopropyl β-d-thiogalactoside at 37°C for 3 h. Cells were collected and broken with a French press. The majority of the recombinant proteins were present in inclusion bodies. Recombinant proteins were purified on a Ni-nitrilotriacetic acid-agarose column (Qiagen) under denaturing conditions and then refolded by dialysis.
Substrates for G/T Mismatch DNA Glycosylase and 5-MCDG.
The substrates were the same as those described by Jost et al. (A and B of table 1 in ref. 5). In the routine assays the substrate was hemimethylated. Additionally we used (only the lower methylated strand is shown): 5′-TCACGGGATCAATGTGTTCTTTCAGGGATmCCTCACGCTGACCAGGAATCC-3′; 5′-AGGATCAATGTGTTCTTTCAGCCCAGmCTCACGCTGACCAGGAATACCCAC-3′; and 5′-AGGATCAATGTGTTCTTTCAGCCCTGmCACACGCTGACCAGGAATACCCAC-3′. Oligonucleotides were labeled at the 5′ end with polynucleotide kinase and γ-32P-ATP to a specific activity of about 3 × 107 cpm/μg duplex.
Enzymatic Tests.
5-MCDG and G/T mismatch DNA glycosylase were tested with labeled nucleotides as described (5). Alternatively the recombinant proteins (0.1–0.25 μg per assay) were tested in 20 μl total volume containing 50 mM Hepes (pH 7.5), 1 mM DTT, 5 mM EDTA, 10 μg nuclease-free BSA, and 5 × 10−8 M of end-labeled substrates.
Upon incubation for 30 min to 1 h at 37°C samples were diluted to 100 μl with H2O and treated as described (5). Before loading the samples onto the DNA sequencing gel they were treated for 10 min at 95°C in 0.1 M NaOH. The alkaline treatment cleaves the abasic sites resulting from the removal of the methylated base or the mispaired base by the glycosylase.
The sensitivity of the recombinant G/T mismatch DNA glycosylase and 5-MCDG activities to micrococcal nuclease was tested as described (13). The formation of abasic sites after enzymatic excision of 5-Me was directly demonstrated by reductive labeling of these sites by radiolabeled sodium borhydride (6). The reaction product was separated on DEAE-Sepharose equilibrated in 20 mM Hepes, pH 7.5 (100 mg resin in Eppendorf tubes). Upon extensive washing with 10 × 1 ml of buffer, the labeled oligonucleotides were eluted from the resin with 1 M ammonium acetate, pH 7.0. The eluate was counted for radioactivity.
Interaction Between RNA Helicase and 5-MCDG.
A reaction mixture of 100 μl composed of 30 mM Hepes (pH 7.5), 5 mM MgCl2, 4 mM ATP, 5 mM DTT, 4 mM Pefabloc, 1 μg poly(C), 50 μg BSA, 10 units of ribonuclease inhibitor, and 5 × 10−8 M labeled oligonucleotide substrate received consecutively (assembled on ice) 40 nM recombinant RNA [clone 1, European Molecular Biology Laboratory (EMBL) accession no. Y14827], 40 nM of recombinant 5-MCDG and variable concentrations of recombinant RNA helicase (EMBL accession no. AF158370). Upon 30 min incubation at 37°C samples were digested with 50 μg of proteinase K (10 min/37°C) followed by phenol chloroform extraction and ethanol precipitation. Sediments were dissolved in 10 μl of 0.1 M NaOH and heated 10 min at 95°C, and upon addition of 10 μl of loading dye samples were separated on a 20% polyacrylamide sequencing gel.
Chemicals and Enzymes.
Polynucleotide kinase and DNA ligase were obtained from Roche Molecular Biochemicals. Restriction enzymes and ribonuclease from BSA were purchased from Biofinex (Praroman, Switzerland). QIA express Type IV kit, QIA quick gel extraction kit, Plasmid Maxi kit and Qiagen Lambda Maxi kit were obtained from Qiagen. Wizard Plus SV Minipreps DNA purification system was from Promega. [γ-32P-ATP] triethylammonium (3,000 Ci/mmol) was from Amersham Pharamica. Sodium borohydride [3H] 521 Ci/mmol was purchased from DuPont/NEN.
Results
Presence of a G/T Mismatch DNA Glycosylase in the Highly Purified 5-MCDG.
