Abstract
JNK proteins have been shown to be involved in liver carcinogenesis in mice, but the extent of their involvement in the development of human liver cancers is unknown. Here, we show that activation of JNK1 but not JNK2 was increased in human primary hepatocellular carcinomas (HCCs). Further, JNK1 was required for human HCC cell proliferation in vitro and tumorigenesis after xenotransplantation. Importantly, mice lacking JNK1 displayed decreased tumor cell proliferation in a mouse model of liver carcinogenesis and decreased hepatocyte proliferation in a mouse model of liver regeneration. In both cases, impaired proliferation was caused by increased expression of p21, a cell-cycle inhibitor, and reduced expression of c-Myc, a negative regulator of p21. Genetic inactivation of p21 in JNK1–/– mice restored hepatocyte proliferation in models of both liver carcinogenesis and liver regeneration, and overexpression of c-Myc increased proliferation of JNK1–/– liver cells. Similarly, JNK1 was found to control the proliferation of human HCC cells by affecting p21 and c-Myc expression. Pharmacologic inhibition of JNK reduced the growth of both xenografted human HCC cells and chemically induced mouse liver cancers. These findings provide a mechanistic link between JNK activity and liver cell proliferation via p21 and c-Myc and suggest JNK targeting can be considered as a new therapeutic approach for HCC treatment.
Introduction
Hepatocellular carcinoma (HCC), the most common type of human liver cancer, is the third leading cause of cancer death worldwide (1). HCC usually develops in association with HBV and HCV infection and other liver diseases, such as liver steatosis and fibrosis (1). Several molecular pathways have been shown to be involved in human HCC development, for example, AP-1 activity and the NF-κB pathway (2–5). AP-1 and NF-κB are stress-responsive transcription factor complexes that are regulated by upstream MAPKs. Of the 3 types of MAPKs, the functions of 2, ERKs and p38s, in human HCCs have been characterized in recent studies (1, 3, 6), while the functions of JNK are less defined.
Among the 3 mammalian JNK genes, JNK1 and JNK2 are ubiquitously expressed, while JNK3 expression is restricted to the brain, heart, and testis. Various cytokines and environmental factors induce JNK activity, which in turn phosphorylates its substrates, such as c-Jun and ATF2. It has been well demonstrated that JNK1 and JNK2 regulate stress-induced apoptosis in different cell types and contexts (7, 8). Recent data also show that increased JNK activities enhance proliferation of mouse embryonic fibroblasts (MEFs) (3, 9). However, the dominant JNK kinase that regulates MEF proliferation appears to be dependent on the experimental system employed (10, 11). The function of JNK in proliferation in vivo is even more complicated. Using a two-thirds partial hepatectomy-induced liver regeneration model, it was found that increased JNK activation enhances hepatocyte proliferation in mice with hepatic deletion of IκB kinase β (Ikkβ) (4). Intriguingly, upregulated JNK activities appear to attenuate proliferation in Gadd45β–/– livers (12). On the other hand, treatment with JNK inhibitor SP600125 reduced hepatocyte proliferation in regeneration (13). However, due to the nonspecific inhibition of this chemical (14), the functions of JNK in hepatocyte proliferation need to be further characterized.
Mouse cancer models have shown that JNKs play a role in multiple tumors. JNK1 appears to play a tumor-suppressive function in 7,12-dimethylbenz(a)anthracene/12-O-tetradecanoylphorbol-13-acetate–induced (DMBA/TPA-induced) skin carcinogenesis (15). It has also been shown that JNK1 suppresses spontaneous colon cancer development in aged mice (16). However, a recent study reported that JNK1 is dispensable for colitis-associated colon cancer development (17), whereas another study revealed that JNK1 has an oncogenic function in N-methyl-N-nitrosourea–induced mouse gastric tumor development (18). These results suggest a context- and tissue-specific function of JNK1 in tumorigenesis. Given the structural homology between JNK1 and JNK2, it is assumed that both JNK kinases may play similar roles in cancer development. Interestingly, it was found that JNK2 promotes papilloma formation instead of repressing tumorigenesis in a DMBA/TPA-induced skin cancer model (19). Therefore, JNK1 and JNK2 might activate distinct pathways in cancer development, although the underlying molecular mechanisms remain largely undefined.
The JNK pathway has also been implicated in regulating liver tumorigenesis. c-Jun, a downstream target of JNKs, promotes chemically induced liver cancer development through suppression of the p53 pathway (2). In addition, enhanced liver cancer development has been shown to be a consequence of increased activity of the JNK/c-Jun pathway in mice lacking p38α in hepatocytes (3, 6). Recently, it was shown that NF-κB attenuates liver carcinogenesis through repressing JNK1 activation and that JNK1–/– mice exhibit reduced liver cancers (5). Decreased tumorigenesis in JNK1–/– mice is correlated with attenuated cytokine production, reduced cyclin D1 and VEGF expression, decreased cell death, and compensatory proliferation (5). However, it remains to be determined by which molecular mechanism JNK1 and/or JNK2 regulate liver carcinogenesis. Furthermore, despite accumulating evidence obtained from mouse models, whether JNK1 or JNK2 is involved in human liver cancers is not yet known.
