Abstract
Nitric oxide (NO) has diverse roles in intercellular communication and (at higher levels) in immune-mediated cell killing. NO reacts with many cellular targets, with cell-killing effects correlated to inactivation of key enzymes through nitrosylation of their iron-sulfur centers. SoxR protein, a redox-sensitive transcription activator dependent on the oxidation state of its binuclear iron-sulfur ([2Fe-2S]) centers, is also activated in Escherichia coli on exposure to macrophage-generated NO. We show here that SoxR activation by NO occurs through direct modification of the [2Fe-2S] centers to form protein-bound dinitrosyl-iron-dithiol adducts, which we have observed both in intact bacterial cells and in purified SoxR after NO treatment. Functional activation through nitrosylation of iron-sulfur centers contrasts with the inactivation typically caused by this modification. Purified, nitrosylated SoxR has transcriptional activity similar to that of oxidized SoxR and is relatively stable. In contrast, nitrosylated SoxR is short-lived in intact cells, indicative of mechanisms that actively dispose of nitrosylated iron-sulfur centers.
Although free radicals are often viewed as toxic, several of these agents and related unstable species function in signal transduction to activate gene expression. Superoxide and hydrogen peroxide mediate stress responses in diverse organisms (1–3) whereas enzymatically generated nitric oxide (NO) plays multiple roles in mammalian cells. At high levels, NO is a cytotoxic weapon against tumor cells or invading pathogens (4). At low (nanomolar) levels, NO modulates vascular muscle tone (5) or mediates neuronal communication (6). The intercellular signaling by NO in the latter cases is effected by a reaction with heme-containing guanylate cyclase in the target cells (5) by the nitrosylation of heme in the enzyme (7). Other types of signaling can occur at higher levels of NO, such as the induction of heme oxygenase 1 and other proteins in mammalian cells (8, 9), or the triggering of the soxRS oxidative stress regulon in bacteria (10, 11). NO is capable of diverse reactions (12–14), however, and the molecular mechanism of these latter signaling pathways has remained unknown.
Study of the Escherichia coli soxRS system has provided opportunities to define the molecular events that occur during free radical signal transduction. The induction of soxRS involves first the activation of SoxR, a transcription factor that contains redox-active binuclear iron-sulfur ([2Fe-2S]) clusters (15, 16). Superoxide-generating agents, such as paraquat, trigger SoxR activity by causing one-electron oxidation to yield [2Fe-2S]2+ clusters; SoxR with reduced [2Fe-2S]1+ centers is transcriptionally inactive (17, 18). Exposure to pure NO gas activates SoxR in E. coli (10). In situ analysis of phagocytosed bacteria within murine macrophages showed that SoxR is activated by NO generated by the inducible NO synthase (11). Direct oxidation of the SoxR [2Fe-2S] centers by NO seems unlikely (14), and we have not detected NO oxidation or reduction products in reactions with SoxR (E. Hidalgo, H.D., J. S. Wishnok, S. R. Tannenbaum, and B.D., unpublished data). Here, we show that, both in intact E. coli and with the purified protein, activation of SoxR by NO occurs through nitrosylation of the [2Fe-2S] clusters in the protein, a reaction usually thought to inactivate protein function (19, 20).
Experimental Procedures
Cell Growth and NO Treatments.
Overnight cultures of E. coli strain XA90 containing the SoxR expression plasmid pKOXR (21) were diluted 100-fold into 50 ml of LB medium containing 100 μg/ml ampicillin in a 300-ml flask. After 2 h of incubation at 37°C with 275-rpm shaking, isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.5 mM, and the incubation continued for 1 h to induce SoxR expression. The cultures were then transferred to sealed 15-ml Falcon tubes and were incubated for 30 min at 37°C without shaking, to achieve anoxic conditions before the NO treatment. NO-saturated solutions were prepared by bubbling pure NO gas (Aga Gas, Cleveland), first passed through a soda lime column (4–8 mesh) to remove other nitrogen oxides, through water under anaerobic conditions. Aliquots of the NO solution (amount indicated in figure legends) were delivered anaerobically to the E. coli cultures by using a gas-tight syringe. After treatment with NO, the sealed cultures were further incubated at 37°C with shaking at 275 rpm and were sampled at different times. Samples for RNA or EPR analysis were immediately frozen in liquid nitrogen and were kept for 1 h at −80°C before RNA extraction. Total RNA was extracted from 1-ml samples by using the RNeasy Mini Kit (Qiagen, Chatsworth, CA), following the manufacturer's instructions. Northern blots (22) of the soxS transcript were quantified by using phosphorimaging (BioImage, Millipore).