We previously have shown that G/T mismatch DNA glycosylase activity copurified with 5-MCDG (5). Throughout the purification of chicken embryo nuclear extracts the ratio of the two enzymes remained constant (5). Tandem mass spectrometry of three peptides derived from the highly purified 5-MCDG showed their identity with mammalian G/T mismatch DNA glycosylase (see sequence underlined in Fig. 1). A chicken cDNA of 3,233 bp was cloned and sequenced. Its ORF codes for a peptide of 408 aa. The amino acids alignment with human G/T mismatch DNA glycosylase is shown in Fig. 1. The amino acids sequence (408 aa) shows 80% identity with the human G/T mismatch DNA glycosylase and the C- and N-terminal parts have about only 50% identity.
Figure 1.
Comparison of the deduced amino acid sequence of the chicken G/T mismatch DNA glycosylase (EMBL accession no. AF202114) with human G/T mismatch DNA glycosylase (EMBL accession no. U51166). The peptide sequences obtained from purified chicken embryo 5-MCDG are underlined.
The Recombinant G/T Mismatch DNA Glycosylase from Chicken Has 5-MCDG Activity.
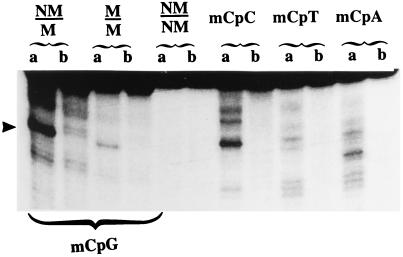
The recombinant G/T mismatch DNA glycosylase produced in E. coli tested in vitro showed significant activity of 5-MCDG (Fig. 2 and see Figs. 4–6). As for the highly purified enzyme from chicken embryos the recombinant 5-MCDG could not make an efficient incision at the abasic site, and the reaction product had to be treated with alkali. Additional evidence for the presence of an abasic site was demonstrated by reductive labeling of the aldehyde group of the open deoxyribose ring with radioactive NaBH4 (results not shown). Fig. 2 shows that the purified recombinant glycosylase has about the same substrate specificity as the chicken embryo enzyme (9). Hemimethylated DNA (NM/M) is the preferred substrate. Symmetrically methylated DNA (M/M) is also to a lesser extent a substrate and the same nonmethylated oligonucleotide duplex (NM/NM) is not a substrate. However, when compared with the enzyme from chicken embryo the recombinant protein, in addition to 5-MeCpGs, could react with 5-MeCpC (Fig. 2).
Figure 2.
Sequence specificity of 5-MCDG. Enzymatic reactions were carried out as described in Materials and Methods. Upon alkaline treatment the reaction product was separated on a 20% polyacrylamide DNA sequencing gel. a is the labeled oligonucleotide incubated with the enzyme, and b is the same oligonucleotide incubated without the enzyme. The arrowhead shows the position of the cleavage product. NM, nonmethylated DNA strand; M, methylated strand.
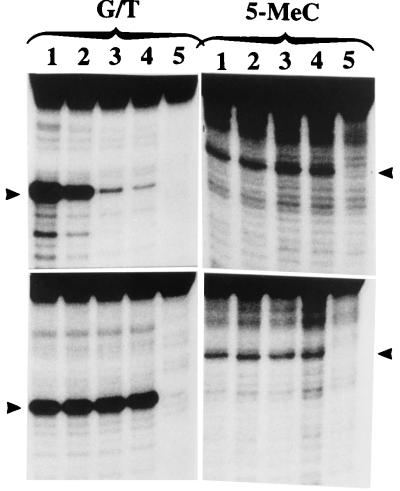
Figure 4.
Assay of the N-terminal mutant recombinant enzyme produced in E. coli. The enzyme preparations shown in Fig. 3B were assayed for G/T mismatch and 5-MCDG activities. The reaction product was treated with NaOH and separated on a 20% polyacrylamide DNA sequencing gel. Lanes 1–5 correspond to FL, ΔC63, ΔC112, ΔC128, and ΔC152, respectively of Fig. 3A, and lane 6 is an incubated blank. The arrowheads show the position of the reaction product.
Figure 6.