In this study, we found that in human HCC, the activation of JNK is increased. Importantly, JNK1 but not JNK2 increases the proliferation of human and mouse liver tumor cells through effects on p21 and c-Myc expression. Loss of p21 or overexpression of c-Myc rescued impaired proliferation of JNK1–/– liver cells. The potential application of JNK inhibition using a peptide inhibitor of JNK, D-JNKI1, for liver cancer therapy was also investigated.
Results
Increased JNK1 activity in human HCCs.
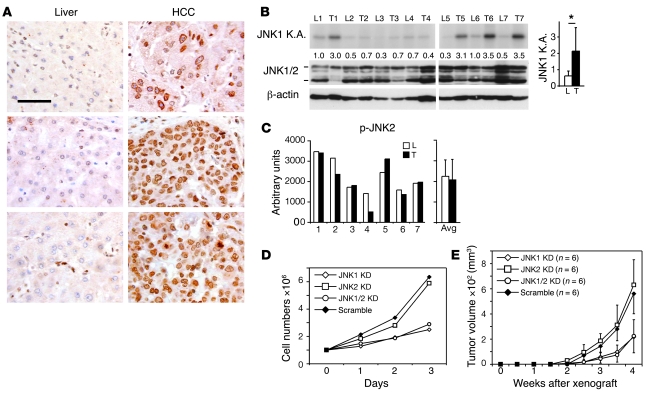
Activation of JNK in human primary HCCs was analyzed by immunohistochemical staining using an antibody detecting both phosphorylated JNK1 and JNK2 (p-JNK) on a tissue microarray. Nuclear staining of p-JNK was detectable in 45 out of 53 HCC tissues. Among these 45 HCC samples, matched nonneoplastic liver tissues from the same patient were available for 41 HCC samples. The p-JNK levels were semiquantified and compared in matched cancerous and nonneoplastic tissues. In 23 (56%) out of 41 matched pairs, p-JNK staining was significantly stronger in HCCs than in nonneoplastic liver tissues (P < 0.05, Wilcoxon-Mann-Whitney test; Figure 1A and Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI37156DS1). To characterize the activity of JNK1, we performed an immunoprecipitation-complex–based kinase assay specific for JNK1 (Supplemental Figure 1A). JNK1-specific kinase assays showed that JNK1 activities were significantly increased in 4 out of 7 HCC samples (P < 0.05, paired Student’s t test; Figure 1B). On the other hand, a p-JNK2–specific ELISA assay showed that p-JNK2 levels remained unchanged in human HCC tissues (Figure 1C and Supplemental Figure 1B). These data suggest that p-JNK levels, most likely due to JNK1, are increased in more than half of human HCCs.
Figure 1. JNK1 activity is increased in human HCC.
(A) Immunohistochemical staining of p-JNK on a tissue microarray of paired human HCCs and nonneoplastic liver tissues. p-JNK staining in HCCs was significantly stronger than that in nonneoplastic liver tissues in 23 out of 41 matched pairs. P < 0.05, Wilcoxon-Mann-Whitney test. Three representative paired samples are shown. Cells with activated JNK show brown nuclear staining. Scale bar: 50 μm. (B) Kinase activities of JNK1 were determined in paired samples of human nonneoplastic livers and HCCs by immunocomplex kinase assay using GST–c-Jun as substrate. Total JNK and β-actin levels were assayed by Western blot. L, liver; T, tumor. Numbers between blots denote JNK kinase activity (K.A.), quantified by normalizing to β-actin levels. Average of JNK1 activities of all samples is shown in the right panel. *P < 0.05, Student’s paired t test. (C) p-JNK2 levels in human HCC samples were analyzed by ELISA assay using total cell extraction. Average of p-JNK2 levels is shown in the right panel. (D) Huh7 cells were infected with lentiviruses expressing shRNAs against JNK1, JNK2, and JNK1/2 as well as scramble control shRNA. Proliferation of cultured cells was analyzed using cells at passage 2 after virus infection. One representative experiment of 3 is shown. KD, knockdown. (E) Tumor formation of JNK1, JNK2, JNK1/2 knockdown, and scramble control Huh7 cells were analyzed after these cells were subcutaneously implanted in nude mice. Data are expressed as mean ± SD.
The specific functions of JNK1 and JNK2 in human HCC cells were further analyzed in a human HCC cell line, Huh7. These cells were infected with lentiviruses expressing shRNAs against JNK1, JNK2, or JNK1/2. Lentiviruses expressing scramble shRNA were used as controls. Efficient JNK1 or JNK2 knockdown was confirmed by Western blots (Supplemental Figure 1C). JNK1 or JNK2 knockdown did not affect the apoptotic rates of these cells as measured by TUNEL staining (data not shown). Interestingly, both JNK1 and JNK1/2 knockdown significantly attenuated the proliferation of cultured Huh7 cells, whereas JNK2 knockdown had no effect on proliferation (Figure 1D). Importantly, when subcutaneously implanted in nude mice, Huh7 cells with JNK1 or JNK1/2 knockdown developed smaller tumors compared with those of JNK2 knockdown or scramble controls (Figure 1E). These results indicate that JNK1 promotes proliferation and tumorigenesis of human Huh7 HCC cells.
