Abstract
Background
The levels of soluble sugars, such as glucose and sucrose, help regulate many plant metabolic, physiological and developmental processes. Genetic screens are helping identify some of the loci involved in plant sugar response and reveal extensive cross-talk between sugar and phytohormone response pathways.
Results
A forward genetic screen was performed to identify mutants with increased resistance to the inhibitory effects of high levels of exogenous sugars on early Arabidopsis seedling development. The positional cloning and characterization of two of these sugar insensitive (sis) mutants, both of which are also involved in abscisic acid (ABA) biosynthesis or response, are reported. Plants carrying mutations in SIS7/NCED3/STO1 or SIS10/ABI3 are resistant to the inhibitory effects of high levels of exogenous Glc and Suc. Quantitative RT-PCR analyses indicate transcriptional upregulation of ABA biosynthesis genes by high concentrations of Glc in wild-type germinating seeds. Gene expression profiling revealed that a significant number of genes that are expressed at lower levels in germinating sis7-1/nced3-4/sto1-4 seeds than in wild-type seeds are implicated in auxin biosynthesis or transport, suggesting cross-talk between ABA and auxin response pathways. The degree of sugar insensitivity of different sis10/abi3 mutant seedlings shows a strong positive correlation with their level of ABA insensitivity during seed germination.
Conclusion
Mutations in the SIS7/NCED3/STO1 gene, which is primarily required for ABA biosynthesis under drought conditions, confer a sugar-insensitive phenotype, indicating that a constitutive role in ABA biosynthesis is not necessary to confer sugar insensitivity. Findings presented here clearly demonstrate that mutations in ABI3 can confer a sugar-insensitive phenotype and help explain previous, mixed reports on this topic by showing that ABA and sugar insensitivity exhibit a strong positive correlation in different abi3 mutants. Expression profiling revealed a potentially novel regulation of auxin metabolism and transport in an ABA deficient mutant, sis7-1/nced3-4/sto1-4.
Background
Plant growth and development are regulated by signal transduction pathways that incorporate environmental stimuli and internal signals such as metabolic status. The absolute levels of soluble sugars, and/or the rate of flux of soluble sugars through certain metabolic pathways, are important indicators of metabolic status. Recent studies have established the role of sugars in regulating, at least partially, a variety of developmental processes from seed development and germination, early seedling development, vegetative growth and senescence to abiotic stress response [1-10].
Genetic screens based on the phenomenon that early seedling development of wild-type Arabidopsis can be arrested by high concentrations of exogenous sucrose (Suc) or glucose (Glc) have yielded sugar-response mutants. Characterization of these mutants has revealed that many also have defects in phytohormone metabolism or response [11-15]. In particular, several of these sugar-response mutants are allelic to abscisic acid (ABA) biosynthesis or response mutants such as aba2, aba3 or abi4. The sugar-resistant mutants sis4 [16], gin1 [15,17] and isi4 [18] are allelic to aba2, an ABA biosynthesis mutant [19]. The gin5 mutant [6,11] is allelic to another ABA deficient mutant, aba3 [19,20]. Similarly, mutations in ABA1 have been shown to confer Glc insensitivity [13]. The sugar-resistant mutants sis5 [16], gin6 [11], isi3 [18] and sun6 [13] are allelic to abi4, an ABA-insensitive mutant [21]. The ABI4 locus encodes an APETALA2 type transcription factor [22]. Mutations in ABI5 have also been shown to confer a weak, but still significant, sugar-insensitive phenotype [11,13,16]. Interestingly, plants carrying mutations in ABI1 or ABI2 have been found to exhibit a wild-type [11,16] or near wild-type [13], response to the inhibitory effects of high sugar concentrations on early seedling development. Previous research has given a mixed report on the role of ABI3 in sugar response. The abi3-1 mutant was found to exhibit an almost wild-type response to high levels of exogenous Glc [11,16] but was also reported to show a less pronounced but significant Glc insensitive phenotype when compared to wild-type plants [13]. In addition, it has been observed that specific abi3 alleles show decreased sensitivity to the inhibitory effects of a combination of ABA and Glc on seedling development [23]. Recently ABI3 has been reported to play an important role in inhibition of seed germination [24] and post-germinative growth [25] by Glc.
In screens of ethyl methane sulphonate (EMS)- or T-DNA mutagenized Arabidopsis populations for mutants with increased tolerance to the inhibitory effects of high levels of exogenous sugars on early seedling development, three sugar insensitive (sis) loci were identified. Map-based cloning experiments resulted in the identification of these loci. The sis9-1 mutation was found to lie in ABA1, a gene previously shown to act in ABA biosynthesis [26], and was not further characterized. The sis7 mutations lie in the same gene as the previously identified nced3/sto1 [27] mutants. The sis10 mutation lies in the same gene as the previously identified abi3 mutants [28]. NCED3/STO1 encodes a 9-cis-epoxycarotenoid dioxygenase [29]. Expression of SIS7/NCED3/STO1 has been suggested to be rate-limiting in the positive feedback regulation of ABA biosynthesis by ABA under stress conditions [17,26]. Expression profiling experiments identified 83 genes as being differentially regulated in the sis7-1 mutant when compared to wild-type seeds upon treatment with 100 mM Glc. ABI3 encodes a B3 domain transcription factor [30]. The sugar- and ABA-insensitivity of multiple abi3 mutant alleles show a strong positive correlation in our assays.
Results
Isolation of sugar-insensitive (sis) mutants sis7 and sis10
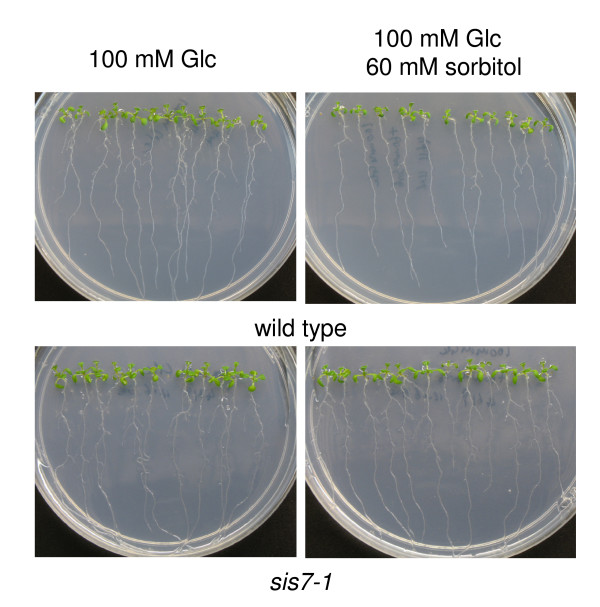
Most wild-type Arabidopsis seeds sown on minimal media [31] supplemented with high levels of Suc or Glc (e.g., 300 mM) germinate but the resulting seedlings fail to develop green expanded cotyledons and true leaves [11-13,15,16,32,33]. Instead, the seedlings develop small, white or purple cotyledons. In contrast, low concentrations (e.g., 30 mM) of exogenous Suc or Glc do not exert this inhibitory effect on normal shoot development. In addition, most wild-type seeds can develop into seedlings with expanded cotyledons and true leaves on media containing high concentrations (300 mM) of sorbitol, which acts as a non-metabolizable sugar analog in Arabidopsis. This observation indicates that the inhibitory effects of high sugar levels on early seedling development are not solely due to osmotic stress [16].
Genetic screens based on the inhibitory effects of high sugar levels on early seedling development led to the discovery of a number of sugar resistant mutants [11-13,15,16]. In one such screen performed previously by our lab, approximately 60,000 M2 seeds derived from an EMS-mutagenized Arabidopsis thaliana var. Col population were screened to identify seedlings that are resistant to 300 mM Suc. The mutants identified via this screen are also resistant to high concentrations of Glc and so were designated sugar insensitive, or sis, mutants [16]. Cloning and characterization of one of these mutants, the sis7-2 mutant, is described below. A similar mutant screen was conducted as part of the work reported here. This new mutant screen was conducted using a T-DNA mutagenized Arabidopsis thaliana var. Col population. The 35 SpBARN binary vector used to generate this population contains random Arabidopsis cDNAs driven by the Cauliflower Mosaic Virus 35S promoter [34]. Approximately 200,000 seeds from ~20,000 independent Arabidopsis thaliana var. Col transgenic lines were screened on media containing 340 mM Suc and ~1,700 seedlings that developed shoot systems with expanded cotyledons and true leaves were transferred to soil. This high rate of seedlings exhibiting an apparent sugar-resistant phenotype is due to the fact that some fraction (~1% in this experiment) of wild-type seedlings are able to escape the developmental arrest caused by high concentrations of exogenous sugars [16]. Therefore, it is necessary to screen seeds harvested from each putative mutant to identify the relatively small percentage that exhibit a heritable sis phenotype. Towards this end, the 1,700 seedlings were allowed to grow to maturity and seeds were harvested from each plant. Re-screening of seeds from each of the 1,700 putative mutants on media containing 340 mM Suc revealed that 18 of the putative mutants exhibit a reproducible sis phenotype. Characterization of these 18 mutants revealed that they appear to represent nine independent mutagenic events. The cloning and characterization of three of these new mutants, sis7-1, sis7-3 and sis10-1, as well as the sis7-2 mutant, is described below. The results of these studies indicate that the sis7 mutations lie in a gene previously shown to affect ABA metabolism, whereas the sis10 mutations lie in ABI3, which acts in ABA response [28].
sis7 mutants exhibit a sugar-insensitive phenotype
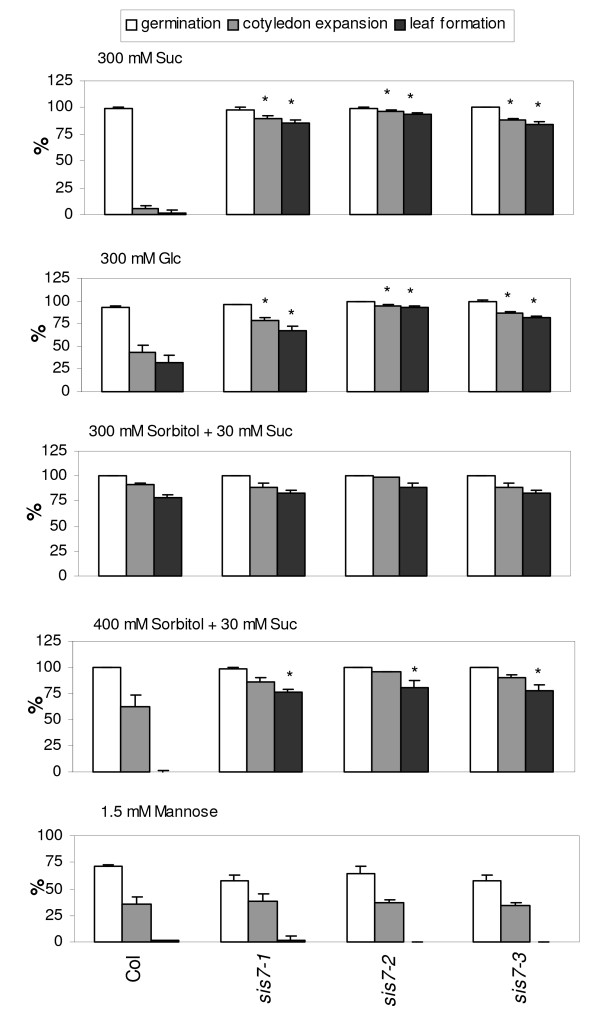
A significantly higher percentage of sis7 than of wild-type seeds develop into seedlings with relatively normal shoot systems on media containing 300 mM Suc or Glc (Figure 1). On media containing 300 mM Suc, more than 80% of all three sis7 mutants form expanded cotyledons and true leaves, whereas less than 6% of wild-type seedlings develop expanded cotyledons and true leaves. The mutants were also assayed for osmotic tolerance using media containing high concentrations of sorbitol. When assayed on equimolar (i.e. 300 mM) sorbitol supplemented with a low (30 mM) amount of Suc, differences are hard to detect between wild-type and mutant lines due to the fact that development of wild-type seedlings is not strongly affected at this concentration of sorbitol. In contrast, when assayed on media containing 400 mM sorbitol and 30 mM Suc, all three mutant alleles confer significantly increased osmotic resistance. Mannose (Man) is a Glc analogue that inhibits seed germination and early seedling development at millimolar concentrations via a mechanism that has been postulated to involve hexokinase [35]. Therefore, the mutants were assayed for Man sensitivity. None of the three sis7 mutant alleles tested cause a significant alteration in sensitivity to 1.5 mM Man compared to wild-type seedlings (Figure 1).
Figure 1.