Effect of increasing concentrations of synthetic oligoribonucleotides on the activity of G/T mismatch and 5-MCDG. (Upper) The enzyme was incubated with 0, 2, 4, and 6 μg (lanes 1–5, respectively) of the oligo GUGACCGGAGC complementary to the target strand. (Lower) Controls incubated with 0, 2, 4, and 6 μg (lanes 1–5, respectively) of GCUCCGGUCAC complementary to the opposite strand. The oligoribonucleotides were protected against ribonucleases by 2-O-methylation of the ribose moity. Lane 6 is a blank. The reaction product was analyzed as for Fig. 5. The arrowheads show the position of the cleavage products.
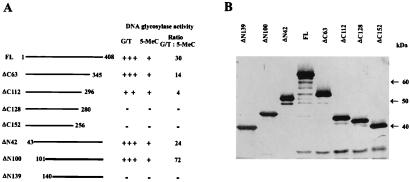
As previously shown (5) throughout the purification from chicken embryos the ratio of G/T mismatch DNA glycosylase over 5-MCDG was about 3. As shown in Fig. 3A for the full-size recombinant enzyme produced in E. coli this ratio is about 30–40.
Figure 3.
Construction of deletion mutants of chicken G/T mismatch DNA glycosylase. The constructs were done as described in Materials and Methods. (A) The relative activities of G/T mismatch and 5-MeC glycosylase are indicated by + or −. The radioactive bands corresponding to the specific reaction product were cut out from the gels and counted for radioactivity. The ratio of the two enzyme activities is given. (B) A silver-stained gel of mutant enzyme isolated on a Ni-nitrilotriacetic acid-agarose column.
G/T and 5-MCDG Activities Are Localized in the Same Regions at the C and N Terminals of the Recombinant Enzyme.
A series of C-terminal deletion construct (Fig. 3 A and B) were expressed in E. coli and tested for G/T mismatch and 5-MCDG activities (Fig. 4). As shown in Figs. 3 and 4, a deletion between amino acid positions 296 and 345 had a profound effect on the activity of G/T mismatch DNA glycosylase without affecting much 5-MCDG activity, thus changing the activity ratio of the two enzymes from 30 to 4 (Fig. 3A). However, further deletions beyond amino acid position 296 abolish both enzyme activities, which means that the two enzymatic activities share a region spanning from amino acid positions 296 to 345. Similarly at the N-terminal region, deletions of up to the first 100 aa had no effect on G/T mismatch and 5-MCDG activities (Fig. 3A), and further deletions abolished the two enzymatic activities.
Effect of a Recombinant CpG-Rich RNA on the Activities of 5-MCDG and G/T Mismatch DNA Glycosylase.
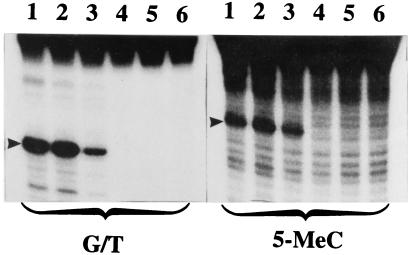
As we have previously shown, DNA demethylation by the purified 5-MCDG needs protein and RNA (11). The cloned RNA isolated from gel purified 5-MCDG is very rich in CpGs and did not show any ORF (12). Fig. 5 shows that increasing concentrations of the recombinant RNA inhibited by about 80% the G/T mismatch DNA glycosylase while stimulating by 20–30% the activity of 5-MCDG. Similar titration experiments carried out with a synthetic oligoribonucleotide complementary to the target site inhibited by over 95% (Fig. 6 Upper Left) the activity of the G/T mismatch DNA glycosylase while stimulating 5-MCDG by about 50% (Fig. 6 Upper Right). In addition, the presence of the RNA in the incubation mixture increases the specificity of the reaction for 5-MeCpG (data not shown). However, an oligoribonucleotide complementary to the opposite strand had no effect on the activities of G/T mismatch glycosylase and 5-MCDG (Fig. 6 Lower), thus ruling out a nonspecific effect of nucleic acids. Similarly the addition of up to 2 μg of polycytidilic acid per 20 μl incubation mixture (as inhibitor of endogenous ribonucleases) did not affect the two enzymic activities. Further experiments clearly indicate that the purified recombinant 5-MCDG produced in E. coli has no RNA and is not sensitive to micrococcal nuclease (results not shown). The above results could possibly explain the high activity ratio of the recombinant G/T mismatch/5-MCDG when compared with the purified enzyme from chicken embryos.