EPR Spectroscopy.
X-band EPR spectra were recorded by using a Bruker model ESP-300 equipped with an Oxford Instruments 910 continuous flow cryostat (courtesy of J. Stubbe and the Chemistry Department, Massachusetts Institute of Technology). Routine EPR measurement conditions were as follows: microwave frequency, 9.47 GHz; microwave power, 1 mW; modulation frequency, 100 kHz; modulation amplitude, 2.0 mT; sweep field, 310–370 mT; sample temperature, 30 K; receiver gain, 105. For direct EPR measurement of intact bacteria, the cells were grown in LB broth to OD600 ≈1 and then were treated with NO. Aliquots of 0.4 ml were transferred directly to EPR tubes and immediately were frozen at −170°C. At the same time, an aliquot of 1-ml culture was frozen for later determination of soxS mRNA as described in Fig. 1. In some cases, the signal intensity was increased by concentrating the bacteria 25-fold (by centrifugation at 900 × g and resuspension with 1/25 the original volume of LB broth) before NO treatment and EPR measurements. As a control, E. coli XA90 containing the empty expression vector pKEN2 was used to generate a baseline EPR spectrum.
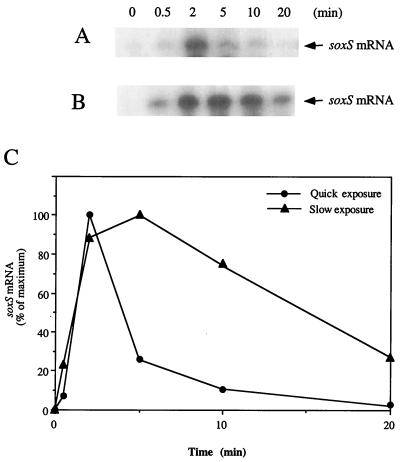
Figure 1.
Rapid activation of SoxR in NO-exposed E. coli. (A) Transient induction of soxS by brief NO exposure. Growing cultures of strain XA90 with the SoxR-expression plasmid pKOXR were treated to induce SoxR and then were transferred to anoxic conditions. Five-milliliter aliquots of anoxic culture were mixed with 100 μl of NO-saturated water (≈2 mM), with the mixture completed within 10 sec. Anoxic incubation was continued, and samples were withdrawn at the times indicated in the figure to analyze for soxS mRNA by Northern blotting. (B) Extended soxS induction by extended NO exposure. Samples were generated as for A, except that the NO solution was added over ≈45 sec. (C) Quantitation of soxS induction by NO. Northern blots as in A and B were quantitated by phosphorimaging.
Analysis of NO-Modified SoxR Protein.
SoxR protein was overproduced and purified, and EPR measurements were conducted as described (17, 23). SoxR activity in soxS transcription in vitro was assayed by using plasmid pBD100 as the template, with the SoxR-independent bla transcript as a control, and the products were quantified by primer extension analysis (24). For in vitro NO treatment, purified SoxR (3 μM in 50 mM Hepes⋅KOH, 0.5 M NaCl) in a sealed cuvette was equilibrated for 1 h at 0°C with pure argon gas, after which samples of NO-saturated water were delivered. NO-treated SoxR was then diluted 5-fold with 50 mM Hepes⋅KOH and was loaded on a 1-ml cation-exchange column (Resource S; Amersham Pharmacia). The column was washed with 50 ml of 50 mM Hepes⋅KOH, 0.2 M NaCl before SoxR elution with 50 mM Hepes⋅KOH, 0.5 M NaCl. SoxR purified after NO treatment was used for EPR measurements and in vitro transcription assays.
Results
We previously monitored the effect of NO on the activity of SoxR in E. coli cells by using reporter fusions to the soxS promoter. In addition to the activation within NO-generating macrophages (11), those studies showed that infusion of pure NO gas activates SoxR. This activation was enhanced by the removal of oxygen (10, 11), which indicates a direct reaction of NO with a cellular target in competition with O2. However, in those experiments, the induction of the reporter occurred more slowly, and appeared to be more limited, than the response to superoxide-generating agents such as paraquat (10, 11).