Mutations in SIS7 confer a sugar-insensitive phenotype. Col wild-type and sis7 seeds were stratified for 3 d and then sown on the indicated media and scored after 14 d at 22°C under continuous light. Data represent the means of three independent assays. The error bars indicate standard deviations. Asterisks indicate results where the mutant phenotype differed from the corresponding wild-type phenotype with a p-value of less than 0.05 according to a Student's t-test.
sis7-1 exhibits resistance to the inhibition of seed germination by ABA and paclobutrazol
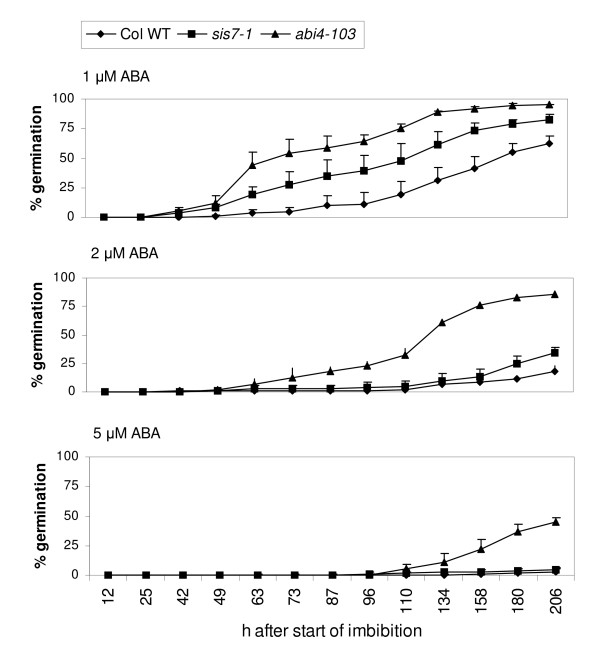
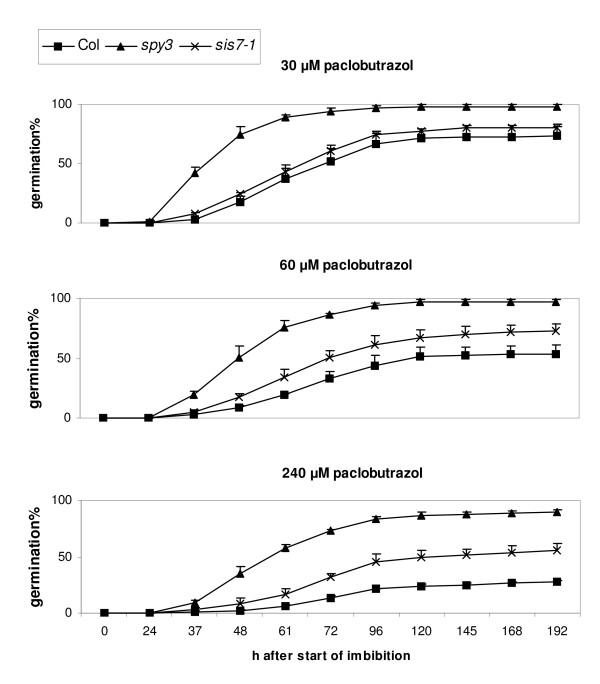
Previous studies have shown that sis4/aba2 mutants are resistant to the inhibitory effects of paclobutrazol, an inhibitor of gibberellin biosynthesis, on seed germination. Similarly, sis5/abi4 mutants are resistant to the inhibitory effects of both paclobutrazol and ABA on seed germination [16]. To determine whether mutations in SIS7 exert similar effects, the germination rates of sis7-1 seeds on media containing ABA or paclobutrazol were determined. Seeds carrying the sis7-1 mutation exhibit significantly faster germination rates than wild-type seeds on media containing 1 μM ABA and slightly faster germination rates on 2 μM ABA, but have wild-type germination rates on higher concentrations (e.g. 5 μM) of ABA (Figure 2). Germination rates of sis7-1 and wild-type seeds were also assayed on media containing several different concentrations of paclobutrazol. The spy3 seeds were included as a positive control as they have previously been shown to be resistant to paclobutrazol [36]. Seeds carrying the sis7-1 mutation display enhanced germination rates compared to wild-type seeds on media containing high levels (e.g. 60 or 240 μM) of paclobutrazol (Figure 3).
Figure 2.
The sis7-1/sto1-4/nced3-4 mutation confers a subtle ABA-insensitive phenotype. Col wild-type and sis7-1/sto1-4/nced3-4 seeds were stratified for 3 d and then sown on media containing 1, 2 or 5 μM ABA and incubated at 25°C under continuous light conditions. Germination rates on minimal media were also checked and found to be consistently high, indicating a high rate of viability in the seeds used for these experiments (data not shown). Seed germination was scored at the indicated time points. Germination is defined as the emergence of the radicle from the seed coat. Data represent the means of three independent assays. The error bars represent standard deviations. The abi4-103/sis5-3 [16] seeds were included as a positive control.
Figure 3.
The sis7-1/sto1-4/nced3-4 mutation confers a paclobutrazol-resistant phenotype. Col wild-type and sis7-1/sto1-4/nced3-4 seeds were stratified for 3 d and then sown on media containing 30, 60 or 240 μM paclobutrazol and incubated at 25°C under continuous light conditions. Germination rates on minimal media were also checked and found to be consistently high, indicating a high rate of viability in the seeds used for these experiments (data not shown). Seed germination was scored at the indicated time points. Germination is defined as the emergence of the radicle from the seed coat. Data represent the means of three independent assays. The error bars represent standard deviations. The spy-3 [36] seeds were included as a positive control.
Cloning of SIS7 and pharmacological complementation of the sis7 phenotype
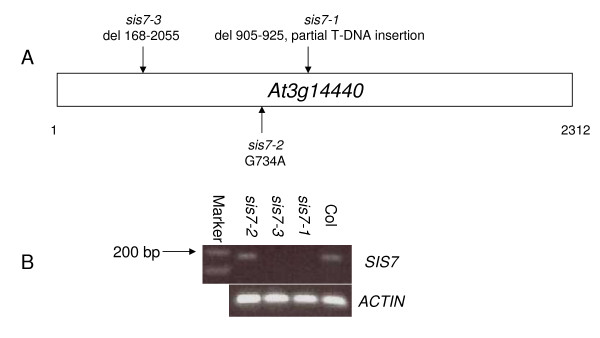
A map-based cloning approach was used to identify the SIS7 gene. The sis7-1 mutant, which is in the Col background, was crossed to wild-type Hi-O plants. F2 progeny of this cross were screened on media containing 340 mM Suc. Those seedlings that formed relatively normal shoot systems (i.e. displayed a sis mutant phenotype) were selected and used to form a mapping population of 940 plants. SSLP and CAPS markers were used to localize sis7 to a 78 kb region between BAC clones MLN21 and MIE1 on chromosome 3. DNA spanning the entire 78 kb region was isolated from sis7-1 by PCR and sequenced. Analysis of this sequencing data reveals that sis7-1 carries a 21 bp deletion and a partial T-DNA insertion in the exon region of At3g14440. No other mutations were found in the region shown by mapping to contain the sis7-1 mutation. Another sis mutation was independently mapped to chromosome 3 between BACs MLN21 and MSJ11, the region which contains the At3g14440 gene. Sequencing of the putative SIS7 gene from this mutant, now named sis7-2, identified a point mutation in the exon region that changes amino acid codon 245 from Gly to Asp. Sequencing of the putative SIS7 gene from a third sis mutant revealed that this mutant, now named sis7-3, contains a large deletion in the SIS7 gene (Figure 4A). The finding that three independent sis mutants all carry mutations in At3g14440 provides convincing evidence that the mutations in this gene are responsible for the sis phenotype observed in these lines. At3g14440 has previously been shown to encode 9-cis-epoxycarotenoid dioxygenase (NCED3), a key enzyme in the biosynthesis of ABA [29,37]. Plants carrying a T-DNA insertion in NCED3 are drought sensitive [29]. An additional mutant of NCED3, named sto1, was identified in a screen for salt-tolerant mutants. These sto1 mutants exhibit resistance to the inhibitory effects of hyperosmotic stress on seed germination, but not on post-germinative growth [27]. A third mutant carrying a T-DNA insertion in NCED3 has also been described [38]. The three mutant alleles identified in this study were accordingly re-designated sis7-1/nced3-4/sto1-4, sis7-2/nced3-5/sto1-5 and sis7-3/nced3-6/sto1-6. To determine whether SIS7 mRNA levels are altered in the sis7 mutants, SIS7 transcript abundance was assayed by RT-PCR in wild-type plants and in all three sis7 mutants. SIS7 transcript was not detected in sis7-1 or sis7-3, whereas in sis7-2 SIS7 transcript abundance is approximately the same as in wild-type plants (Figure 4B).
Figure 4.
Characterization of sis7 molecular lesions. (A) Mutations in SIS7/NCED3/STO1. The molecular lesions associated with the three sis7/nced3/sto1 mutants identified as part of this work are shown. Nucleotides are numbered with respect to the start codon. (B) SIS7 transcript levels in wild-type and sis7 plants. RT-PCR was used to amplify SIS7/NCED3/STO1 transcripts from 4-week old wild-type (Col) and sis7 mutant plants. 2 μL cDNA was used as a template for the first PCR reaction (35 cycles). ACTIN transcripts were amplified as a positive control.
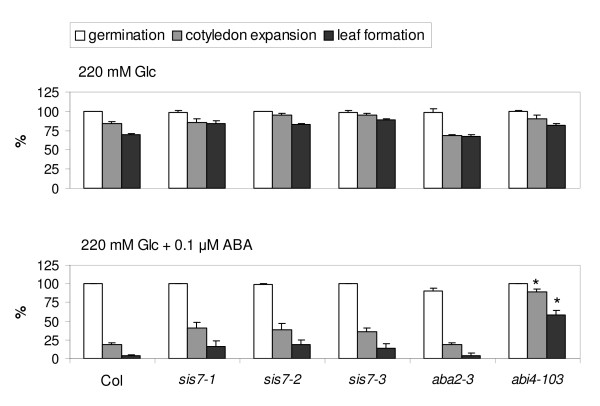
Consistent with other aba mutants, sis7-1/nced3-4/sto1-4 exhibits a wilty phenotype when subject to water deprivation (data not shown). To determine if the sugar insensitivity of the sis7/nced3/sto1 mutants is affected by ABA deficiency, pharmacological complementation of sis7/nced3/sto1 mutant seedlings was conducted as described [11,17]. Early seedling development of Col wild type, three sis7 mutant alleles, the ABA deficient mutant aba2-3 and an ABA insensitive mutant abi4-103 were not affected by 220 mM Glc media. Addition of a noninhibitory level of 0.1 μM ABA to the media containing 220 mM (4%) Glc restored Glc sensitivity of sis7-1/nced3-4/sto1-4, sis7-2/nced3-5/sto1-5 and sis7-3/nced3-6/sto1-6, as well as of the ABA deficient mutant aba2-3, but not that of the ABA insensitive mutant abi4-103 (Figure 5). These results indicate that the sis phenotype of all three sis7 mutant alleles is caused by ABA deficiency.
Figure 5.
Exogenous ABA causes sis7 mutants to exhibit approximately wild-type sugar sensitivity. Wild-type (Col) and mutant seeds were cold treated for 3 d and then sown on minimal media supplemented with 220 mM Glc, with or without 0.1 μM ABA. Additional mutants deficient in ABA biosynthesis (aba2-3) or ABA response (abi4-103) were included as controls. Percent seed germination, cotyledon expansion and true leaf formation were scored after an additional 14 d incubation under continuous light at 22°C. Data represent the means of three independent assays. The error bars represent standard deviations. Asterisks indicate results where the phenotype of one of the mutants differed from that of the wild type with a p-value of less than 0.05 according to a Student's t-test.