JNK1 but not JNK2 is required for proliferation in mouse liver cancers.
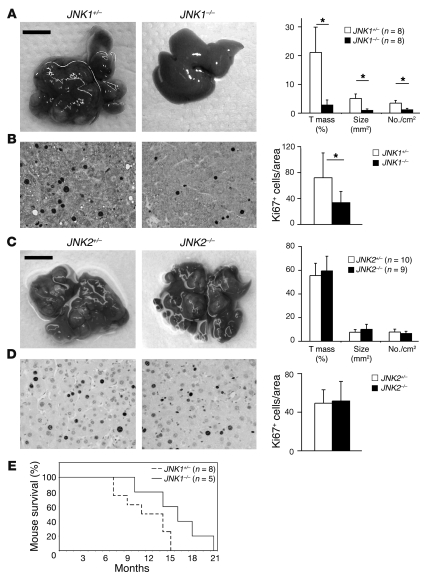
Due to the shortage of human HCC samples and the limitation of in vitro HCC cell cultures, we next investigated the specific roles of JNK1 and JNK2 in proliferation of liver cancer cells using a diethylnitrosamine-phenobarbital–induced (DEN-Pb–induced) mouse liver cancer model (2). Fully developed liver cancers were induced in JNK1+/– mice 7 months after DEN treatment. JNK1–/– mice exhibited impaired liver tumorigenesis with reduced tumor masses, tumor sizes, and tumor numbers (Figure 2A). JNK1–/– mice also showed a longer life span (15.8 ± 4.1 months) compared with controls (11.4 ± 3.4 months) following tumor induction (Figure 2E). Ki67 staining revealed that the number of proliferating cells in JNK1–/– liver cancers was reduced to half of that in control mice (Figure 2B). In contrast, JNK2–/– mice showed DEN-induced liver carcinogenesis similar to that of controls 9 months after DEN treatment, with comparable proliferation rates (Figure 2, C and D). Intriguingly, the apoptotic rates in JNK1–/– and JNK2–/– liver cancers were similar to those of controls as determined by TUNEL staining (Supplemental Figure 2, A and B). Vessel densities in both JNK1–/– and JNK2–/– cancers were also unaffected, as assessed using von Willebrand factor immunohistochemical staining (Supplemental Figure 2C). Moreover, slot-blot analyses using O6-ethyldeoxyguanosine–specific (O6-Et-dG–specific) and O2-ethyldeoxythymidine–specific (O2-Et-dT–specific) antibodies showed that these DEN-induced DNA adducts were not altered in JNK1–/– livers, suggesting that JNK1 does not affect genotoxic damage induced by DEN at the initiation stages (Supplemental Figure 2D). Together, these data indicate that JNK1 is required for liver cancer development, most likely because of its promotion of proliferation of liver cancer cells.
Figure 2. JNK1 but not JNK2 enhances proliferation of mouse liver cancer cells.
(A) DEN-induced liver cancers in JNK1+/– and JNK1–/– mice are shown. Quantification of tumor mass (percentage of total tumor size compared with total liver tissue size), average tumor size (mm2), and tumor numbers per cm2 on H&E-stained liver sections are shown in the right panel. (B) Proliferation of liver cancer cells in JNK1+/– and JNK1–/– mice was analyzed by immunostaining for Ki67. Ki67-positive cells are indicated by a brown-stained nucleus. Ki67-positive cells in cancers were quantified. In total, 21 liver tumors from 6 JNK1+/– mice and 15 tumors from 5 JNK1–/– mice were analyzed. *P < 0.05, Student’s t test. (C) Liver cancers from DEN-treated JNK2+/– and JNK2–/– mice are shown. Quantification of tumor mass, average tumor size, and tumor numbers are shown in the right panel. Scale bars: 1 cm (A and C). (D) Proliferation of liver cancer cells in JNK2+/– and JNK2–/– mice was determined by Ki67 staining. Ki67-positive cells were quantified. Original magnification, ×200 (B and D). (E) Kaplan-Meier survival curve of JNK1+/– and JNK1–/– mice subjected to DEN-Pb liver carcinogenesis protocol. P < 0.05, Gehan’s test. Data are expressed as mean ± SD.
Reduced liver cell proliferation and tumorigenesis in JNK1–/– mice are caused by increased p21 expression.
Excessive production of growth factors and proinflammatory cytokines induce enhanced proliferation during liver carcinogenesis (20, 21). However, quantitative RT-PCR (qRT-PCR) revealed no alteration in HGF, EGF, IGF-1, TGF-α, and TGF-β expression between JNK1–/– and control liver cancers (Supplemental Figure 3A). TNF-α, IL-1α, IL-1β, IL-6, and IL-12 cytokine levels were also comparable as measured by ELISA (Supplemental Figure 3B). Moreover, the infiltration of inflammatory cells appeared to be unchanged in JNK1–/– liver cancers, as estimated by immunohistochemical staining of markers for neutrophilic granulocytes, macrophages, and T lymphocytes (data not shown).