Limited induction might be explained either by inefficient activation of SoxR by NO, or by instability of the activated form. We examined this point by exposing E. coli “instantaneously” to NO by introducing aliquots of a saturated solution of the pure gas, and we directly monitored the induction of the soxS gene by Northern blotting. These experiments (Fig. 1) showed rapid but transient induction of soxS mRNA to a high level (maximal within 2 min after NO exposure). Extending the NO exposure (from <10 sec to 45 sec) gave more prolonged soxS transcription, which peaked at 5 min and decayed over the next 15 min (Fig. 1). These results are consistent with the formation of an activated form of SoxR, the lifetime of which is extended as the NO exposure continues.
The predominant known product of NO with iron-sulfur centers is the dinitrosyl-iron-dithiol complex, in which the sulfide ligands are displaced by formation of Fe-NO bonds, and the iron atoms remain bound to the protein by cysteine ligands (the thiols) (25, 26). This type of modification typically inactivates protein function, as in the NO-mediated inhibition of aconitase (27) and ribonucleotide reductase (28). To determine whether such adducts form in SoxR during cellular exposure to NO, we used electron paramagnetic resonance (EPR) spectroscopy. EPR spectroscopy of intact E. coli cells has been used to monitor oxidation of the [2Fe-2S] centers of SoxR during its oxygen-dependent activation by paraquat (15, 21, 24). We now find that 2-min NO exposure of intact, SoxR-expressing bacteria produces a new EPR spectrum (Fig. 2A; cells concentrated 25-fold to facilitate EPR measurements). The g value for the new signals (g = 2.03) was similar to that reported for other cases of nitrosylated iron-sulfur centers (29–32) whereas the signature of the reduced [2Fe-2S] centers was greatly diminished (Fig. 2A). By comparing the amplitudes of the signals for SoxR in control and NO-treated cells, we estimate that ≤25% of the original, reduced [2Fe-2S] centers remain in the NO-treated cells, and that the g = 2.03 signal accounts for ≥70% of the [2Fe-2S] centers present initially.
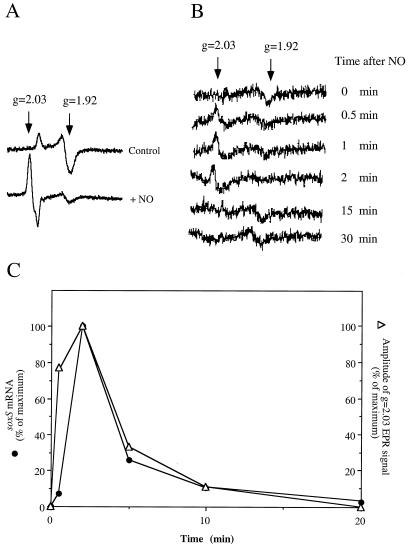
Figure 2.
EPR spectroscopy of nitrosylated SoxR in intact cells. (A) E. coli XA90/pKOXR (21) were grown to OD600 ≈1 and were concentrated 25-fold. The concentrated cells (1 ml) were mixed with 0.2 ml of a NO-saturated solution in water (as described in Fig. 1A) and after 2 min were frozen at −170°C. EPR spectroscopy of the intact cells was carried out as described (21). The g values used to model the EPR signals are indicated (g = 1.92, reduced [2Fe-2S] SoxR; g = 2.03, NO-SoxR). (B) Kinetics of nitrosyl-SoxR formation and removal in intact cells. XA90/pKOXR cells were exposed to NO as indicated in Fig. 1A. At the indicated times after NO addition, 0.4-ml aliquots were collected into EPR tubes and immediately were frozen at −170°C. EPR analysis of these cells was as described (21). The profiles shown are the net signal after subtraction of spectra for control cells not overexpressing SoxR. (C) Correlation of nitrosyl-SoxR and transcription in intact cells. The amplitude of the g = 2.03 signal was measured and is plotted in comparison with the soxS mRNA level in the same cells (Fig. 1C).