Glc regulation of ABA biosynthesis gene expression in germinating seeds
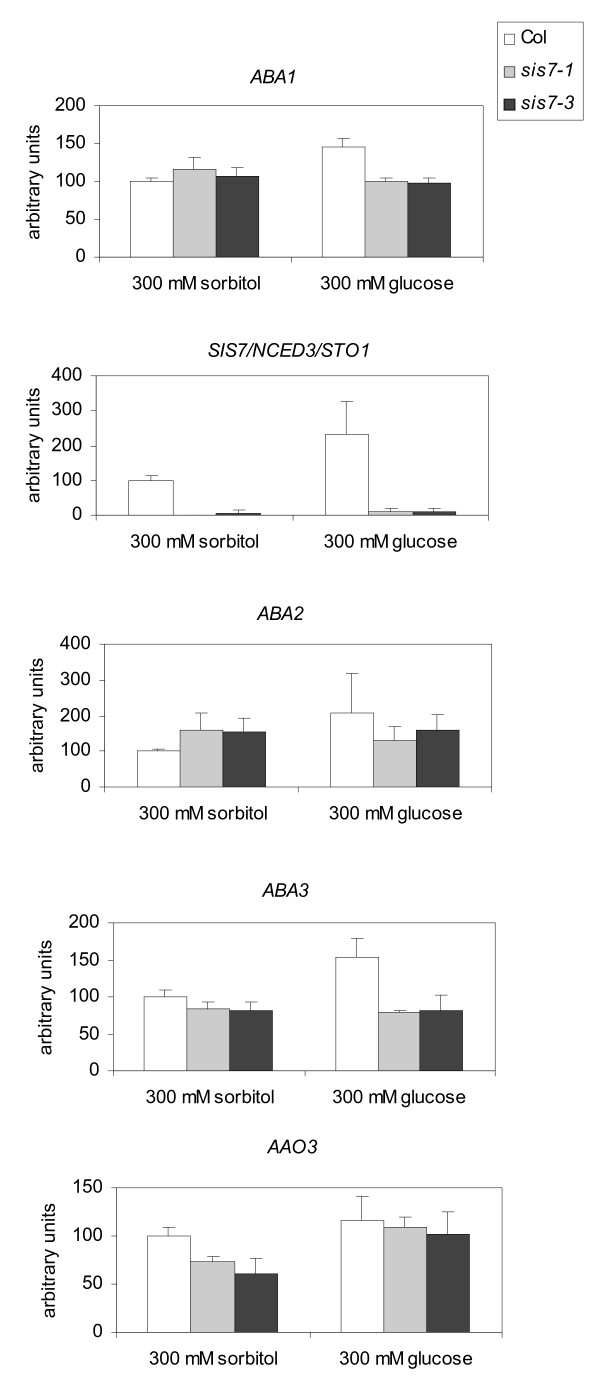
Previously it has been shown that 7% Glc causes ABA accumulation (or decreased metabolism) in wild-type seedlings and that an increase in ABA levels is required for Glc-dependent developmental arrest [11]. The expression levels of the ABA biosynthesis genes, ABA1, ABA2, AAO3 and ABA3 were found to be increased by 2 and 6% Glc. In contrast, NCED3 expression was not activated by Glc [17]. However, as these studies were conducted using young seedlings, the mechanism of high Glc-induced ABA accumulation in germinating seeds remains unclear. Furthermore, it has been shown that high concentrations of exogenous sugars inhibit early seedling development only during a narrow temporal window (less than 40 hours after the start of imbibition) [12]. We therefore performed quantitative real-time PCR (qRT-PCR) analysis of ABA biosynthesis gene expression in both sis7 and wild-type germinating seeds. In brief, two biologically independent batches of Col wild-type, sis7-1 and sis7-3 seeds were sown on minimal media for 20 h and transferred to media supplemented with 300 mM Glc or 300 mM sorbitol for 13 h under continuous light before harvest. The total time of treatment was within the temporal window during which exogenous sugars can arrest seedling development [12]. Total RNA was extracted for qRT-PCR analysis. As shown in Figure 6, ABA1, SIS7/NCED3, ABA2 and ABA3 exhibit higher steady-state mRNA levels in wild-type seeds germinating in the presence of 300 mM Glc than in the presence of 300 mM sorbitol. In contrast, AAO3 steady-state mRNA levels are not significantly different in wild-type seeds germinating on Glc than on sorbitol. Glc induction of SIS7/NCED3, as well as ABA1, ABA2 and ABA3, is abolished in the sis7-1 and sis7-3 null mutants, suggesting a certain level of endogenous ABA may be required for Glc induction of ABA biosynthesis gene expression in germinating seeds.
Figure 6.
Quantitative RT-PCR analysis of ABA biosynthesis genes in sis7 and wild-type germinating seeds. Relative expression levels were measured by qRT-PCR in Col, sis7-1 and sis7-3 germinating seeds treated with 300 mM Glc or 300 mM sorbitol. Data were obtained from two biologically independent experiments and relative mRNA levels were determined by using UBQ6 as a reference. The relative mRNA level of each gene in sorbitol-treated Col was set to 100. Bars indicate standard deviations.
Transcriptional profiling identifies mis-regulated genes in the sis7/nced3/sto1 mutant
To identify genes that are differentially expressed in sis7-1/nced3-4/sto1-4 versus wild type, Arabidopsis Affymetrix ATH1 GeneChips were used for transcriptional profiling of germinating sis7-1/nced3-4/sto1-4 and wild-type seeds. Col wild-type and sis7-1/nced3-4/sto1-4 seeds were sown on minimal media for 20 h and transferred to 100 mM Glc or equimolar sorbitol (as an osmotic control) media for 13 h under continuous light to allow induction or repression of Glc-responsive genes. At the end of this incubation period 0–4% of the seeds had germinated. The reason that seeds were collected after a total of 33 h is based on the observation that exogenous sugars inhibit seedling development during a narrow temporal window (less than 40 hours after start of imbibition) [12]. Data analysis using the Expressionist software package is as detailed in "Materials and methods". To identify genes that are expressed at significantly different levels in sis7-1/nced3-4/sto1-4 versus wild-type germinating seeds grown in the presence of 100 mM Glc, the results of three independent GeneChip experiments using mutant seeds were compared with the results of six independent biological replicates of wild-type samples. Significant differences in expression values were defined as those where the average expression levels between mutant and wild-type seeds differ by at least two fold and have a Student's t-test p value of less than 0.05. Using these cutoffs, the levels of 83 transcripts were found to be altered in sis7-1/nced3-4/sto1-4 versus wild-type seeds incubated in the presence of 100 mM Glc (Table 1). Of these genes, 20 had higher mRNA levels in the mutant than in the wild type, whereas 63 genes had lower mRNA levels in the mutant.
Table 1.
Genes with altered expression in sis7-1 versus wild-type (WT) seeds incubated on 100 mM Glc media.
| Description | AGI | sis7-1 | WT | sis7-1/WT | P-Value |
| genes with higher transcript levels in sis7-1 | |||||
| expressed protein | AT5G58375 | 116965 | 3030 | 38.60 | 9.47E-09 |
| 2S seed storage protein 4 | AT4G27170 | 15028 | 874.6 | 17.18 | 0.002 |
| cytosolic small heat shock protein | AT5G12030 | 6968 | 977.5 | 7.13 | 0.020 |
| protease inhibitor/seed storage family protein | AT5G54740 | 9475 | 1824 | 5.19 | 0.033 |
| heat shock protein 70, putative | AT3G12580 | 36044 | 9615 | 3.75 | 0.016 |
| expressed protein | AT2G32210 | 5544 | 1555 | 3.56 | 0.004 |
| gibberellin-responsive protein 3 | AT4G09600 | 2995 | 1066 | 2.81 | 0.008 |
| alternative oxidase 1a, mitochondrial (AOX1A) | AT3G22370 | 4803 | 1982 | 2.42 | 0.024 |
| low temperature and salt responsive protein | AT4G30650 | 4272 | 1769 | 2.42 | 0.029 |
| putative GTP-binding protein | AT1G09180 | 1816 | 758.1 | 2.40 | 0.026 |
| AP2 domain transcription factor | AT2G40350 | 1848 | 787.9 | 2.35 | 0.031 |
| steroid 5alpha-reductase-like protein | AT5G16010 | 10002 | 4287 | 2.33 | 0.001 |
| expressed protein | AT5G24570 | 3708 | 1594 | 2.33 | 0.028 |
| arabinogalactan-protein | AT5G64310 | 33863 | 14633 | 2.31 | 0.011 |
| CDC48 – like protein | AT3G53230 | 21244 | 9218 | 2.30 | 0.007 |
| pathogenesis-related thaumatin family protein | AT4G36010 | 10479 | 4607 | 2.27 | 0.013 |
| mitochondrial heat shock 22 kd protein-like | AT5G51440 | 3154 | 1492 | 2.11 | 0.041 |
| expressed protein | AT5G64230 | 2056 | 982.4 | 2.09 | 0.026 |
| glycine-rich protein/oleosin | AT3G18570 | 3873 | 1872 | 2.07 | 0.006 |
| salt-tolerance zinc finger protein | AT1G27730 | 4463 | 2200 | 2.03 | 0.017 |
| genes with lower transcript levels in sis7-1 | |||||
| cytochrome P450 79B2, putative (CYP79B2) | AT4G39950 | 1389 | 7597 | 0.18 | 0.001 |
| RuBisCO activase | AT2G39730 | 1489 | 4706 | 0.32 | 0.007 |
| auxin-responsive GH3 family protein | AT4G27260 | 2324 | 6679 | 0.35 | 0.047 |
| cytochrome P450 83B1 (CYP83B1) | AT4G31500 | 15681 | 44352 | 0.35 | 0.003 |
| S1 RNA-binding domain-containing protein | AT1G12800 | 1259 | 3475 | 0.36 | 0.007 |
| pectin methylesterase | AT1G11580 | 4380 | 11956 | 0.37 | 0.009 |
| DegP2 protease | AT2G47940 | 1759 | 4776 | 0.37 | 0.007 |
| AMP-binding protein | AT3G23790 | 878 | 2291 | 0.38 | 0.040 |
| putative activator subunit of SNF1-related protein kinase, SNF4 | AT1G09020 | 1759 | 4584 | 0.38 | 0.035 |
| WPP-domain interacting protein 3 | AT3G13360 | 627 | 1614 | 0.39 | 0.014 |
| MATE efflux family protein | AT1G71870 | 1830 | 4621 | 0.40 | 0.016 |
| glutathione S-transferase | AT2G30860 | 36776 | 92522 | 0.40 | 0.002 |
| hydrolase family protein | AT2G25870 | 981 | 2438 | 0.40 | 0.000 |
| endo-beta-1,4-glucanase | AT1G64390 | 2208 | 5381 | 0.41 | 0.011 |
| heat shock protein 100, putative | AT5G15450 | 1817 | 4398 | 0.41 | 0.026 |
| eIF-2 family protein | AT1G76810 | 748 | 1809 | 0.41 | 0.035 |
| auxin transport protein, putative (PIN3) | AT1G70940 | 1147 | 2735 | 0.42 | 0.009 |
| male sterility MS5 family protein | AT5G48850 | 1109 | 2638 | 0.42 | 0.032 |
| phosphatidylinositolglycan class family protein | AT3G01380 | 1150 | 2691 | 0.43 | 0.006 |
| epsilon-adaptin, putative | AT1G31730 | 1767 | 4134 | 0.43 | 0.016 |
| PSII low MW protein | ATCG00220 | 11808 | 27598 | 0.43 | 0.026 |
| protein kinase family protein | AT1G75640 | 393 | 897.5 | 0.44 | 0.021 |
| guanylate-binding family protein | AT5G46070 | 607 | 1382 | 0.44 | 0.018 |
| proteaseI (pfpI)-like protein (YLS5) | AT2G38860 | 4039 | 9166 | 0.44 | 0.008 |
| sulfotransferase family protein | AT1G74100 | 11829 | 26477 | 0.45 | 0.002 |
| chlorophyll A-B binding protein 4, chloroplast | AT3G47470 | 532 | 1175 | 0.45 | 0.042 |
| senescence-associated protein-related | AT5G20700 | 633 | 1395 | 0.45 | 0.028 |
| early dehydration-induced gene ERD13 | AT2G30870 | 3684 | 8114 | 0.45 | 0.046 |
| squamosa promoter-binding protein-like 1 | AT2G47070 | 976 | 2147 | 0.45 | 0.011 |