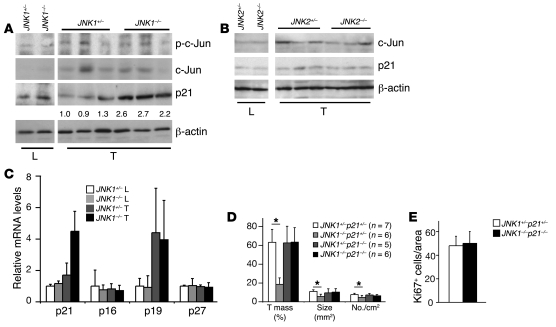
We next characterized the activities of downstream effectors of the JNK pathway. The crosstalk between JNK and the ERK and AKT pathways was found to be important in regulating proliferation of melanoma and prostate cancer cells (22, 23). However, Western blot analyses showed that the activation of the ERK and AKT pathways was unchanged in JNK1–/– liver cancers (Supplemental Figure 3C). c-Jun was previously shown to mediate hepatocyte proliferation during liver regeneration and carcinogenesis (3, 24). Interestingly, p–c-Jun levels were unchanged in JNK1–/– liver cancers (Figure 3A). These results are consistent with our previous findings that c-Jun phosphorylation is not required for liver carcinogenesis and regeneration (2, 24). JNK has also been shown to affect c-Jun stability in cultured mouse fibroblasts (10). However, c-Jun protein levels were not changed in either JNK1–/– or JNK2–/– liver tumors (Figure 3, A and B), suggesting that JNK1 likely regulates liver cancer development independently of c-Jun.
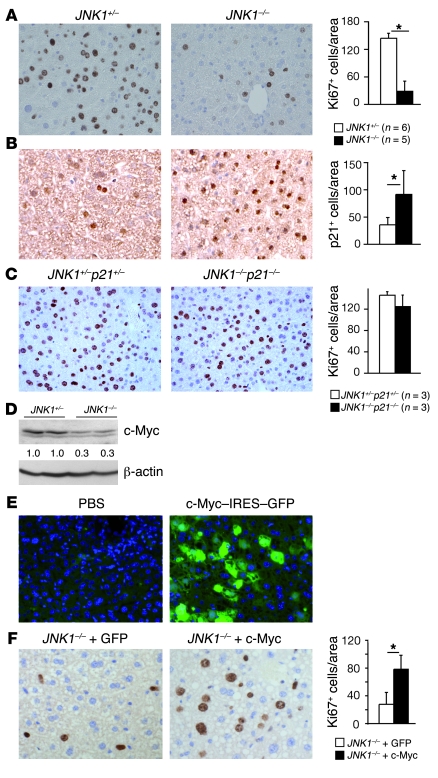
Figure 3. Upregulation of p21 causes reduced proliferation of JNK1–/– liver cancer cells.
(A) Protein levels of p–c-Jun, c-Jun, and p21 were determined by Western blot in JNK1+/– and JNK1–/– liver and cancer tissues. β-actin levels were used as loading control. Numbers below p21 blot represent quantification as normalized to β-actin. (B) c-Jun and p21 protein levels were determined by Western blot in JNK2+/– and JNK2–/– liver cancer tissues. β-actin was used as loading control. (C) mRNA levels of p21, p16, p19, and p27 were determined by qRT-PCR in cancer and normal liver tissues from JNK1+/– and JNK1–/– mice. (D) DEN-induced liver cancers in JNK1+/–p21+/–, JNK1–/–p21+/–, JNK1+/–p21–/–, and JNK1–/–p21–/– mice were quantified in terms of tumor mass, average tumor size, and tumor numbers. (E) Proliferation of cancer cells was analyzed by Ki67 staining on liver sections from DEN-treated JNK1+/–p21+/– and JNK1–/–p21–/– mice. Quantification of Ki67-positive cells in cancers is shown. *P < 0.05, Student’s t test. Data are expressed as mean ± SD.
p21, a well-established cyclin-dependent kinase (CDK) inhibitor, has been implicated in regulating TGF-β–induced proliferation of cultured hepatocytes by interacting with the JNK pathway (25). p21 overexpression has been also shown to reduce hepatocyte proliferation during postnatal development and liver regeneration (26). Moreover, impaired hepatocyte proliferation in mice lacking c-Jun is completely restored in c-Jun and p21 double-knockout mice after partial hepatectomy (27). Indeed, p21 was found to be upregulated at the protein and mRNA levels in JNK1–/– cancers (Figure 3, A and C). The expression levels of p16 and p27 CDK inhibitors were not changed as measured by qRT-PCR, whereas p19 levels were upregulated in liver cancers independently of JNK1 (Figure 3C). In contrast, p21 levels remained unchanged in JNK2–/– liver cancers (Figure 3B). These results suggest that elevated p21 levels may be responsible for decreased proliferation of JNK1–/– cancer cells.