To follow the kinetics of these changes, we subjected unconcentrated E. coli cell samples directly to EPR spectroscopy (21). This analysis revealed the rapid formation of the EPR signal at g = 2.03 representing nitrosylated iron-sulfur clusters (within 0.5 min after single injection of NO gas), and its decay after 2 min (Fig. 2B). The kinetics of formation and decay of nitrosylated SoxR [2Fe-2S] clusters (from the spectroscopic data) showed a direct and quantitative correlation with soxS mRNA induction (Fig. 2C). The short delay between the rise in the EPR signal for modified SoxR and the increased level of soxS mRNA is expected because of the time needed to synthesize the transcript. The tight correlation between the spectroscopic and mRNA analyses provides strong evidence that formation of dinitrosyl-iron-dithiol centers in SoxR activates the protein in NO-treated cells.
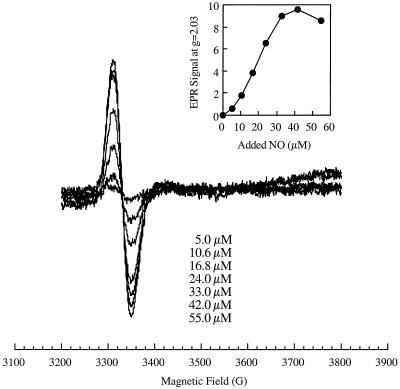
To define the properties of nitrosylated SoxR more precisely, we prepared samples by anaerobic exposure of the purified protein to solutions of pure NO. A titration of purified SoxR with a NO solution showed the formation of a single EPR-detectable species (Fig. 3) with g = 2.03 as seen in the intact cells. Thus, the reaction with NO apparently converted SoxR protein to the nitrosylated form independent of other cellular components. Samples were also treated with NO and then were repurified by ion-exchange chromatography (Fig. 4A, traces a and b). By spin quantification of the sample shown in Fig. 4, we estimated that the g = 2.03 signal represented at least 18% of the original [2Fe-2S] centers. It appears that the NO-modified metal centers are unstable during the purification because most of the SoxR corresponded to this form both in vivo (Fig. 2A) and in vitro before purification (Fig. 3). Less then 1% unmodified [2Fe-2S]1+ centers remained in the NO-treated, repurified protein as shown by EPR of a sample treated with the chemical reductant dithionite (Fig. 4A, trace c).
Figure 3.
NO modification of SoxR in vitro. Purified SoxR protein (23, 33) (2.5 ml, 3 μM) was mixed with increasing amounts of a NO-saturated solution in water to yield the indicated final concentrations of NO (nominal; some is lost to the gas phase and some by reaction with residual oxygen or other components present in the sample). The samples were frozen and subjected to EPR spectroscopy. The inset shows the quantitation of the amplitude of the g = 2.03 signal as a function of NO treatment.
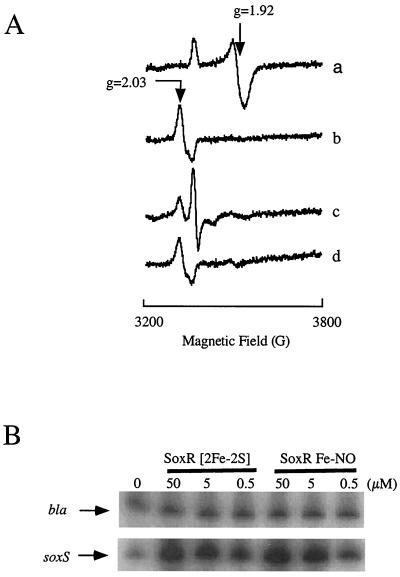
Figure 4.
Properties and activity of re-purified, NO-treated SoxR. (A) EPR spectroscopy. Purified SoxR protein (23, 33) (2.5 ml, 3 μM) was mixed with 100 μl of a NO-saturated solution in water. The NO-treated protein was then reduced with sodium dithionite (20 mM), followed by reoxidation with 1 mM potassium ferricyanide. Trace a, dithionite-reduced, purified SoxR; trace b, NO-treated SoxR, showing d7 signal; trace c, NO-treated SoxR reduced with dithionite, showing d9 signal; trace d, reoxidation of protein from trace c. A small signal at ≈3,500 G may be attributable to dithionite, as it persists in the reoxidized sample (compare traces c and d). (B) In vitro transcription by NO-treated SoxR. In vitro transcription reactions (23, 24) used the indicated amounts of purified, oxidized SoxR (SoxR [2Fe-2S]), or purified, NO-treated SoxR (SoxR Fe-NO). The transcripts were quantified by primer extension (40). bla, SoxR-independent transcript; soxS, transcript from the SoxR-dependent soxS gene.