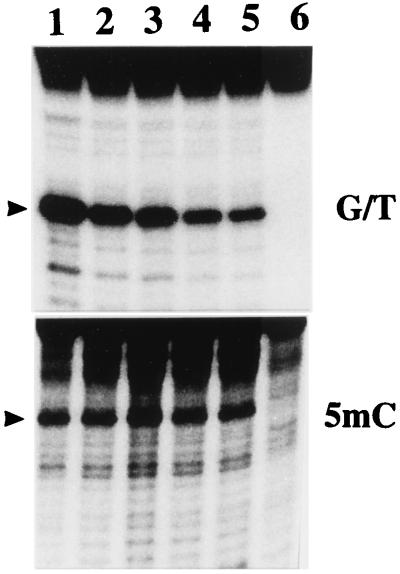
Figure 5.
Effect of increasing concentrations of recombinant RNA (CpG-rich RNA, clone 1) on the activity of G/T mismatch and 5-MCDG. Experiments were carried out as described in Materials and Methods. Lanes 1–5 received 0, 1, 2, 4, and 6 μg of recombinant RNA, respectively, and lane 6 is a blank. The reaction product was treated with NaOH and separated on a 20% polyacrylamide DNA sequencing gel. The arrowheads show the position of the cleavage product.
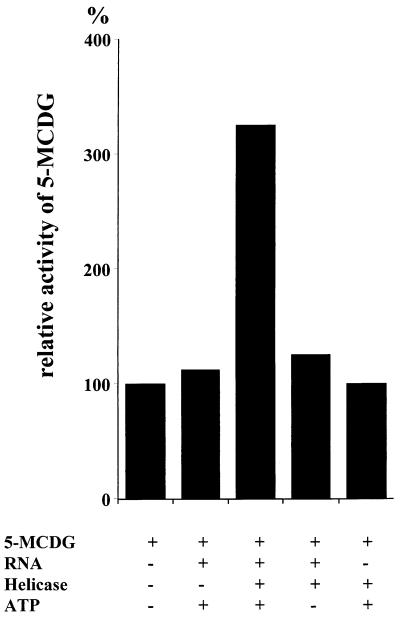
A Combination of Recombinant RNA and RNA Helicase in the Presence of ATP Potentiates the Activity of 5-MCDG.
The results of Fig. 7 show that an incubation of RNA or RNA helicase in combination with recombinant 5-MCDG does not have any significant effect on the activity of 5-MCDG. However, a combination of RNA helicase RNA and ATP potentiates the activity of 5-MCDG and this potentiation is ATP dependent.
Figure 7.
Potentiation of 5-MCDG by RNA helicase, RNA, and ATP. Incubations were assembled in duplicates and carried out as described in Materials and Methods. Bands corresponding to the specific reaction products were cut out from the gel and counted for radioactivity. Blanks were subtracted from the test values and expressed in cpm/μg enzyme. The value obtained for the enzyme incubated without any further addition was expressed as 100%.
Discussion
As we already have shown, throughout the purification of 5-MCDG there is a copurification of the G/T mismatch glycosylase and the ratio of the two enzymes activities remains constant (5). All attempts made so far to separate the two activities have failed, suggesting that the two activities may be associated to the same protein. Moreover, on DNA affinity matrix the behavior of 5-MCDG from chicken embryos was different from the G/T mismatch glycosylase purified from HeLa cells. For example, the G/T mismatch glycosylase from HeLa cells binds specifically to oligonucleotides containing a G/T mismatch (15), whereas no such specificity was observed for chicken embryo 5-MCDG, which bound to single-stranded, double-stranded methylated, or not methylated DNA and DNA containing a G/T mismatch. An explanation for such a difference could be the association of chicken embryo 5-MCDG with other macromolecules. This turned out to be the case. The highly purified enzyme (over 100,000 × fold purification) was associated with RNA and different proteins. Both RNA and protein(s) were required for the full activity of 5-MCDG (11–13). The RNA associated with the DNA demethylation complex consists of CpG-rich RNA (12). Among the 11 different cDNA clones sequenced so far none had an ORF and none had any sequence homology with any other known RNA (12). Recently we have shown by in situ hybridization in chicken embryos that one of the CpG-rich RNA is located mainly in dividing cells undergoing differentiation where the level of DNA demethylation activity is the highest (S.S., unpublished work). In developing embryos this RNA is not tissue specific but developmental stage specific, and it shows a very complex secondary structure. This RNA may have several functions such as the inhibition of DNA methyltransferase by its stem loop structures (31), the activation and the targeting of DNA demethylation (11–12). Among the proteins so far identified in the DNA demethylation complex we have an RNA helicase (14) that may modulate the structure and function of the CpG-rich RNA. As shown by in situ hybridization, the RNA helicase is found in the same tissues as the CpG-rich RNA (ref. 14 