To determine whether increased p21 expression is responsible for decreased proliferation of JNK1–/– liver cancer cells, we generated JNK1–/–p21–/– double-mutant mice and subjected them to DEN-induced liver carcinogenesis. JNK1–/–p21–/– mice were viable and exhibited no obvious liver dysfunction, as evaluated by histomorphology and serum levels of glutamate pyruvate transaminase (GPT) and glutamate oxaloacetate transaminase (GOT) (data not shown). Upon induction of liver cancers by DEN, JNK1–/–p21–/– mice showed levels of tumor masses, sizes, and numbers similar to those of JNK1+/–p21+/– controls, whereas liver cancer development was reduced in JNK1–/–p21+/– mice and unchanged in JNK1+/–p21–/– mice (Figure 3D). Importantly, proliferation rates of liver cancer cells were completely restored in JNK1–/–p21–/– mice (Figure 3E). These data demonstrate that increased p21 levels are responsible for reduced proliferation of JNK1–/– cancer cells, which further leads to impaired liver carcinogenesis.
c-Myc expression is reduced in JNK1–/– liver cancer cells.
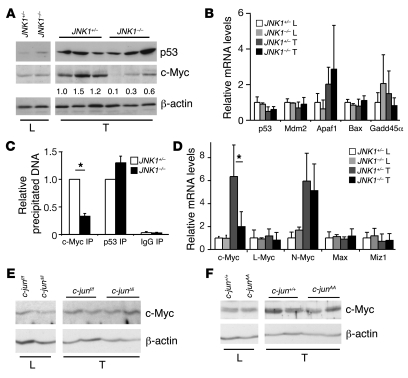
p21 transcription can be activated by p53 (28) and repressed by c-Myc in various cell types (29–31). Expression levels of p53 and its downstream targets appeared to be unchanged in JNK1–/– liver cancers (Figure 4, A and B). ChIP confirmed that p53 binding to the p21 promoter was unaffected in JNK1–/– liver cancers (Figure 4C). However, c-Myc protein and mRNA levels were decreased in JNK1–/– liver cancers (Figure 4, A and D), whereas expression levels of c-Myc–related genes L-Myc, N-Myc, Max, and Miz1 were unaltered as measured by qRT-PCR (Figure 4D). ChIP assay also revealed that binding of c-Myc to the p21 promoter was reduced in JNK1–/– liver cancers (Figure 4C). In contrast, c-Myc levels were unchanged in JNK2–/– liver cancers, as determined by Western blots (Supplemental Figure 3D). These results, consistent with previous reports (29–31), suggest that increased p21 levels in JNK1–/– hepatocytes are most likely caused by reduced c-Myc expression.
Figure 4. c-Myc expression levels are reduced in JNK1–/– liver cancers.
(A) Protein levels of p53 and c-Myc were analyzed by Western blots in JNK1–/– cancer and normal liver tissues. β-actin levels were used as loading control. c-Myc levels were quantified. (B) mRNA levels of p53 and its target genes Mdm2, Apaf1, Bax, and Gadd45α were determined by qRT-PCR in JNK1+/– and JNK1–/– liver and cancer tissues. (C) The binding of c-Myc and p53 to the p21 promoter (–216 to –108) was analyzed in pooled DEN-induced liver cancers using ChIP assays (c-Myc IP and p53 IP). Rabbit IgG was used as negative control (IgG IP). The amount of c-Myc or p53 binding p21 promoter DNA was quantified by qRT-PCR assays. (D) mRNA levels of c-Myc, L-Myc, N-Myc, Max, and Miz1 were analyzed by qRT-PCR in JNK1–/– liver cancers. (E and F) c-Myc protein levels were analyzed by Western blot in liver cancers from c-junΔli and c-junAA mice. *P < 0.05, Student’s t test. Data are expressed as mean ± SD.
Next, we analyzed c-Myc protein levels in liver cancers from mice with hepatic deletion of c-Jun (Alfp-cre, c-junf/f, designated as c-junΔli). Western blot analyses revealed that c-Myc was not changed in c-junΔli liver cancers (Figure 4E). Moreover, c-Myc protein levels were also unaltered in liver cancers in mice harboring c-jun alleles with serines 63 and 73 mutated to alanines (c-junAA; Figure 4F). These results indicate that c-Myc expression is regulated in a c-Jun and p–c-Jun–independent manner in liver cancers.
JNK1 facilitates hepatocyte proliferation through regulating expression of p21 and c-Myc in liver regeneration.
We next characterized the proliferation of JNK1–/– and JNK2–/– hepatocytes during liver regeneration. In adult mice, quiescent hepatocytes synchronously enter the cell cycle and reach the peak of proliferation at 48 hours after two-thirds partial hepatectomy. This event is highly regulated, providing an excellent model system for studying liver cell proliferation. Hepatocyte proliferation was previously found to be increased in JNK2–/– livers at 30 hours after one-third hepatectomy (32). Following two-thirds hepatectomy, JNK2–/– mice displayed hepatocyte proliferation rates similar to those of controls at 48 hours (data not shown). In contrast, the number of Ki67-positive proliferating hepatocytes in JNK1–/– mice was reduced by 80% compared with that in controls at 48 hours after two-thirds partial hepatectomy (Figure 5A). These data demonstrate that JNK1 is a positive regulator of hepatocyte proliferation during liver regeneration.
Figure 5. JNK1 facilitates hepatocyte proliferation by regulating expression of p21 and c-Myc.