Nitrosylated SoxR was not fully reduced, even by excess amounts of dithionite (1 mM; Eh 64–500 mV), although up to 60% of the d9 form was produced (Fig. 4A, trace c). The nitrosylated iron centers were stable on reoxidation (Fig. 4A, trace d). This resistance to reduction is consistent with the very low midpoint potentials reported for model compounds (29). Moreover, the contrasting stabilities of nitrosylated SoxR in vitro and in vivo suggests that the modified form is actively eliminated in intact cells.
The critical function of activated SoxR is the specific and powerful stimulation of transcription from the soxS promoter (16). A quantitative comparison showed that, for soxS transcription in vitro, nitrosylated SoxR was nearly as potent as SoxR with oxidized [2Fe-2S]2+ centers (Fig. 4B). As noted above, such oxidized centers are virtually gone from the NO-treated protein. Moreover, the transcriptional activity was not attributable to small amounts of apo-SoxR that might be present in the sample, as the apo-form is not active in this assay (23, 33). These results indicate that the NO-modified protein binds its target DNA with high affinity and exerts structural effects on the soxS promoter similar to those of oxidized SoxR (34).
Discussion
The paradigm for NO signaling pathways, in which NO binds the heme moiety of “soluble” guanylate cyclase, has remained the predominant model for some years (5, 35). Recently, alternative pathways have been proposed that involve nitrosothiols (36, 37) or destructive reactions of NO with iron-sulfur centers (19, 20), but without a detailed mechanistic underpinning. One stumbling block has been the breadth of NO chemistry, with diverse oxygen-dependent and -independent products, and many possible targets for modification by NO (12–14). An early example of such a reaction was the adduction by NO of protein iron-sulfur centers and the consequent inactivation of key functions such as the [4Fe-4S] enzyme aconitase (20, 26). For the aconitase homolog iron-reponse-protein 1, this modification leads indirectly to activation of RNA binding activity through disassembly of the damaged metal center (20).
The studies presented here reveal a novel, direct role for NO in signal transduction: modification of iron-sulfur centers in SoxR directly activates its transcriptional activity (Fig. 5). This observation is also remarkable in that the only other known activated form of this protein has one-electron oxidized [2Fe-2S]2+ centers (Fig. 5A). The similar transcriptional activity of the two forms suggests that they may have unexpectedly similar structures, but the structure of SoxR protein has not yet been determined. The fact that nitrosylation (Fig. 5B) is an activating rather than inactivating reaction for SoxR suggests that the protein evolved to exploit this signal of cellular damage by NO. Nitrosylated iron-sulfur centers might also function in signal transduction in mammalian cells, where they are certainly formed on NO exposure. To date, only global measurements have been made with NO-treated mammalian cells (20), but the EPR and chemical data from these studies may contain features corresponding to NO-modified signaling proteins. More direct approaches will be needed to search for potential mammalian counterparts to SoxR in NO-mediated signal transduction.
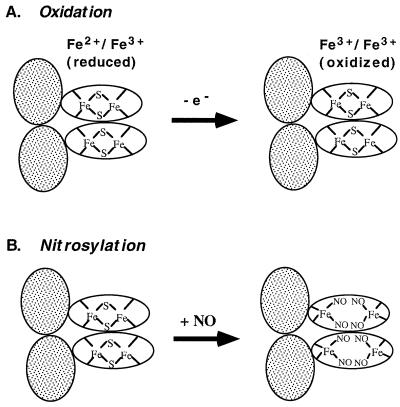
Figure 5.
Distinct activation mechanisms for SoxR. Shaded ovals: DNA binding domains; unshaded ovals, iron-binding domains. SoxR is shown as a homodimer (16). (A) Redox-regulation. When the [2Fe-2S] centers of SoxR are in the reduced state, the protein does not activate soxS transcription. One-electron oxidation converts the protein to a potent transcription activator. (B) Model for activation of SoxR by nitrosylation. The intact [2Fe-2S] clusters of SoxR are directly modified by NO to generate dinitrosyl-iron-dithiol clusters. This modification disrupts the iron-sulfur clusters; the remaining thiols are provided by the cysteine residues of SoxR protein. The model shows all iron atoms remaining bound to the protein, but may not be the case, particularly in the protein subjected to repurification.