and S.S., unpublished work). The exact nature of the interactions between the RNA, the RNA helicase, and 5-MCDG presently is being studied, and preliminary results indicate that RNA helicase and RNA in the presence of ATP may potentiate the activity of 5-MCDG (Fig. 7). The effect of the recombinant CpG-rich RNA on the recombinant G/T mismatch DNA glycosylase (see Figs. 5 and 6) could possibly explain the differences in the activities ratios of G/T mismatch glycosylase and 5-MCDG obtained for the recombinant enzyme and the purified enzyme from chicken embryos. Alternatively the absence of RNA in the recombinant enzyme could explain the high activity ratio of the two enzymes. In addition, one cannot exclude the possibility that the specificity of the enzyme for the G/T mismatch or 5-Me depends on some covalent modifications of the protein. For example, we found that the addition of β-glycerophosphate (an inhibitor of protein phosphatases) to the buffers during the purification of chicken embryo 5-MCDG, increased up to 3-fold the specific activity of the enzyme (J.-P.J., unpublished results). The chicken G/T mismatch DNA glycosylase has a high sequence homology with its human homologue with the exception of its C- and N-terminal parts (16). It is hardly surprising that 5-MCDG activity is part of a protein linked to a DNA repair system. We know that methylated DNA is a hot spot of mutations (17–19). The presence of the two enzymatic activities in the same molecule could have a dual function: preventing the mutations and maintaining the CpG islands free of methylation (20).
A third protein, which is part of the DNA demethylation complex, has just been identified by tandem mass spectrometry. This protein is identical to the nuclear matrix protein ARBP (attachment region binding protein), which has homology with MeCP2 (21). In this context it is interesting to note that the Ig κ enhancer element and the nuclear matrix attachment region MAR are important for the specific demethylation of Ig κ gene during B-cell maturation (22). Similar results also have been obtained for the demethylation of the Ig Mu enhancer (23). In another system we have shown that the association of the 5′ and 3′ flanking region of avian vitellogenin II gene to the nuclear matrix is under the control of estrogen (24). Once bound to the nuclear matrix this region becomes demethylated (25). These results imply that the nuclear matrix may be the site of DNA demethylation. However, an important question remains: are there still other alternative reactions involved in the enzymic DNA demethylation? 5-MCDG may not be the only enzyme responsible for DNA demethylation. For example, the group of Szyf recently has identified a protein MBD2b that can remove the methyl group from 5-Me in an energy-independent reaction (7, 26, 27). However, Wade et al. (28) could not show any demethylation activity of human MBD2b produced by the in vitro-translated protein. MBD2b may have other additional functions; for example, in HeLa cells, MBD2b is associated with a histone deacetylase in the MeCP1 repressor complex (29, 30). Therefore, one cannot exclude that the DNA demethylation reaction by MBD2b is more complex than anticipated and in the interim one should remain open to the existence of alternative functions of this protein.
Acknowledgments
We thank Mrs. Y. C. Jost for typing the manuscript. Many thanks also go to Mr. P. Müller for the synthesis of the oligonucleotides and Mrs. I. Obergfoell for the photographic work. This work was sponsored by Novartis AG, Basel, Switzerland.