| heavy-metal-associated domain protein | AT5G19090 | 1141 | 2502 | 0.46 | 0.010 |
| expressed protein | AT3G50370 | 1209 | 2652 | 0.46 | 0.004 |
| methyltransferase MT-A70 family protein | AT4G09980 | 850 | 1840 | 0.46 | 0.005 |
| SET domain-containing protein | AT2G19640 | 2024 | 4381 | 0.46 | 0.008 |
| UV-damaged DNA-binding protein, putative | AT4G05420 | 2387 | 5151 | 0.46 | 0.000 |
| expressed protein | AT4G27450 | 7573 | 16323 | 0.46 | 0.010 |
| zinc finger protein-related | AT3G18290 | 1140 | 2444 | 0.47 | 0.005 |
| uroporphyrinogen decarboxylase, putative | AT2G40490 | 1408 | 2997 | 0.47 | 0.017 |
| molybdenum cofactor sulfurase family protein | AT5G44720 | 1619 | 3427 | 0.47 | 0.020 |
| expressed protein | AT1G36990 | 1152 | 2437 | 0.47 | 0.008 |
| kinesin motor family protein | AT5G66310 | 255 | 538.8 | 0.47 | 0.019 |
| pentatricopeptide repeat-containing protein | AT1G71060 | 419 | 883 | 0.47 | 0.035 |
| protein kinase-related | AT3G03930 | 441 | 927.2 | 0.48 | 0.008 |
| exportin1 (XPO1) | AT5G17020 | 2877 | 6035 | 0.48 | 0.010 |
| auxin efflux carrier protein, putative (PIN1) | AT1G73590 | 2463 | 5154 | 0.48 | 0.021 |
| pentatricopeptide repeat-containing protein | AT5G46580 | 3046 | 6365 | 0.48 | 0.007 |
| pentatricopeptide repeat-containing protein | AT5G28370 | 3099 | 6453 | 0.48 | 0.019 |
| expressed protein | AT5G63135 | 902 | 1875 | 0.48 | 0.011 |
| male sterility MS5 family protein | AT1G04770 | 2548 | 5292 | 0.48 | 0.004 |
| MAPK, putative (MPK8) | AT1G18150 | 749 | 1541 | 0.49 | 0.046 |
| expressed protein | AT5G64460 | 769 | 1580 | 0.49 | 0.043 |
| importin beta-2 subunit family protein | AT2G31660 | 2429 | 4987 | 0.49 | 0.007 |
| defense-related protein, putative | AT4G30530 | 3242 | 6650 | 0.49 | 0.003 |
| calcium-transporting ATPase 4 | AT2G41560 | 6059 | 12419 | 0.49 | 0.001 |
| ubiquitin-transferase family protein | AT4G38600 | 5327 | 10886 | 0.49 | 0.011 |
| sulfite reductase/ferredoxin (SIR) | AT5G04590 | 4299 | 8781 | 0.49 | 0.020 |
| VARIEGATED 3 | AT5G17790 | 646 | 1317 | 0.49 | 0.005 |
| heat shock protein, putative | AT1G11660 | 1120 | 2280 | 0.49 | 0.030 |
| pentatricopeptide repeat-containing protein | AT2G17033 | 716 | 1452 | 0.49 | 0.013 |
| nuclease family protein | AT5G61780 | 8110 | 16406 | 0.49 | 0.027 |
| pentatricopeptide repeat-containing protein | AT2G31400 | 3807 | 7676 | 0.50 | 0.040 |
| patched family protein | AT4G38350 | 2104 | 4230 | 0.50 | 0.015 |
| Involved in the biosynthesis of brassinosteroids | AT2G07050 | 3755 | 7538 | 0.50 | 0.022 |
| ATP phosphoribosyl transferase | AT1G09795 | 1402 | 2805 | 0.50 | 0.000 |
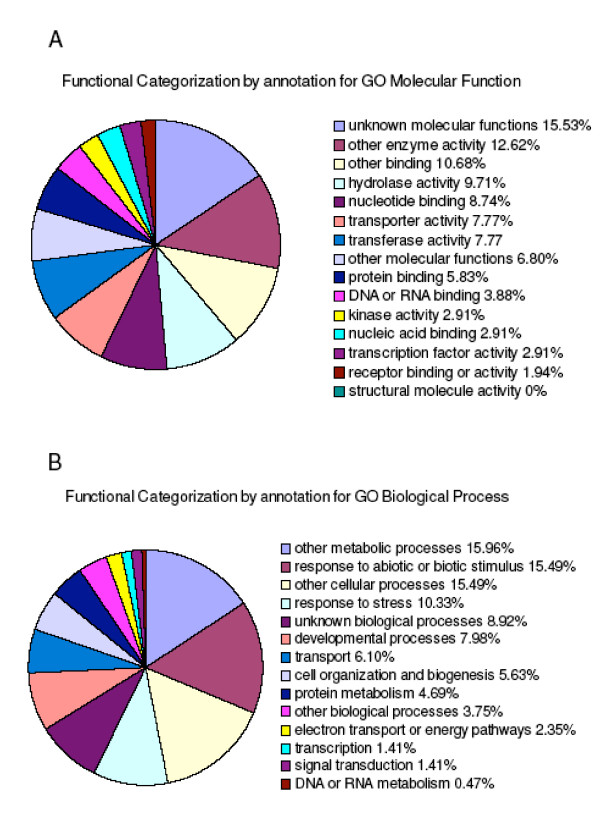
Functional categorization of these mis-regulated genes was performed using the Gene Ontology (GO) Annotations tool from TAIR [39] and the results are presented in Figure 7. Notably, a significant percentage of the mis-regulated genes are involved in response to abiotic or biotic stimulus and to stress (a combined 25.82%). Over-representation analysis (ORA) was also performed. ORA is a useful tool for analyzing gene expression data obtained from microarray experiments as it can determine whether a set of genes is statistically over-represented among the total genes expressed on the microarray. An online version of the GeneMerge program [40] was used for ORA to identify whether particular biological processes or molecular functions, as defined by GO annotations, are statistically enriched in the set of differentially expressed genes in the sis7-1 mutant compared to the wild type [41]. Genes involved in response to stimulus (GO:0050896) are significantly enriched (16/83; p = 0.0095 after Bonferroni correction for multiple tests) and genes involved in response to abiotic stimulus (GO:0009628, a child term of GO:0050896) are also significantly over-represented (10/83; p = 0.0167). Both of these sets of genes include PIN1, PIN3 and CYP83B1. Manual inspection of the data identified two other genes involved in auxin metabolism: CYP79B2 and At4g27260. Thus, interestingly, genes involved in auxin biosynthesis and transport represent 6% of the genes expressed at altered levels in sis7-1 versus wild-type seeds grown on 100 mM Glc, and so are significantly over-represented amongst these genes, as auxin-related genes comprise far less than 6% of the Arabidopsis genome. CYP79B2 is involved in Trp metabolism and converts Trp to indole-3-acetaldoxime (IAOx), a precursor to indole acetic acid (IAA) [42,43]. Expression of CYP79B2 is reduced more than five-fold in mutant seeds relative to wild-type seeds grown in the presence of 100 mM Glc. At4g27260 encodes an IAA-amido synthase that conjugates Asp and other amino acids to auxin in vitro. An insertional mutation in this gene causes increased sensitivity to IAA in seedling roots [44]. CYP83B1 is required for red light signal transduction and auxin homeostasis [45]. PIN1 and PIN3 are auxin efflux transporters and regulate root development [46]. Interplay between auxin and ABA signaling to regulate lateral root development has been suggested by previous studies [47-49]. The decreased mRNA levels of these auxin-related genes in Glc-treated sis7-1/nced3-4/sto1-4 seeds provide further evidence of cross-talk between these pathways.
Figure 7.
Functional categorization of genes mis-regulated in sis7-1 on 100 mM Glc. Affymetrix ATH1 GeneChips were used for transcriptional profiling of germinating sis7-1/nced3-4/sto1-4 and wild-type seeds. Col wild-type and sis7-1/nced3-4/sto1-4 seeds were sown on minimal media for 20 h and transferred to 100 mM Glc media for 13 h under continuous light. The Expressionist software package was used to identify genes with transcript levels that differ by at least two fold and with a p value of less than 0.05 when comparing the results of mutant versus wild-type samples. Using these cutoffs, the levels of 83 transcripts were found to be altered in sis7-1/nced3-4/sto1-4 versus wild-type seeds (Table 1). Functional categorization of these mis-regulated genes was performed using the GO (Gene Ontology) Annotations tool from TAIR [39]. Panel A shows the distribution of annotations for molecular function and panel B shows the distribution for annotations for biological process. Percentages were calculated as: (number of annotations to terms in this GOslim category/number of total annotations to terms in this ontology) × 100.
To determine which of the genes listed in Table 1 are glucose regulated, the data from GeneChip experiments performed using seeds grown in the presence of 100 mM Glc were compared with data from GeneChip experiments performed using seeds grown in the presence of equimolar sorbitol. Based on these comparisons, the fold change in expression level (Glc/sorbitol) was determined for each of the genes listed in Table 1 for both sis7-1 and wild-type seeds (Table 2). Based on their glucose-responsiveness in sis7-1 and wild type, these genes can be classified into three categories. The first category includes genes that show similar glucose induction or repression in both sis7-1 and wild type. For example, the photosynthesis-related genes encoding CAB4 and RuBisCo activase are repressed by 100 mM Glc in wild-type as well as in sis7-1 seeds (Table 2). Similarly, the expression of PIN3 is Glc-repressed in both sis7-1 and wild type (Table 2 and Figure 8). The second category represents the majority of the genes on the list, which are not significantly regulated by Glc in either wild-type or sis7-1 seeds. The third category contains a few genes that are Glc-regulated in sis7-1 but that exhibit little to no Glc regulation in wild type. For example, At4g30650 transcript levels are increased 3.6 fold by 100 mM Glc in sis7-1 seeds but only 1.3 fold in wild-type seeds. Similarly, the expression of CYP79B2 is repressed 2-fold by Glc in sis7-1 seeds but only 1.4 fold in wild-type seeds. At5g46070 transcript levels are decreased approximately 2-fold by 100 mM Glc in sis7-1 seeds but are unchanged by Glc in wild-type seeds (Table 2 and Figure 8). The identification of this third category of genes, combined with the fact that sis7-1 seeds are ABA-deficient [27,29], suggests that the Glc-responsiveness of the genes in the third category is affected by endogenous ABA levels.
Table 2.
Glc-responsiveness of genes identified in Table I in sis7-1 and wild-type (WT) seeds. ND = not determined.