The relative stability of nitrosylated iron-sulfur centers in SoxR in vitro contrasts with their rapid disappearance in vivo. This difference suggests that active pathways may exist to reverse or destroy this modification in SoxR. It is noteworthy that iron-nitrosyl adducts are released rapidly from NO-exposed mammalian cells (38). The resistance of the NO-modified metal centers to chemical reduction caused us to test other compounds for such activity. Thiols can influence the stability of oxidized SoxR (23) or of other nitrosylated iron-sulfur centers (39). The thiol proteins thioredoxin and glutaredoxin (at ≈100-fold molar excess) had only a modest inhibitory effect on the transcriptional activity of nitrosylated SoxR in vitro (data not shown); more work is required to establish the relevance and mechanism of this effect. Other compounds or enzymatic pathways that reverse or eliminate the nitrosylated iron-sulfur centers remain possible. In view of the toxic effects of NO on various iron-sulfur proteins, an ability of cells to eliminate such products could make an important contribution to cellular defenses. In addition, the utility of NO modification as a signaling mechanism would depend on its reversibility. SoxR will provide an important means to reveal the pathways of nitrosylation of iron-sulfur centers and the reversal of this modification.
Acknowledgments
We are grateful to our colleagues in the Demple laboratory for discussions during this work, and to Armen H. Tashjian, Jr., Steven R. Tannenbaum, and Thomas Michel for helpful comments on the manuscript. We are indebted to Prof. J. Stubbe and the Massachusetts Institute of Technology Chemistry Department for allowing us access to their EPR spectrometer. This work was supported by grants from the National Institutes of Health (Grants CA37831 to B.D. and F32-ES05726 to H.D.).
Abbreviation
- NO
nitric oxide
References
- 1.Hidalgo E, Demple B. In: Regulation of Gene Expression in Escherichia coli. Lin E C C, Lynch A S, editors. Austin, TX: Landes; 1996. pp. 435–452. [Google Scholar]
- 2.Godon C, Lagniel G, Lee J, Buhler J M, Kieffer S, Perrot M, Boucherie H, Toledano M B, Labarre J. J Biol Chem. 1998;273:22480–22489. doi: 10.1074/jbc.273.35.22480. [DOI] [PubMed] [Google Scholar]
- 3.Muller J M, Rupec R A, Baeuerle P A. Methods. 1997;11:301–312. doi: 10.1006/meth.1996.0424. [DOI] [PubMed] [Google Scholar]
- 4.MacMicking J, Xie Q W, Nathan C. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 5.Hobbs A J, Ignarro L J. Methods Enzymol. 1996;269:134–148. doi: 10.1016/s0076-6879(96)69016-8. [DOI] [PubMed] [Google Scholar]
- 6.Brenman J E, Bredt D S. Methods Enzymol. 1996;269:119–129. doi: 10.1016/s0076-6879(96)69014-4. [DOI] [PubMed] [Google Scholar]
- 7.Stone J R, Sands R H, Dunham W R, Marletta M A. Biochemistry. 1996;35:3258–3262. doi: 10.1021/bi952386+. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi K, Hara E, Ogawa K, Kimura D, Fujita H, Shibahara S. J Biochem (Tokyo) 1997;121:1162–1168. doi: 10.1093/oxfordjournals.jbchem.a021710. [DOI] [PubMed] [Google Scholar]
- 9.Marquis J C, Demple B. Cancer Res. 1998;58:3435–3440. [PubMed] [Google Scholar]
- 10.Nunoshiba T, deRojas-Walker T, Wishnok J S, Tannenbaum S R, Demple B. Proc Natl Acad Sci USA. 1993;90:9993–9997. doi: 10.1073/pnas.90.21.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nunoshiba T, deRojas-Walker T, Tannenbaum S R, Demple B. Infect Immun. 1995;63:794–798. doi: 10.1128/iai.63.3.794-798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beckman J S, Koppenol W H. Am J Physiol. 1996;271:C1424–C1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 13.Singh S P, Wishnok J S, Keshive M, Deen W M, Tannenbaum S R. Proc Natl Acad Sci USA. 1996;93:14428–14433. doi: 10.1073/pnas.93.25.14428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Squadrito G L, Pryor W A. Free Radical Biol Med. 1998;25:392–403. doi: 10.1016/s0891-5849(98)00095-1. [DOI] [PubMed] [Google Scholar]