(A) Proliferating hepatocytes were analyzed by Ki67 staining in JNK1–/– livers 48 hours after partial hepatectomy. Quantification of Ki67-positive cells is shown. (B) Immunohistochemical staining of p21 on liver sections of JNK1+/– and JNK1–/– mice during liver regeneration (brown nuclear staining). (C) Hepatocyte proliferation in JNK1+/–p21+/– and JNK1–/–p21–/– mice was analyzed by Ki67 immunostaining at 48 hours after partial hepatectomy. (D) c-Myc protein levels were analyzed by Western blot in livers from JNK1+/– and JNK1–/– mice after partial hepatectomy. (E) Mice were injected with PBS or 30 μg of c-Myc–IRES–GFP bicistronic plasmid through tail veins. Two days after injection, transfection of hepatocytes was monitored by GFP expression in liver cryosections. Nuclei were stained blue by DAPI. (F) Hepatocyte proliferation was analyzed 48 hours after partial hepatectomy in JNK1–/– mice with hydrodynamics-based GFP or c-Myc–IRES–GFP (c-Myc) transfection. *P < 0.05, Student’s t test. Original magnification, ×200. Data are expressed as mean ± SD.
The production of growth factors of HGF, EGF, IGF-1, TGF-α, and TGF-β and proinflammatory cytokines of TNF-α, IL-1α, IL-1β, IL-6, and IL-12 appeared to be unchanged during liver regeneration, as measured by qRT-PCR and ELISA assays (Supplemental Figure 3, E and F). Notably, the number of p21-positive cells was increased in JNK1–/– regenerating livers, as determined by immunohistochemical staining (Figure 5B). Furthermore, Ki67 staining revealed that hepatocyte proliferation was reverted in JNK1–/–p21–/– mice compared with that in controls 48 hours after two-thirds partial hepatectomy (Figure 5C). These results suggest that decreased proliferation rate of JNK1–/– hepatocytes is also due to increased p21 levels during liver regeneration.
Interestingly, c-Myc expression was reduced in JNK1–/– regenerating livers as well (Figure 5D), correlating with increased p21 levels. To analyze whether increased c-Myc levels could restore impaired proliferation of JNK1–/– liver cells, we overexpressed c-Myc in mouse livers using hydrodynamics-based transfection and subjected the mice to partial hepatectomy. Two days after tail vein injection of the c-Myc–IRES–GFP (IRES, internal ribosome entry site) bicistronic plasmid, the transfection rate of hepatocytes reached 60%–70% as determined by GFP expression (Figure 5E). Strikingly, 48 hours after partial hepatectomy, the number of proliferating cells in JNK1–/– mice with c-Myc–IRES–GFP transfection was increased 2.9-fold compared with that in GFP-transfected JNK1–/– mice (Figure 5F). The number of proliferating hepatocytes in JNK1–/– mice with c-Myc–IRES–GFP transfection was also increased to 60% of that in control JNK1+/– mice with c-Myc–IRES–GFP transfection (data not shown). These data strongly indicate that JNK1 facilitates mouse liver cell proliferation through regulation of p21 and c-Myc.
JNK1 affects p21 and c-Myc expression in human HCC cells.
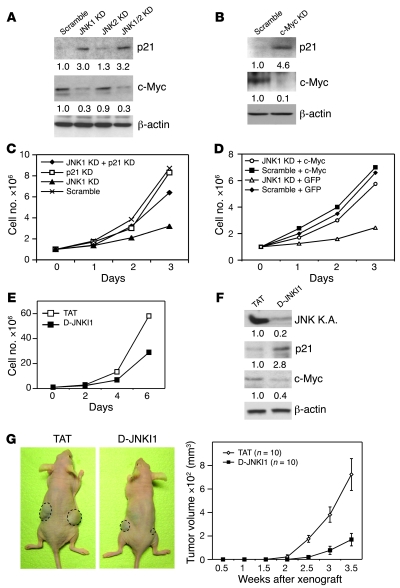
Next, we determined whether JNK1 controls the expression of p21 and c-Myc in human HCC cells. shRNA-mediated knockdown of JNK1 or JNK1/2 but not JNK2 led to elevated p21 and decreased c-Myc expression in human Huh7 HCC cells (Figure 6A). This indicates that JNK1 modulates p21 and c-Myc expression in these cells. Importantly, siRNA-mediated c-Myc knockdown led to significant p21 upregulation in Huh7 human HCC cells (Figure 6B). In addition, impaired proliferation rate of JNK1-knockdown Huh7 cells was restored to 75% of control cells by p21 knockdown (Figure 6C). Finally, overexpression of c-Myc significantly increased proliferation of JNK1 knockdown Huh7 cells (Figure 6D). These data demonstrate that JNK1 also promotes cell proliferation through regulation of p21 and c-Myc expression in human HCC cells.
Figure 6. JNK1 regulates p21 and c-Myc expression in human Huh7 HCC cells.