Abbreviations
- 5-Me
5-methylcytosine
- 5-MCDG
5-methylcytosine DNA glycosylase
- EMBL
European Molecular Biology Laboratory
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF202114).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100107597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100107597
References
- 1.Russo V E A, Martienssen R A, Riggs A D. Epigenetic Mechanisms of Gene Regulation. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. [Google Scholar]
- 2.Jost J P, Bruhat A. Prog Nucleic Acids Res Mol Biol. 1997;57:217–248. doi: 10.1016/s0079-6603(08)60282-2. [DOI] [PubMed] [Google Scholar]
- 3.Paroush Z, Keshet I, Yisraeli J, Cedar H. Cell. 1990;63:1229–1237. doi: 10.1016/0092-8674(90)90418-e. [DOI] [PubMed] [Google Scholar]
- 4.Shemer R, Kafri T, O'Connell A, Eisenberg S, Breslow J L, Razin A. Proc Natl Acad Sci USA. 1991;88:11300–11304. doi: 10.1073/pnas.88.24.11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jost J P, Siegmann M, Sun L, Leung R. J Biol Chem. 1995;270:9734–9739. doi: 10.1074/jbc.270.17.9734. [DOI] [PubMed] [Google Scholar]
- 6.Vairapandi M, Duker N J. Nucleic Acids Res. 1993;21:5323–5327. doi: 10.1093/nar/21.23.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharya S K, Ramchandani S, Cervoni N, Szyf M. Nature (London) 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh C L. Mol Cell Biol. 1999;19:46–56. doi: 10.1128/mcb.19.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jost J P. Proc Natl Acad Sci USA. 1993;96:4684–4688. doi: 10.1073/pnas.90.10.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jost J P, Jost Y C. J Biol Chem. 1994;269:10040–10043. [PubMed] [Google Scholar]
- 11.Frémont M, Siegmann M, Gaulis S, Matthies R, Hess D, Jost J P. Nucleic Acids Res. 1997;25:2375–2380. doi: 10.1093/nar/25.12.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jost J P, Frémont M, Siegmann M, Hofsteenge J. Nucleic Acids Res. 1997;25:4545–4550. doi: 10.1093/nar/25.22.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jost J P, Siegmann M, Thiry S, Jost Y C, Benjamin D, Schwarz S. FEBS Lett. 1999;449:251–254. doi: 10.1016/s0014-5793(99)00454-8. [DOI] [PubMed] [Google Scholar]
- 14.Jost J P, Schwarz S, Hess D, Angliker H, Fuller-Pace F V, Stahl H, Thiry S, Siegmann M. Nucleic Acids Res. 1999;27:3245–3252. doi: 10.1093/nar/27.16.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neddermann P, Jiricny J. J Biol Chem. 1993;268:21218–21224. [PubMed] [Google Scholar]
- 16.Neddermann P, Gallinari P, Lettieri T, Schmid D, Truong O, Hsuan J J, Wiebauer K, Jiricny J. J Biol Chem. 1996;271:12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- 17.Jones P A. Cancer Res. 1996;56:163–167. [PubMed] [Google Scholar]
- 18.Toyota M, Ahuja N, Ohe-Toyota M, Herman J G, Baylin S B, Issa J P. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones P A, Laird P W. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 20.Antequera F, Bird A. In: DNA Methylation: Molecular Biology and Biological Significance. Jost J P, Saluz H P, editors. Basel: Birkhäuser; 1993. pp. 169–185. [Google Scholar]
- 21.Weitzel J M, Buhrmester H, Strätling W H. Mol Cell Biol. 1997;17:5656–5666. doi: 10.1128/mcb.17.9.5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirillov A, Kistler B, Mostoslavsky R, Cedar H, Wirth T, Bergman Y. Nat Genet. 1996;13:435–441. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez L A, Winkler M, Forrester W, Jenuwein T, Grossschedl R. Cold Spring Harbor Symp Quant Biol. 1998;LXIII:515–524. doi: 10.1101/sqb.1998.63.515. [DOI] [PubMed] [Google Scholar]
- 24.Jost J P, Seldran M. EMBO J. 1984;3:2005–2008. doi: 10.1002/j.1460-2075.1984.tb02082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saluz H P, Jiricny J, Jost J P. Proc Natl Acad Sci USA. 1986;83:7167–7171. doi: 10.1073/pnas.83.19.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolffe A P, Jones P L, Wade P A. Proc Natl Acad Sci USA. 1999;96:5894–5896. doi: 10.1073/pnas.96.11.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cedar H, Verdine G L. Nature (London) 1999;397:568–569. doi: 10.1038/17492. [DOI] [PubMed] [Google Scholar]
- 28.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 29.Ng H H, Zhang Y, Hendrich B, Johnson C A, Turner B M, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Ng H H, Erdjament-Bromage H, Tempst P, Bird A. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frémont M. Ph.D. thesis. Paris-Grignon, France: Institut National Agronomique; 1998. [Google Scholar]