- 15.Gaudu P, Moon N, Weiss B. J Biol Chem. 1997;272:5082–5086. doi: 10.1074/jbc.272.8.5082. [DOI] [PubMed] [Google Scholar]
- 16.Hidalgo E, Ding H, Demple B. Trends Biochem Sci. 1997;22:207–210. doi: 10.1016/s0968-0004(97)01068-2. [DOI] [PubMed] [Google Scholar]
- 17.Ding H, Hidalgo E, Demple B. J Biol Chem. 1996;271:33173–33175. doi: 10.1074/jbc.271.52.33173. [DOI] [PubMed] [Google Scholar]
- 18.Gaudu P, Weiss B. Proc Natl Acad Sci USA. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pantopoulos K, Weiss G, Hentze M W. Mol Cell Biol. 1996;16:3781–3788. doi: 10.1128/mcb.16.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drapier J-C. Methods. 1997;11:319–329. doi: 10.1006/meth.1996.0426. [DOI] [PubMed] [Google Scholar]
- 21.Ding H G, Demple B. Proc Natl Acad Sci USA. 1997;94:8445–8449. doi: 10.1073/pnas.94.16.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ausubel F M, Roger B, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley Interscience; 1997. [Google Scholar]
- 23.Ding H, Demple B. Biochemistry. 1998;37:17280–17286. doi: 10.1021/bi980532g. [DOI] [PubMed] [Google Scholar]
- 24.Hidalgo E, Ding H, Demple B. Cell. 1997;88:121–129. doi: 10.1016/s0092-8674(00)81864-4. [DOI] [PubMed] [Google Scholar]
- 25.Pellat C, Henry Y, Drapier J C. Biochem Biophys Res Commun. 1990;166:119–125. doi: 10.1016/0006-291x(90)91919-j. [DOI] [PubMed] [Google Scholar]
- 26.Kennedy M C, Antholine W E, Beinert H. J Biol Chem. 1997;272:20340–20347. doi: 10.1074/jbc.272.33.20340. [DOI] [PubMed] [Google Scholar]
- 27.Hibbs J B, Jr, Taintor R R, Vavrin Z. Science. 1987;235:473–476. doi: 10.1126/science.2432665. [DOI] [PubMed] [Google Scholar]
- 28.Kwon N S, Stuehr D J, Nathan C F. J Exp Med. 1991;174:761–767. doi: 10.1084/jem.174.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler A R, Glidewell C, Hyde A R, Walton J C. Polyhedron. 1985;4:797–809. [Google Scholar]
- 30.Reddy D, Lancaster J R, Jr, Cornforth D P. Science. 1983;221:769–770. doi: 10.1126/science.6308761. [DOI] [PubMed] [Google Scholar]
- 31.Lancaster J R, Jr, Hibbs J B., Jr Proc Natl Acad Sci USA. 1990;87:1223–1227. doi: 10.1073/pnas.87.3.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henry Y, Ducrocq C, Drapier J C, Servent D, Pellat C, Guissani A. Eur Biophys J. 1991;20:1–15. doi: 10.1007/BF00183275. [DOI] [PubMed] [Google Scholar]
- 33.Hidalgo E, Demple B. EMBO J. 1994;13:138–146. doi: 10.1002/j.1460-2075.1994.tb06243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hidalgo E, Demple B. EMBO J. 1997;16:1056–1065. doi: 10.1093/emboj/16.5.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt H H H W, Walter U. Cell. 1994;78:919–925. doi: 10.1016/0092-8674(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 36.Stamler J S. Cell. 1994;78:931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 37.Stamler J S, Jia L, Eu J P, Mcmahon T J, Demchenko I T, Bonaventura J, Gernert K, Piantadosi C A. Science. 1997;276:2034–2037. doi: 10.1126/science.276.5321.2034. [DOI] [PubMed] [Google Scholar]
- 38.Mulsch A, Mordvintcev P I, Vanin A F, Busse R. Biochem Biophys Res Commun. 1993;196:1303–1308. doi: 10.1006/bbrc.1993.2394. [DOI] [PubMed] [Google Scholar]
- 39.Oliveira L, Bouton C, Drapier J C. J Biol Chem. 1999;274:516–521. doi: 10.1074/jbc.274.1.516. [DOI] [PubMed] [Google Scholar]
- 40.Hidalgo E, Demple B. J Biol Chem. 1996;271:7269–7272. doi: 10.1074/jbc.271.13.7269. [DOI] [PubMed] [Google Scholar]