(A) p21 and c-Myc protein levels in JNK1, JNK2, and JNK1/2 knockdown Huh7 cells and scramble controls were determined by Western blot. (B) p21 and c-Myc levels were assayed by Western blot in human Huh7 cells transfected with scramble control siRNA or c-Myc siRNA. Quantification is normalized to β-actin levels. (C) Proliferation rate of Huh7 cells with both JNK1 and p21 knockdown was analyzed. (D) Huh7 cells with scramble or JNK1 shRNAs were transfected with either pIRES-GFP (GFP) or c-Myc–IRES2–AcGFP (c-Myc). Proliferation of these cells was analyzed for 3 days. (E) Proliferation of cultured Huh7 cells was reduced upon treatment with D-JNKI1 (20 μM). One representative experiment of 3 is shown. (F) JNK kinase activity in D-JNKI1–treated (20 μM) Huh7 cultures was analyzed by immunocomplex kinase assay using GST–c-Jun as substrates. p21 and c-Myc protein levels were determined by Western blot. Quantification was normalized to β-actin. (G) Tumor growth of subcutaneously implanted Huh7 cells was analyzed following 4 weeks of treatment with TAT or D-JNKI1 (5 mg/kg) in nude mice. Data are expressed as mean ± SD.
We then determined whether blocking JNK activity using D-JNKI1, a specific JNK1/2 dual inhibitor, has an inhibitory effect on human HCC cells. D-JNKI1 treatment significantly reduced proliferation and JNK activity of cultured Huh7 HCC cells, which were associated with increased p21 and reduced c-Myc levels (Figure 6, E and F). Importantly, D-JNKI1 treatment strikingly attenuated tumor growth of subcutaneously implanted Huh7 cells in nude mice (Figure 6G).
JNK inhibitor treatment reduces liver cancer development.
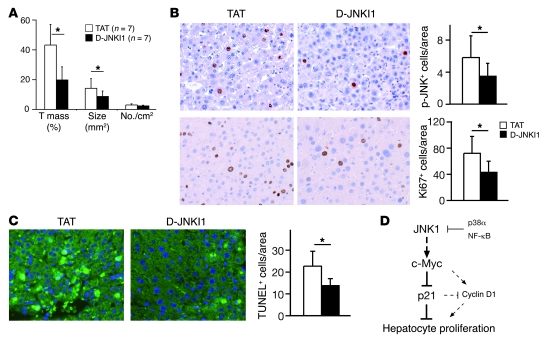
The results obtained from D-JNKI1 treatment of HCC cells in vitro and in xenografted tumors prompted us to analyze its therapeutic effect on liver tumors developed under physiological conditions. We treated wild-type mice bearing DEN-induced liver cancers with D-JNKI1 for 3 months. Tumor masses and sizes in D-JNKI1–treated mice were decreased by half compared with those in controls, whereas tumor numbers remained unchanged (Figure 7A). Notably, D-JNKI1 treatment reduced the number of p-JNK–positive cells in liver cancers, correlating with decreased proliferation as assayed by Ki67 staining (Figure 7B). These results unequivocally establish that JNK, most likely JNK1, activity is required at late stages of liver carcinogenesis in mice.
Figure 7. JNK inhibitor treatment reduces liver cancer development.
(A) Five months after DEN injection, mice were treated with JNK inhibitor peptide D-JNKI1 or control peptide TAT (5 mg/kg) twice a week for 3 months. Quantification of tumor mass, tumor size, and tumor number from D-JNKI1– or TAT-treated mice is shown. (B) Phosphorylation of JNK in DEN-induced liver cancers from D-JNKI1– or TAT-treated mice was analyzed by immunohistochemical staining on liver sections (brown nuclear staining). Proliferation of cancer cells was analyzed by immunostaining for Ki67. (C) Wild-type mice at 4 weeks of age were injected with D-JNKI1 or TAT at 5 mg/kg 2 days before DEN treatment and killed 24 hours after. DEN-induced cell death was analyzed by TUNEL staining on liver sections. TUNEL-positive cells (green-stained nucleus) were quantified. *P < 0.05, Student’s t test. Original magnification, ×200. (D) Schematic model of JNK1-dependent hepatocyte proliferation. JNK1 activation and decreased p21 levels are apparently important for cell proliferation during liver carcinogenesis and regeneration. p21, repressed by c-Myc, attenuates proliferation of liver cells, probably via suppressing cyclin D1. Importantly, this pathway is negatively regulated by p38α and NF-κB stress-signaling pathways and appears to be independent of c-Jun. Data are expressed as mean ± SD.
It was previously proposed that JNK1 promotes hepatocarcinogenesis by enhancing liver cell death at early stages (4–6), suggesting that JNK1 may play a dual, stage-dependent function in regulating hepatocarcinogenesis. We therefore analyzed the inhibitory effect of D-JNKI1 on DEN-induced cell death. TUNEL staining revealed that D-JNKI1 treatment significantly reduced cell death in wild-type mice 24 hours after DEN treatment (Figure 7C). This is similar to the phenotype of reduced hepatocyte death in JNK1–/– mice (refs. 5 and 6 and data not shown), implying that D-JNKI1 treatment at the initiation stages might also be beneficial (4–6).