| AGI | Description | sis7-1 glc/sorbitol | WT glc/sorbitol |
| AT1G04770 | male sterility MS5 family protein | 0.54 | 0.64 |
| AT1G09020 | putative activator subunit of SNF1-related protein kinase, SNF4 | 0.35 | 0.69 |
| AT1G09180 | putative GTP-binding protein | 0.94 | 1.54 |
| AT1G09795 | ATP phosphoribosyl transferase | 0.62 | 1.11 |
| AT1G11580 | pectin methylesterase | 0.21 | 0.38 |
| AT1G11660 | heat shock protein, putative | 0.82 | 1.20 |
| AT1G12800 | S1 RNA-binding domain-containing protein | 0.55 | 0.84 |
| AT1G18150 | MAPK, putative (MPK8) | 0.52 | 0.93 |
| AT1G27730 | salt-tolerance zinc finger protein | 3.28 | 2.04 |
| AT1G31730 | epsilon-adaptin, putative | 0.51 | 0.85 |
| AT1G36990 | expressed protein | 0.56 | 0.81 |
| AT1G64390 | endo-beta-1,4-glucanase | 0.39 | 0.65 |
| AT1G70940 | auxin transport protein, putative (PIN3) | 0.53 | 0.66 |
| AT1G71060 | pentatricopeptide repeat-containing protein | 0.64 | 0.92 |
| AT1G71870 | MATE efflux family protein | 0.36 | 0.69 |
| AT1G73590 | auxin efflux carrier protein, putative (PIN1) | 0.77 | 1.09 |
| AT1G74100 | sulfotransferase family protein | 0.73 | 0.89 |
| AT1G75640 | protein kinase family protein | 0.57 | 1.10 |
| AT1G76810 | eIF-2 family protein | 0.64 | 1.05 |
| AT2G07050 | Involved in the biosynthesis of brassinosteroids | 0.66 | 0.99 |
| AT2G17033 | pentatricopeptide repeat-containing protein | 0.61 | 0.71 |
| AT2G19640 | SET domain-containing protein | 0.51 | 0.93 |
| AT2G25870 | hydrolase family protein | 0.63 | 1.14 |
| AT2G30860 | glutathione S-transferase | 0.70 | 0.91 |
| AT2G30870 | early dehydration-induced gene ERD13 | 0.91 | 1.08 |
| AT2G31400 | pentatricopeptide repeat-containing protein | 0.53 | 0.73 |
| AT2G31660 | importin beta-2 subunit family protein | 0.76 | 1.11 |
| AT2G32210 | expressed protein | 3.32 | 1.93 |
| AT2G38860 | proteaseI (pfpI)-like protein (YLS5) | 1.02 | 1.17 |
| AT2G39730 | RuBisCO activase | 0.42 | 0.52 |
| AT2G40350 | AP2 domain transcription factor | 0.86 | 1.11 |
| AT2G40490 | uroporphyrinogen decarboxylase, putative | 0.55 | 0.67 |
| AT2G41560 | calcium-transporting ATPase 4 | 0.44 | 0.68 |
| AT2G47070 | squamosa promoter-binding protein-like 1 | 0.52 | 1.10 |
| AT2G47940 | DegP2 protease | 0.58 | 0.91 |
| AT3G01380 | phosphatidylinositolglycan class family protein | 0.42 | 0.79 |
| AT3G03930 | protein kinase-related | 0.78 | 0.90 |
| AT3G12580 | heat shock protein 70, putative | 1.08 | 1.09 |
| AT3G13360 | WPP-domain interacting protein 3 | 0.56 | 1.48 |
| AT3G18290 | zinc finger protein-related | 0.39 | 0.66 |
| AT3G18570 | glycine-rich protein/oleosin | 3.23 | N/D |
| AT3G22370 | alternative oxidase 1a, mitochondrial (AOX1A) | 1.40 | 1.38 |
| AT3G23790 | AMP-binding protein | 0.40 | 0.75 |
| AT3G47470 | chlorophyll A-B binding protein 4, chloroplast | 0.17 | 0.41 |
| AT3G50370 | expressed protein | 0.52 | 0.86 |
| AT3G53230 | CDC48 – like protein | 0.81 | 0.81 |
| AT4G05420 | UV-damaged DNA-binding protein, putative | 0.53 | 0.84 |
| AT4G09600 | gibberellin-responsive protein 3 | 2.53 | 1.90 |
| AT4G09980 | methyltransferase MT-A70 family protein | 0.51 | 0.78 |
| AT4G27170 | 2S seed storage protein 4 | 1.31 | 1.25 |
| AT4G27260 | auxin-responsive GH3 family protein | 0.89 | 1.11 |
| AT4G27450 | expressed protein | 0.24 | 0.40 |
| AT4G30530 | defense-related protein, putative | 0.78 | 0.81 |
| AT4G30650 | low temperature and salt responsive protein | 3.62 | 1.33 |
| AT4G31500 | cytochrome P450 83B1 (CYP83B1) | 0.60 | 0.77 |
| AT4G36010 | pathogenesis-related thaumatin family protein | 1.61 | 1.79 |
| AT4G38350 | patched family protein | 0.68 | 0.87 |
| AT4G38600 | ubiquitin-transferase family protein | 0.67 | 0.95 |
| AT4G39950 | cytochrome P450 79B2, putative (CYP79B2) | 0.49 | 0.70 |
| AT5G04590 | sulfite reductase/ferredoxin (SIR) | 0.72 | 1.01 |
| AT5G12030 | cytosolic small heat shock protein | 1.57 | 1.16 |
| AT5G15450 | heat shock protein 100, putative | 0.93 | 1.51 |
| AT5G16010 | steroid 5alpha-reductase-like protein | 1.12 | 1.16 |
| AT5G17020 | exportin1 (XPO1) | 0.61 | 0.86 |
| AT5G17790 | VARIEGATED 3 | 0.43 | 0.85 |
| AT5G19090 | heavy-metal-associated domain protein | 0.72 | 0.98 |
| AT5G20700 | senescence-associated protein-related | 0.66 | 0.87 |
| AT5G24570 | expressed protein | 1.11 | 0.96 |
| AT5G28370 | pentatricopeptide repeat-containing protein | 0.73 | 1.07 |
| AT5G44720 | molybdenum cofactor sulfurase family protein | 0.71 | 0.89 |
| AT5G46070 | guanylate-binding family protein | 0.46 | 1.02 |
| AT5G46580 | pentatricopeptide repeat-containing protein | 0.60 | 0.80 |
| AT5G48850 | male sterility MS5 family protein | 1.39 | 1.76 |
| AT5G51440 | mitochondrial heat shock 22 kd protein-like | 1.25 | 0.98 |
| AT5G54740 | protease inhibitor/seed storage family protein | 0.82 | 0.94 |
| AT5G58375 | expressed protein | 0.85 | 1.02 |
| AT5G61780 | nuclease family protein | 0.62 | 0.76 |
| AT5G63135 | expressed protein | 0.65 | 1.23 |
| AT5G64230 | expressed protein | 1.26 | 1.24 |
| AT5G64310 | arabinogalactan-protein | 5.52 | 3.36 |
| AT5G64460 | expressed protein | 0.63 | 0.70 |
| AT5G66310 | kinesin motor family protein | 0.75 | N/D |
| ATCG00220 | PSII low MW protein | 1.05 | 1.06 |
Figure 8.
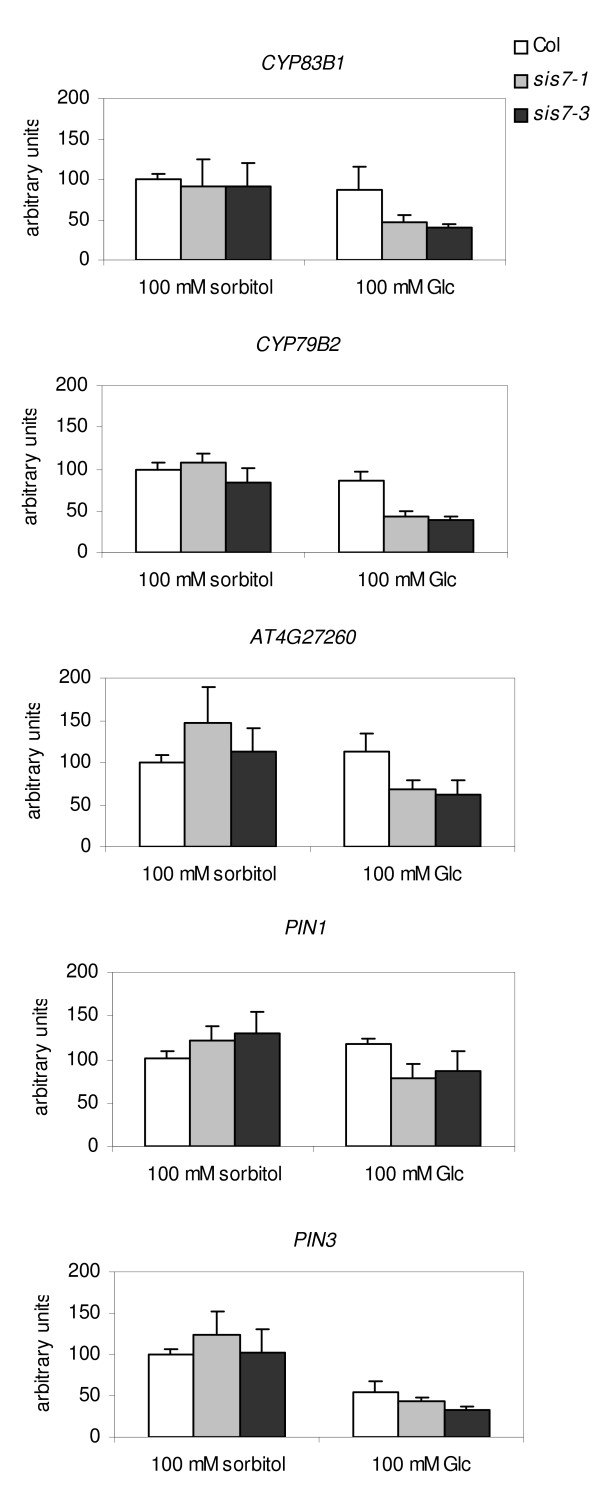
Quantitative RT-PCR analysis of auxin-related genes. Genes involved in auxin metabolism or transport were chosen for analysis. Relative expression levels of these genes were measured by qRT-PCR in wild-type Col, sis7-1 and sis7-3 germinating seeds treated with 100 mM Glc or 100 mM sorbitol. Data were obtained from two biologically independent experiments and relative mRNA levels were determined by using UBQ6 as a reference. The relative mRNA level of each gene in sorbitol-treated wild-type Col was set to 100. Bars indicate standard deviations.
Quantitative real-time RT-PCR validation of microarray data
To re-test the results obtained through microarray experiments, qRT-PCR analysis was performed on selected genes, with an emphasis placed on genes related to auxin transport and metabolism. Two biologically independent batches of Col wild-type, sis7-1 and sis7-3 seeds were sown on minimal media, incubated for 20 h and then transferred to 100 mM Glc or 100 mM sorbitol media and incubated for an additional 13 h under continuous light before harvest. Total RNA was extracted for qRT-PCR analysis to monitor the relative expression levels of an array of genes, including PIN1, PIN3, CYP83B1, CYP79B2 and AT4G27260 (an IAA-amido synthase). On 100 mM sorbitol (osmotic control) media, the expression levels of these genes are similar in sis7-1, sis7-3 and Col wild type. In contrast, on 100 mM Glc media, the transcript levels of each gene are lower in the sis7-1 and sis7-3 mutants than in the Col wild type (Figure 8). These qRT-PCR results confirm the findings from microarray data that the steady-state mRNA levels of genes involved in auxin transport and metabolism are lower in the sis7 mutants than in the wild type on 100 mM Glc.
sis7-1 mutants exhibit increased lateral root systems
The decreased mRNA levels of auxin-related genes observed in Glc-treated sis7-1/nced3-4/sto1-4 seeds provide further evidence of cross-talk between these pathways. To test whether the osmotic regulation of lateral root development is altered in the sis7 mutants, wild-type and sis7-1/nced3-4/sto1-4 seeds were germinated and grown on sorbitol media. The sis7-1 seedlings exhibit increased lateral root systems on sorbitol, when compared to the wild-type plants (Figure 9). These observations are consistent with previous findings that aba2-1 and aba3-1 display increased root system size on mannitol [48].
Figure 9.
sis7-1 exhibits increased root system size on sorbitol media. Wild-type Col and sis7-1 seedlings were grown for 12 d on vertically-oriented Petri plates containing the indicated media. Root systems of sis7-1 are less repressed by osmotica than are the root systems of wild-type seedlings.
Evidence for an expanded role for ABI3 in sugar response through mapped-based cloning of SIS10
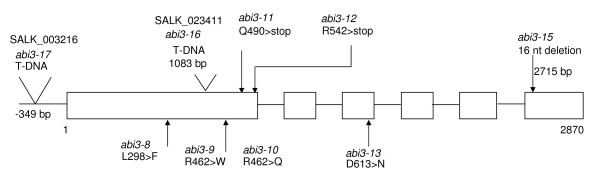
The sis10-1 mutant was isolated during the same screen of a T-DNA mutagenized population, described above, that was used to isolate the sis7-1 and sis7-3 mutants. The sis10-1 mutant has strong Glc and Suc insensitive phenotypes during early seedling development. A map-based approach was used to identify the SIS10 gene. Towards this end, sis10-1 (in the Col-0 ecotype) was crossed with a wild-type plant of the Hi-O ecotype. F2 seeds from this cross were selected using Arabidopsis media supplemented with 320 mM Suc to identify seedlings with a sis mutant phenotype. An F2 population of 320 sis plants was then analyzed using SSLP markers. The sis10-1 mutation was mapped to chromosome 3, approximately 0.15 cM above marker 470509 on BAC MSD24. Examination of this region of the genome revealed that ABI3 (At3g24650) is present in the region believed to contain the sis10 locus. Sequencing of ABI3 DNA isolated from sis10-1 revealed that the sis10-1 mutation results in deletion of 16 nucleotides between base pairs 2715 and 2730 of the genomic DNA (with respect to the start codon) in the 6th exon of ABI3. Accordingly, the sis10-1 mutant has been renamed abi3-15.
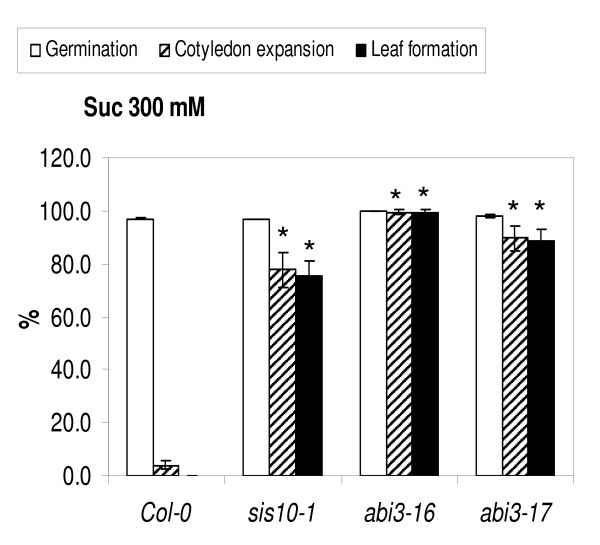
Previous work from several labs [11,13,16] characterizing the abi3-1 mutant, which is in the Landsberg erecta background, found only slight to no effect of this mutation on sugar response. Therefore, the finding that the sis10-1 mutation, which causes a strong sis phenotype, lies in the ABI3 gene was unexpected. To test whether the mutation in ABI3 is truly the cause of the sis10-1 sugar insensitive phenotype, two additional abi3 mutant lines were obtained and characterized. SALK_023411 carries a mutation, here designated as abi3-16, consisting of a T-DNA insertion in the first exon of the ABI3 gene (Figure 10). SALK_003216 carries a mutation, here designated as abi3-17, consisting of a T-DNA insertion in the ABI3 promoter region [50]. Sugar-insensitivity assays on plants homozygous for the sis10-1/abi3-15, abi3-16 and abi3-17 mutations reveal that all three mutations confer significant resistance to the inhibitory effects of 300 mM Suc on early seedling development, compared to wild-type plants (Figure 11). These results confirm that mutations in ABI3 can lead to a strong sugar-insensitive phenotype.
Figure 10.
Molecular lesions present in different abi3 mutants. Nucleotides are numbered with respect to the start codon. Exons are depicted with boxes and introns are depicted with single lines.
Figure 11.
Suc-response assays of three abi3 mutants. Seeds were sown on media containing 300 mM Suc and incubated at 22°C for 14 d under 2200-3400 Lux continuous light prior to scoring. Seed germination, cotyledon expansion and true leaf formation were scored as previously described [16]. Error bars indicate standard deviations. Asterisks indicate results where the mutant phenotype differed from the corresponding wild-type phenotype with a p-value of less than 0.05 according to a Student's t-test.
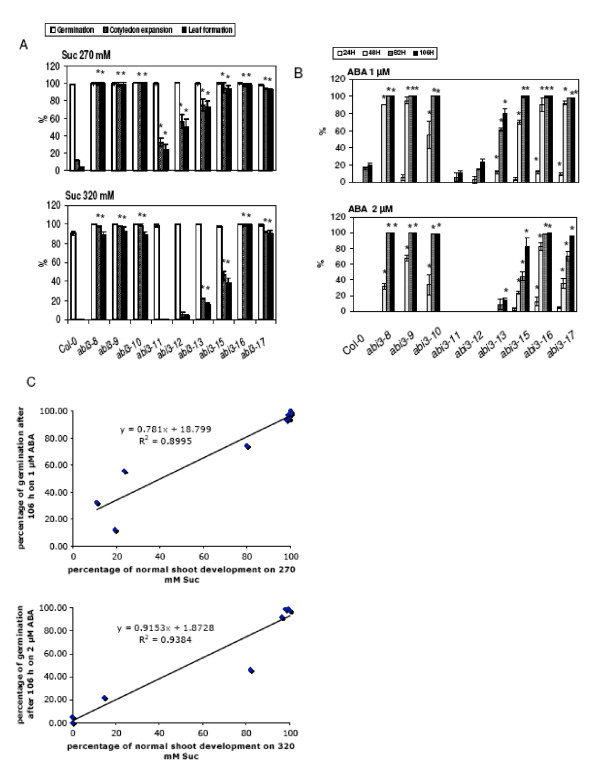
Previous work indicated that abi3-1 has a relatively strong ABA insensitive seed germination phenotype [28], but displays only a very subtle sugar insensitive phenotype during early seedling development [11,13,16]. The finding that other abi3 mutants, such as sis10-1/abi3-15, abi3-16 and abi3-17, can have strong sugar-insensitive phenotypes during early seedling development raised the possibility that different mutations in ABI3 may predominantly affect either ABA or sugar response. To test this hypothesis a careful analysis was conducted to determine the relative degrees of sensitivity to the inhibitory effects of ABA on seed germination and of Suc on early seedling development of nine different abi3 mutant lines (Figure 12). These nine abi3 lines included the sis10-1/abi3-15, abi3-16 and abi3-17 mutants described above, as well as the abi3-8, abi3-9, abi3-10, abi3-11, abi3-12 and abi3-13 mutants [23]. All of these mutants are in the Col ecotype. The molecular lesions of the different abi3 mutants are depicted in Figure 10. To measure sugar response, seeds of all nine abi3 mutants plus Col wild-type were sown on minimal Arabidopsis media [31] supplemented with 270 or 320 mM Suc and grown in continuous light for 14 d before scoring. These Suc concentrations were chosen to allow detection of differences in the degree of Suc insensitivity. The 270 mM Suc concentration is sufficient to allow detection of mutants with relatively weak sugar-insensitive phenotypes, as these mutants will still exhibit a significantly greater ability than wild-type plants to develop expanded cotyledons and true leaves on 270 mM Suc. However, a 270 mM concentration of Suc may not be sufficient to distinguish between mutations that confer moderate versus strong sugar-insensitive phenotypes. This distinction can be made using the 320 mM Suc concentration. Selection on media supplemented with 320 mM Suc revealed that five of the abi3 mutations (abi3-8, abi3-9, abi3-10, abi3-16 and abi3-17) confer strong sugar-insensitive phenotypes. The other abi3 mutations tested conferred lesser degrees of sugar-insensitivity. The relative degree of sugar insensitivity of these other four mutants, from highest to lowest, is: abi3-15, abi3-13, abi3-12 and abi3-11 (Figure 12A). To assess the relative effects of the different abi3 mutations on ABA sensitivity, seed germination was assayed at different time points and in the presence of different concentrations of ABA (Figure 12B). A regression analysis of the results indicates a strong positive correlation between sugar insensitivity and ABA insensitivity of tested abi mutants (Fig 12C). In other words, mutations that confer a high degree of insensitivity to ABA also confer a high degree of insensitivity to Suc. Conversely, mutations that confer only weak ABA insensitivity also confer weak insensitivity to Suc.
Figure 12.
Quantification of Suc and ABA insensitivity in nine abi3 mutants. (A) For Suc-response assays seeds were sown on media containing 270 or 320 mM Suc and incubated at 22°C for 14 d under 2200-3400 Lux continuous light prior to scoring. Seed germination, cotyledon expansion and true leaf formation were scored as previously described [16]. Error bars indicate standard deviations. Asterisks indicate results where the mutant phenotype differed from the corresponding wild-type phenotype with a p-value of less than 0.05 according to a Student's t-test. (B) For ABA response assays seeds were imbibed on media with the indicated ABA concentrations and scored for germination after 24, 48, 82 and 106 h. Seed germination is defined as the emergence of the radicle from the seed coat. Error bars indicate standard deviations. Asterisks indicate results where the mutant phenotype differed from the corresponding wild-type phenotype with a p-value of less than 0.05 according to a Student's t-test. (C) Regression analysis of data on Suc and ABA response assays reveals strong positive correlation between sugar- and ABA- insensitivity. Each data point on the scatterplot represents the percentage of normal shoot development (defined as seedlings with green expanded cotyledons) on Suc media and the percentage of seeds that germinated after 106 h on ABA media for each genotype. Correlation coefficients were calculated for two assay conditions: 1 μM ABA and 270 mM Suc representing relatively non-stringent assay conditions and 2 μM ABA and 320 mM Suc representing more stringent assay conditions.
Discussion
Molecular genetic studies on Arabidopsis sugar response mutants have revealed extensive evidence for cross-talk between sugar and phytohormone response pathways [11-13,15-18,51,52]. For example, exogenous Glc has been proposed to slow the decrease in ABA concentrations that occurs during seed germination [53]. Glc has also been shown to help regulate expression of a number of genes involved in ABA metabolism in seedlings. Several ABA biosynthetic genes, including ABA1, AAO3 and ABA3 are upregulated by 110 mM and 330 mM Glc via a mechanism that requires that a certain endogenous ABA level is maintained [17]. Interestingly, these same genes are downregulated by 330 mM mannitol, via a mechanism that does not appear to require wild-type levels of endogenous ABA. These results suggest that regulation of these three genes by Glc is distinct from their regulation by osmotic stress. Similarly to ABA1, AAO3 and ABA3, the ABA biosynthetic gene ABA2 is also upregulated by 110 mM and 330 mM Glc via a mechanism that requires that endogenous ABA levels be maintained above a certain level. However, unlike ABA1, AAO3 and ABA3, expression of ABA2 is also upregulated by 110 mM and 330 mM mannitol via a mechanism that does require maintenance of endogenous ABA levels [17]. So, although regulation of all four genes by Glc appears similar, ABA2 exhibits significant differences in expression in response to osmotic stress.
The ABA biosynthetic gene SIS7/NCED3/STO1 also exhibits an expression pattern that is distinct from that of the other ABA biosynthetic genes discussed above. In wild-type seedlings, the expression of SIS7/NCED3/STO1 is not activated by Glc but is strongly induced by salt or osmotic stress [17,26]. In fact, SIS7/NCED3/STO1 may act primarily in the biosynthesis of ABA during stress conditions [29]. Due to these regulatory and functional differences, it was not clear whether mutations in SIS7/NCED3/STO1 would cause a sugar-insensitive phenotype, as had been previously shown for mutations in other ABA biosynthetic genes [11,13,16]. Results presented here, showing that the sis7 mutations lie in NCED3/STO1, demonstrate that mutations in SIS7/NCED3/STO1 do confer a strong sugar-insensitive phenotype.
Expression of SIS7/NCED3/STO1 has been suggested to be rate-limiting in the positive feedback regulation of ABA biosynthesis by ABA under stress conditions [54,55]. Transcriptional upregulation of SIS7/NCED3/STO1 by ABA has been found in certain ABA-deficient mutants and ecotypes [17,26]. However, the transcriptional regulation of NCED3 as well as other ABA biosynthesis genes by Glc in germinating seeds is unclear. Results presented here (Figure 6) show that in wild-type germinating seeds high concentrations of exogenous Glc result in increased SIS7/NCED3 steady-state mRNA levels. Steady-state mRNA levels of ABA1, ABA2 and ABA3 in wild-type germinating seeds are also upregulated by high concentrations of exogenous Glc. In contrast, in germinating seeds of the sis7-1 and sis7-3 mutants, Glc induction of ABA1, SIS7/NCED3, ABA2 and ABA3 is abolished. Previously it has been shown that 333 mM Glc induces ABA accumulation (or decreases ABA metabolism) in imbibed seeds [53]. The results of experiments presented here, analyzing Glc regulation of ABA biosynthesis genes in sis7 and wild-type germinating seeds, indicate that high Glc levels activate ABA biosynthesis by increasing the steady-state mRNA levels of ABA biosynthesis genes. These increases may, in turn, subsequently lead to ABA accumulation.
Besides transcriptional regulation, translational and post-translational mechanisms may also be involved in sugar-mediated regulation of ABA levels and/or signaling in germinating seeds. For example, loss-of-function of the KEEP ON GOING (KEG) gene, which is involved in ABI5 degradation, causes the mutant seedlings to exhibit a sugar hypersensitive phenotype [56]. In wild-type germinating seeds, the Glc-mediated increases in ABA levels may induce SIS7/NCED3 expression, as well as the expression of other ABA biosynthetic genes, thus resulting in sustained increases in ABA levels [26,55]. In the sis7/nced3/sto1 mutant, this positive feedback control loop is likely broken as is evidenced by the loss of Glc induction of ABA1, SIS7/NCED3, ABA2 and ABA3 in sis7-1 and sis7-3 germinating seeds. Thus the failure to accumulate ABA under 'sugar stress' may render mutant seedlings unable to carry out a normal sugar response. Consistent with this possibility are findings that the nced3/sto1 mutant accumulates only one-third as much ABA as wild type under sorbitol stress [27]. In addition, it has been reported that ABA levels in sto1/nced3 plants are somewhat lower than those found in unstressed wild-type plants [27]. These results may explain the enhanced germination rate of sis7-1 seeds on very low (1 μM), but not on higher (2-5 μM), levels of ABA. Also, addition of exogenous ABA to the 220 mM Glc media eliminated the sugar-insensitive phenotype of sis7/nced3/sto1 seedlings, indicating that the sis phenotype is caused by a deficiency in endogenous ABA.
As some mutants that are resistant to Glc and Suc also exhibit resistance to Man, the ability of the sis7-1 mutant to germinate on media containing 1.5 mM and higher concentrations of Man was tested. Man inhibition of seed germination has been postulated to act through a hexokinase-dependent pathway. This hypothesis is based on the fact that the hexokinase inhibitor mannoheptulose can reverse the inhibitory effects of Man on germination of wild-type seeds [35]. The results of experiments testing the ability of sis7-1 seeds to germinate in the presence of Man indicate that sis7-1 does not exhibit increased resistance to the inhibitory effects of exogenous Man on seed germination. This finding is consistent with the results of Man seed germination assays conducted on other aba mutants, where aba1-1 and aba3-2 were found to be unable to germinate in the presence of Man and aba2-1 [13] and aba2-3 and aba2-4 [16] were found to be only very slightly resistant to Man. These results suggest that response to the inhibitory effects of Man on seed germination may not require ABA accumulation. In contrast, mutations in ABI4 and ABI3 cause reduced seed sensitivity to Man [24,35], indicating that defects in ABA signaling components can affect Man sensitivity. The molecular mechanism by which defects in some ABA signaling components, but not defects in components of the ABA biosynthetic pathway, affects inhibition of seed germination by Man remains to be elucidated. Further complicating analyses of the role of SIS7/NCED3/STO1 in seed germination and other processes are findings that SIS7/NCED3/STO1 belongs to a multigene family with nine members in Arabidopsis, five of which are postulated to be involved in ABA biosynthesis [37,57].
Transcriptional profiling analyses identified 83 genes with altered mRNA levels in sis7-1 germinating seeds grown on 100 mM Glc. Functional categorization and over-representation analyses by GO annotations revealed significant enrichment of genes involved in response to stimulus, particularly abiotic stimulus. An interesting finding from these analyses is the decreased steady-state mRNA levels of auxin biosynthesis and transport genes in sis7-1 treated with Glc when compared to the wild type. These findings were re-tested by qRT-PCR analyses. The results of the qRT-PCR experiments confirm that the steady-state mRNA levels of these auxin-related genes are indeed lower in sis7-1 and sis7-3 germinating seeds compared to wild-type seeds (Figure 8). Previous studies have suggested cross-talk between the ABA and auxin response pathways in regulating Arabidopsis root system development [47-49]. The sis7-1 mutant exhibits increased lateral root development on sorbitol media when compared to the wild type (Figure 9). In addition, AtNCED3::GUS expression has been observed at lateral root initiation sites, suggesting control of lateral root development by both ABA and auxin [37]. The findings reported here suggest that part of the molecular mechanism(s) by which ABA/auxin cross-talk occurs during regulation of root development may involve regulation of auxin biosynthesis and transport genes in response to ABA levels.
To determine which of the genes listed in Table 1 are glucose regulated, the results from GeneChip experiments performed using seeds grown in the presence of 100 mM Glc were compared with data from GeneChip experiments performed using seeds grown in the presence of equimolar sorbitol (Table 2). Interestingly, a subset of the 83 genes listed in Table 1 exhibit only slight or no obvious Glc response in wild-type seeds, but exhibit significant Glc regulation in sis7-1 seeds, which have decreased ABA levels [27,29]. This result suggests that changes in endogenous ABA levels modulate the expression of this subset of genes. Modulation of Glc response by ABA at transcriptional or tissue levels has been demonstrated. Rook et al. (2001) have shown that ABA does not activate ApL3 expression alone, but greatly enhances ApL3 induction by sugars [18]. Glucose has been shown to delay Arabidopsis seed germination [53,58]. Dekkers et al. (2004) have demonstrated that low ABA concentrations combined with Glc enhance the inhibitory effect of Glc on seed germination [58]. Together, these results indicate that Glc-response may be modulated by ABA levels.
Different abscisic acid insensitive (abi) mutants display different responses to high levels of exogenous Glc and Suc. Previous studies showed that mutations in ABI4 confer significant resistance to the inhibitory effects of high concentrations of exogenous Glc or Suc on early seedling development, whereas the abi1-1, abi2-1, abi3-1 and abi5-1 mutants exhibit only slight to no resistance to Glc or Suc [11,13,16,18]. Interestingly, abi3-6 mutants, which lack one third of the ABI3 gene and are thus believed to be null mutants [59], have been shown to be resistant to the inhibitory effects of 330 mM Glc on early seedling development [25]. However, quantitative data indicating the degree to which abi3-6 mutants are resistant to Glc are lacking. Therefore, the finding that the sis10-1 mutant, which has a strong sis phenotype, lies in the ABI3 gene was unexpected. In addition, the findings reported here that sis10-1/abi3-15 has significant sis and abi phenotypes, whereas abi3-1 has been reported to have a significant abi phenotype [28] but little to no sis phenotype [11,13,16], suggested that it might be possible to separate genetically the role of ABI3 in ABA and sugar response. To test this hypothesis, quantitative assays were conducted to determine the sensitivities of nine different abi3 mutant lines, all in the Col background, at two Suc and two ABA concentrations. The results of these experiments indicate that the degree of Suc resistance shows a close, positive correlation with the degree of ABA resistance. These results therefore do not provide evidence for a genetically separable role for ABI3 in sugar and ABA resistance, although such a possibility cannot be ruled out at this time. The previous findings that abi3-1 has only a very slight effect on sugar response may be at least partially explained by the fact that the abi3-1 mutation is in the Landsberg erecta rather than the Col background. The Landsberg erecta ecotype exhibits a significantly greater level of sugar sensitivity than that of the Col ecotype (data not shown and [16]). A role for ABI3 in both sugar and ABA response is also consistent with results indicating that mutants carrying specific abi3 alleles exhibit increased resistance to Glc in the presence of ABA [23] and that overexpression of ABI3 confers hypersensitivity to Glc [60].
Conclusion
In this paper the map-based cloning and characterization of two genes that affect sugar response at the early seedling developmental stage are reported. These studies resulted in identification of new sis mutants which carry defects in the ABA biosynthesis gene NCED3/STO1 or the ABA response gene ABI3. The SIS7/NCED3/STO1 gene is rate-limiting in the positive feedback regulation of ABA biosynthesis by ABA under stress conditions [17,26]. Transcriptional upregulation of ABA biosynthesis genes by 300 mM Glc has been shown to occur in germinating wild-type seeds, but not in germinating sis7-1 or sis7-3 seeds. Transcriptional profiling experiments resulted in identification of 83 transcripts with altered steady-state mRNA levels in sis7-1 versus wild-type germinating seeds treated with 100 mM Glc. Over-representation analysis revealed that these 83 genes are significantly enriched for genes involved in response to abiotic stimulus. Of particular interest are findings that auxin metabolism and transport genes are significantly over-represented in genes with lower transcript levels in the sis7-1/nced3-4/sto1-4 mutant. These results have been verified by qRT-PCR analyses. These findings suggest that part of the molecular mechanism(s) by which ABA and auxin interact during regulation of Arabidopsis root development may involve regulation of auxin homeostasis and/or transport by ABA levels. Previous reports that abi3-1 has a significant abi phenotype but little to no sis phenotype, together with results presented here showing that sis10-1/abi3-15 has significant sis and abi phenotypes, suggested that it might be possible to separate genetically the role of ABI3 in sugar and ABA response. However, quantitative analyses of the magnitudes of the defects in Suc and ABA response of nine different abi3 mutants, all in the Col background, show that the strengths of these two phenotypes exhibit a strong positive correlation. The relatively weak sis phenotype shown by the abi3-1 mutant may be due, at least in part, to the fact that it is in the Landsberg erecta background, as different ecotypes have been shown to exhibit significantly different levels of endogenous sugar sensitivity (data not shown and [16]).
Methods
Plant materials, growth conditions and media
The sis7-1, sis7-3 and sis10-1 mutants were isolated from a mutant population generated by insertion of a T-DNA library carrying random Arabidopsis cDNAs into the genome of Arabidopsis thaliana of the Columbia (Col) ecotype [34]. The sis7-2 mutant was obtained by screening ethyl methyl sulfonate (EMS)-mutagenized Arabidopsis thaliana var. Col from Lehle Seeds (Tucson, AZ, USA). The gl1 mutation was removed from the sis7-2 mutant line by backcrossing this line to wild-type plants of the Col ecotype. The abi3-16 (SALK_023411) and abi3-17 (SALK_003216) mutants were identified from a T-DNA mutagenized population of Arabidopsis thaliana var. Col [50]. Seeds that are homozygous for the abi3-16 mutation were obtained from Drs. Laurie Host and Mauricio Bustos (University of Maryland, Baltimore County). Seeds segregating for the abi3-17 mutation were obtained from the Arabidopsis Biological Resource Center. PCR was then used to screen for plants that are homozygous for the abi3-17 mutation. The abi3-8, abi3-9, abi3-10, abi3-11, abi3-12 and abi3-13 mutants [23] of Arabidopsis thaliana var. Col were obtained from Dr. Eiji Nambara (RIKEN Plant Science Center, Japan). Isolation of abi4-103, which is in the Col background, has previously been described [16]. Seeds of spy-3 [36], which is also in the Col background, were obtained from Dr. Neil Olszewski. Wild-type Hi-O (CS6736) and Col-0 (CS6000) seeds were obtained from the Arabidopsis Biological Resource Center at Ohio State University. Seeds to be sown on plates were surface-sterilized and stratified in the dark at 4°C for 3 d. Plants were grown on plates under continuous white fluorescent light at 22°C unless otherwise noted. Minimal Arabidopsis media was prepared as described [31].
Sugar-insensitive mutant screen and genetic analyses
To isolate sugar-insensitive mutants, M2 seeds derived from EMS-mutagenized Col seeds were sown on Petri plates containing minimal media supplemented with 300 mM Suc. Seeds derived from a T-DNA insertion library [34] were screened on minimal media supplemented with 340 mM Suc. Subsequent screening procedures were performed as described [16]. All sis mutations were caused by single locus recessive mutations (data not shown). The sis7-1/nced3-4/sto1-4 mutant was backcrossed twice and the sis7-2/nced3-5/sto1-5 mutant was backcrossed once to wild-type Col before physiological assays were conducted.
Sugar response assays
The sugar sensitivity of the mutants was assayed as described previously [16].
Abscisic acid and paclobutrazol response assays
To measure ABA sensitivity, seeds were sown on minimal media supplemented with 0, 1, 2 or 5 μM ABA. Plates were incubated at 25°C under continuous light. The percentages of seeds that had germinated were determined at time points of up to 9 d. Germination is defined as the emergence of the radicle from the seed coat. To measure paclobutrazol sensitivity, seeds were sown on minimal media supplemented with 0, 30, 60 or 240 μM paclobutrazol. Plates were incubated at 25°C under continuous light. The percentages of seeds that had germinated were determined until the eighth d.
Map-based cloning
Mapping populations for sis7-1 and sis10-1 were generated as described in the Results section. Each of these F2 mapping populations was screened on media containing 300-340 mM Suc to identify plants that are homozygous for the sis mutation carried within that population. Genomic DNA was prepared from leaves of these seedlings via alkaline lysis [61]. Other mapping procedures were performed as described [62,63]. The molecular markers used to identify recombinants in the mapping experiments were simple sequence length polymorphisms (SSLPs), cleaved amplified polymorphic sequences (CAPS) and single nucleotide polymorphisms (SNPs) that were obtained from the The Arabidopsis Information Resource (TAIR) and Monsanto SNP and Ler sequence collections [62] and tested for polymorphisms between Col and Hi-O wild-type plants.
Reverse Transcriptase (RT) PCR Assays
The Qiagen RNeasy plant mini kit (Qiagen, Valencia, CA) was used to isolate total RNA from leaves of 4-week-old wild-type Col and sis7 mutant plants grow on soil under continuous light. First strand cDNA was synthesized from total RNA (0.5 μg) using the Promega Improm-II RT system kit (Promega, Madison, WI). Two μl of cDNA were used for PCR reactions. Primers used were: SIS7, 5'-TAAAAACCGTTGGTCGGTTC-3' and 5'-CGACGTCCGGTGATTTAGTT-3'; ACTIN, 5'-TGCTGACCGTATGAGCAAAG-3' and 5'-GATTGATCCTCCGATCCAGA-3'.
Quantitative Real-Time PCR
For Glc regulation of ABA biosynthesis gene expression, seeds of Col, sis7-1 and sis7-3 were stratified and sown on nytex screens on minimal media and incubated for 20 h under continuous light and then transferred to 300 mM Glc or sorbitol media for an additional 12 h under continuous light before harvesting. For validation of microarray data, seeds of Col, sis7-1 and sis7-3 were treated as described above, except that the seeds were transferred to 100 mM Glc or sorbitol media. Seeds from two biologically independent batches were used to minimize the variations in gene expression engendered by possible changes in plant growth conditions. Total RNA was isolated using the Spectrum™ Plant Total RNA Kit (Sigma). First strand cDNA was synthesized with the Promega Improm-II RT system kit (Promega) in a reaction volume of 20 μl. Real-time PCR was performed on an Applied Biosystems 7500 Real-Time PCR Machine using the SYBR Green JumpStart Taq ReadyMix (Sigma, St. Louis, MO). UBQ6 (AT2G47110) was used as an internal reference. The following primers were used: UBQ6, 5'-CCATCGACAATGTCAAGGCC-3' and 5'-GGTACGTCCGTCTTCGAGCT-3'; ABA1, 5'-GAACGTACTATAAAGGGAGAATGG-3' and 5'- CTGAGACGAAGGGATCACAAT-3'; SIS7/NCED3, 5'-TAAAAACCGTTGGTCGGTTC-3' and 5'-CGACGTCCGGTGATTTAGTT-3'; ABA2, 5'-CTAAACTCGCTTTGGCTCATT-3' and 5'-CGCTACATCATCAACCGTCAG-3'; ABA3, 5'-TTTGCTGGGATGACAATGAT-3' and 5'-CCTTCCACTGACGACGGTTC-3'; AAO3, 5'-GTCAGCGAGGTGGAAGTGGA-3' and 5'-CAAATGCTCCTTCGGTCTGT-3'; CYP83B1, 5'-CCCTAACCGCCCTAAACAAGA-3' and 5'-CATACCACCACTGCAGCCG-3'; CYP79B2, 5'-GACAATCCATCAAACGCCGT-3' and 5'-GCGGAGGATAGCTTTGACGTA-3'; AT4G27260, 5'-TGTTGTCACCACTTACGCCG-3' and 5'-CGTTCTGAAGCTCAACCTCGT-3'; PIN1, 5'-CGGTGGGAACAACATAAGCA-3' and 5'-TGAAGGAAATGAGGGACCAGG-3' and PIN3, 5'-TGGAGATTTCGGAGGAGAACA-3' and 5'-TTCGTTGACTTGCTTCGGC-3'.
Gene expression analysis using Affymetrix ATH1 GeneChips
For transcriptional profiling experiments, Col wild-type and sis7-1 seeds were sown on nytex screens on minimal media and incubated for 20 h under continuous light and then transferred to 100 mM Glc or sorbitol media for an additional 13 h under continuous light before harvesting. Seeds were pooled from different batches to minimize the variation in gene expression profiles engendered by possible changes in plant growth conditions. Total RNA extraction was performed as described [64]. For reproducibility, three independent seed tissue collections and RNA extractions were performed for sis7-1 treated with Glc or sorbitol and Col wild type treated with sorbitol and six independent experiments were performed for Col wild type treated with Glc. The generation of biotinylated cRNA from total RNA and subsequent hybridization to Arabidopsis ATH1 genome arrays and quantification of fluorescent signals were performed by the Molecular Genomics Core Facility at the University of Texas Medical Branch at Galveston. Data analyses were performed using Expressionist (Genedata AG, Switzerland). Gene expression values and presence/absence calls were computed using the Affymetrix Statistical Analysis (MAS5) algorithm. Student's t-test values and fold changes of individual transcript signal intensities from sis7-1 and wild-type replicates were determined. The mis-regulated gene list was generated by identifying transcripts that varied between mutant and wild-type samples with a p value of less than 0.05 and a fold change value of greater than 2.0. For over-representation analysis using GeneMerge, the population genes consist of 8791 genes called present on the ATH1 arrays by the Expressionist program. P value was calculated after Bonferroni correction.
Authors' contributions
YH identified the SIS7/NCED3/STO1 gene, performed most of the characterization of the sis7/nced3/sto1 mutants and drafted the major part of the manuscript. CL identified the SIS9/ABA1 and SIS10/ABI3 genes, performed the characterization of the sis10/abi3 mutants and aided in drafting the manuscript. KB identified the sis7-1, sis7-3 and sis10-1 mutants and performed a preliminary characterization of those mutants. SG conceived of the study, assisted in its design and the interpretation of the data and aided in drafting the manuscript. All authors approved of the manuscript.
Note
During submission of this manuscript, a manuscript appeared (Plant Mol Biol 67: 151) that also describes findings that mutations in ABI3 can confer a sugar-insensitive phenotype.
Acknowledgments
Acknowledgements
We thank Drs. Laurie Host and Mauricio Bustos (University of Maryland, Baltimore County) for seeds homozygous for the abi3-16 (SALK_023411) mutation and Dr. Eiji Nambara (RIKEN Plant Science Center, Japan) for providing abi3-8, abi3-9, abi3-10, abi3-11, abi3-12 and abi3-13 mutants. Dr. Donna Pattison and Melissa Moon are thanked for assistance with early characterization of the sis7-1, sis7-3 and sis10-1 mutants and Jason Daniels and Mary Larson for assistance in cloning SIS10/ABI3. Michael Sean Kincaid is thanked for isolating the sis7-2 and sis9-1 mutants. We thank three anonymous reviewers for their comments and suggestions. We also thank the Arabidopsis Biological Resource Center (ABRC) and SALK Institute for providing T-DNA insertional lines and mutant seed stocks. The Minnesota Supercomputing Institute at the University of Minnesota is thanked for providing computational support. Research was supported by the United States Department of Energy, Energy Biosciences Program grant DE-FG02-03ER15417 and the Consortium for Plant Biotechnology Research grant DE-FG36-02GO12026-216.
Contributor Information
Yadong Huang, Email: huan0277@umn.edu.
Chun Yao Li, Email: lixxx265@umn.edu.
Kelly D Biddle, Email: kbiddle@rice.edu.
Susan I Gibson, Email: gibso043@tc.umn.edu.
References
- Gibson SI. Plant sugar-response pathways. Part of a complex regulatory web. Plant Physiol. 2000;124:1532–1539. doi: 10.1104/pp.124.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI. Control of plant development and gene expression by sugar signaling. Curr Opin Plant Biol. 2005;8:93–102. doi: 10.1016/j.pbi.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Graham IA. Carbohydrate control of gene expression in higher plants. Res Microbiol. 1996;147:572–580. doi: 10.1016/0923-2508(96)84014-9. [DOI] [PubMed] [Google Scholar]
- Koch KE. Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J. Sugar sensing and signaling in plants. Plant Cell. 2002;14:S185–S205. doi: 10.1105/tpc.010455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F, Bevan MW. Genetic approaches to understanding sugar-response pathways. J Exp Bot. 2003;54:495–501. doi: 10.1093/jxb/erg054. [DOI] [PubMed] [Google Scholar]
- Smeekens S. Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:49–81. doi: 10.1146/annurev.arplant.51.1.49. [DOI] [PubMed] [Google Scholar]
- Wobus U, Weber H. Sugars as signal molecules in plant seed development. Biol Chem. 1999;380:937–944. doi: 10.1515/BC.1999.116. [DOI] [PubMed] [Google Scholar]
- Yu S-M. Cellular and genetic responses of plants to sugar starvation. Plant Physiol. 1999;121:687–693. doi: 10.1104/pp.121.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, León P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 2000;14:2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Gibson SI, Laby RJ, Kim D. The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem Biophys Res Commun. 2001;280:196–203. doi: 10.1006/bbrc.2000.4062. [DOI] [PubMed] [Google Scholar]
- Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S. The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. Plant J. 2000;23:577–586. doi: 10.1046/j.1365-313x.2000.00822.x. [DOI] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA. 1998;95:10294–10299. doi: 10.1073/pnas.95.17.10294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laby RJ, Kincaid MS, Kim D, Gibson SI. The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 2000;23:587–596. doi: 10.1046/j.1365-313x.2000.00833.x. [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen H-C, Arroyo A, Leon P, Nambara E, Asami T, Seo M, Koshiba T, Sheen J. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002;14:2723–2743. doi: 10.1105/tpc.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2001;26:421–433. doi: 10.1046/j.1365-313X.2001.2641043.x. [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 1996;10:655–661. doi: 10.1046/j.1365-313X.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu J-K. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell. 2001;13:2063–2083. doi: 10.1105/tpc.13.9.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 1994;5:765–771. doi: 10.1046/j.1365-313X.1994.5060765.x. [DOI] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. The Arabidopsis abscisic acid response locus ABI4 encodes an Apetala2 domain protein. Plant Cell. 1998;10:1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Suzuki M, Abrams S, McCarty DR, Kamiya Y, McCourt P. A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics. 2002;161:1247–1255. doi: 10.1093/genetics/161.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K, Wysocka-Diller J. Phytohormone signalling pathways interact with sugars during seed germination and seedling development. J Exp Bot. 2006;57:3359–3367. doi: 10.1093/jxb/erl096. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Kermode AR. A gymnosperm ABI3 gene functions in a severe abscisic acid-insensitive mutant of Arabidopsis (abi3-6) to restore the wild-type phenotype and demonstrates a strong synergistic effect with sugar in the inhibition of post-germinative growth. Plant Mol Biol. 2004;56:731–746. doi: 10.1007/s11103-004-4952-y. [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Zhu J-K. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J Biol Chem. 2002;277:8588–8596. doi: 10.1074/jbc.M109275200. [DOI] [PubMed] [Google Scholar]
- Ruggiero B, Koiwa H, Manabe Y, Quist TM, Inan G, Saccardo F, Joly RJ, Hasegawa PM, Bressan RA, Maggio A. Uncoupling the effects of abscisic acid on plant growth and water relations. Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in Arabidopsis. Plant Physiol. 2004;136:3134–3147. doi: 10.1104/pp.104.046169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant. 1984;61:377–383. doi: 10.1111/j.1399-3054.1984.tb06343.x. [DOI] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis -epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge B, Valon C, Smalle J, Parcy F, Goodman H. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz AR, Kirchheim B. Genetic Resources in Arabidopsis. Arabidopsis Inf Serv. 1987;24:1–111. [Google Scholar]
- Jang JC, León P, Zhou L, Sheen J. Hexokinase as a sugar sensor in higher plants. Plant Cell. 1997;9:5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kálmán Z, Stankovic-Stangeland B, Bakó L, Mathur J, Ökrész L, Stabel S, Geigenberger P, Stitt M, Rédei GP, Schell J, Koncz C. Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 1998;12:3059–3073. doi: 10.1101/gad.12.19.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere S, Bartel B. A library of Arabidopsis 35S-cDNA lines for identifying transcripts that cause phenotypes when overexpressed or cosuppressed. Plant Mol Biol. 2001;46:695–703. doi: 10.1023/A:1011699722052. [DOI] [PubMed] [Google Scholar]
- Pego JV, Weisbeek PJ, Smeekens SCM. Mannose inhibits Arabidopsis germination via a hexokinase-mediated step. Plant Physiol. 1999;119:1017–1023. doi: 10.1104/pp.119.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003;35:44–56. doi: 10.1046/j.1365-313X.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- Wan X-R, Li L. Regulation of ABA level and water-stress tolerance of Arabidopsis by ectopic expression of a peanut 9-cis-epoxycarotenoid dioxygenase gene. Biochem Biophys Res Commun. 2006;347:1030–1038. doi: 10.1016/j.bbrc.2006.07.026. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Information Resource http://www.arabidopsis.org/tools/bulk/go/index.jsp
- GeneMerge http://genemerge.cbcb.umd.edu/online/
- Castillo-Davis CI, Hartl DL. GeneMerge – post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem. 2000;275:33712–33717. doi: 10.1074/jbc.M001667200. [DOI] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci USA. 2000;97:2379–2384. doi: 10.1073/pnas.040569997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17:616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Toledo-Ortiz G, Bender J, Quail PH. The photomorphogenesis-related mutant red1 is defective in CYP83B1, a red light-induced gene encoding a cytochrome P450 required for normal auxin homeostasis. Planta. 2004;219:195–200. doi: 10.1007/s00425-004-1211-z. [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- Brady SM, Sarkar SF, Bonetta D, McCourt P. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 2003;34:67–75. doi: 10.1046/j.1365-313X.2003.01707.x. [DOI] [PubMed] [Google Scholar]
- Deak KI, Malamy J. Osmotic regulation of root system architecture. Plant J. 2005;43:17–28. doi: 10.1111/j.1365-313X.2005.02425.x. [DOI] [PubMed] [Google Scholar]
- Rock CD, Sun X. Crosstalk between ABA and auxin signaling pathways in roots of Arabidopsis thaliana (L.) Heynh. Planta. 2005;222:98–106. doi: 10.1007/s00425-005-1521-9. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:1849. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Arroyo A, Bossi F, Finkelstein RR, León P. Three genes that affect sugar sensing (abscisic acid insensitive 4, abscisic acid insensitive 5, and constitutive triple response 1) are differentially regulated by glucose in Arabidopsis. Plant Physiol. 2003;133:231–242. doi: 10.1104/pp.103.021089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P. Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr Opin Plant Biol. 2001;4:387–391. doi: 10.1016/S1369-5266(00)00190-4. [DOI] [PubMed] [Google Scholar]
- Price J, Li T-C, Kang SG, Na JK, Jang J-C. Mechanisms of glucose signaling during germination of Arabidopsis. Plant Physiol. 2003;132:1424–1438. doi: 10.1104/pp.103.020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Zeevaart JA. The 9-cis -epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci USA. 1999;96:15354–15361. doi: 10.1073/pnas.96.26.15354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu JK. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003;133:29–36. doi: 10.1104/pp.103.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell. 2006;18:3415–3428. doi: 10.1105/tpc.106.046532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006;45:309–319. doi: 10.1111/j.1365-313X.2005.02622.x. [DOI] [PubMed] [Google Scholar]
- Dekkers BJW, Schuurmans JAMJ, Smeekens SCM. Glucose delays seed germination in Arabidopsis thaliana. Planta. 2004;218:579–588. doi: 10.1007/s00425-003-1154-9. [DOI] [PubMed] [Google Scholar]
- Nambara E, Keith K, McCourt P, Naito S. Isolation of an internal deletion mutant of the Arabidopsis thaliana ABI3 gene. Plant Cell Physiol. 1994;35:509–513. [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimyuk VI, Carroll BJ, Thomas CM, Jones JD. Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J. 1993;3:493–494. doi: 10.1046/j.1365-313X.1993.t01-26-00999.x. [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 2002;129:440–450. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR. Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 2000;123:795–805. doi: 10.1104/pp.123.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]