Discussion
Our findings demonstrate that JNK1 is critically involved in human and mouse liver cancers, while JNK2 appears to be dispensable. Elevated activity of total p-JNK was observed in 50% of characterized HCCs in this study. Epidemiological studies show that human HCC develops with HBV or HCV infection and liver steatosis and fibrosis (1). All of these diseases are associated with chronic liver damage and inflammation, which activate stress-associated kinases, such as JNKs. Interestingly, retrospective analysis of the disease histories of the patients recruited in this study showed that JNK activities in HCC tissues are not correlated with these pathological conditions (data not shown), suggesting that there may be other stimuli responsible for enhanced JNK activity. In addition, JNK1 activities are specifically increased in these human HCC samples, implying that these stimuli may selectively activate JNK1. Although JNK1 and JNK2 are commonly regulated by upstream kinases and phosphatases, selective activation of JNK1 has been shown to be regulated in a TAK1-dependent manner by the X chromosome–linked inhibitor of apoptosis protein (XIAP) (33). Moreover, JNK1 is specifically activated in mouse osteoclast progenitors by RANKL (34), which also activates TAK1 through the TAB2/TRAF6 pathway (35). Therefore, we speculate that the RANKL/TAK1 pathway may deserve further investigation in human HCCs.
Our findings demonstrate that JNK1 is essential for proliferation of hepatocytes and liver cancer cells in vivo. While impaired hepatocyte proliferation is evident in JNK1–/– mice, altered apoptosis of liver cancer cells was not observed in JNK1–/–, JNK2–/–, or D-JNKI1–treated mice. Interestingly, inflammation- or ROS-induced apoptosis of hepatocytes was reduced in JNK1–/– mice and D-JNKI1–treated wild-type mice at early stages of liver carcinogenesis (refs. 4, 5, Figure 7C, and data not shown). It is therefore likely that the JNK1 pathway has a dual, stage-dependent function. JNK1 activation at late stages leads to excessive cell proliferation by repression of p21 and enhancement of c-Myc expression (Figure 7D). JNK1 probably exerts its oncogenic function on proliferation independent of c-Jun, since phosphorylation and protein levels of c-Jun were unchanged in JNK1–/– liver tumors and c-Jun phosphorylation appears to be dispensable for liver carcinogenesis and regeneration (2, 5, 24). On the other hand, JNK1 may enhance c-Jun–mediated cell death during liver tumor initiation (4, 5). However, the function of c-Jun may be even more complex, since c-Jun can also mediate hepatocyte survival in TNF-α–dependent acute hepatitis (36). Since activation of JNK is believed to be important in various chronic liver diseases, a careful characterization of the functions of JNK and its targets at specific disease stages along with a better understanding of JNK interaction with other stress-signaling pathways, such as p38 and NF-κB (3, 5), will provide novel and important insights into the molecular links between inflammation and liver cancer (Figure 7D).
At the molecular level, our findings demonstrate that increased p21 expression is responsible for attenuated proliferation of JNK1–/– cells (Figure 7D). Genetic inactivation of p21 in JNK1–/– mice completely rescued impaired proliferation, indicating an important role for p21 in JNK1-dependent liver carcinogenesis. A recent study also showed that JNK inhibitor SP600125 treatment upregulated p21 levels and increased CD95-mediated G2/M cell-cycle arrest in cultured hepatoma cells (37). The function of p21 in suppressing liver cell proliferation has been previously shown using transgenic mice overexpressing p21 (26). Moreover, increased p21 levels were found to cause impaired hepatocyte proliferation in mice lacking c-Jun during liver regeneration (27). Increased p21 protein levels negatively regulate cyclin D1 expression in liver cell proliferation following partial hepatectomy (26, 27). Reduced cyclin D1 expression was also found in JNK1–/– liver tumors (ref. 5 and data not shown) and was proposed to be responsible for decreased cancer cell proliferation (5). Interestingly, cyclin D1 levels were completely restored in JNK1–/–p21–/– liver cancers, as measured by Western blot (data not shown), suggesting that cyclin D1 may be one of the mediators in controlling proliferation of JNK1–/– cancer cells (Figure 7D).
Both mRNA and protein levels of p21 are upregulated in JNK1–/– liver cancer cells. Intriguingly, JNK was shown to stabilize p21 protein through phosphorylation or deubiquitination (38, 39). We speculate that increased p21 might be regulated at the transcriptional level. It has been well documented that p21 transcription is repressed by c-Myc in normal and cancer cells (29–31). Our results revealed that both c-Myc levels and binding of c-Myc protein to the p21 promoter are decreased in JNK1–/– cells. Moreover, c-Myc knockdown increased p21 expression in human Huh7 HCC cells. Therefore, we propose that enhanced p21 expression is most likely caused by decreased c-Myc levels in both liver cancers and regenerating livers (Figure 7D). It is also worth noting that expression of p21 and c-Myc is unchanged in JNK2–/– liver cancer cells, which suggests that the expression of p21 and c-Myc is specifically regulated by JNK1. We speculate that JNK1 may directly stabilize c-Myc protein levels by phosphorylation of serine 71, as shown in the previous study (40). On the other hand, JNK1 may indirectly enhance the transcription of c-Myc mRNA through activating transcription factors of STAT3, NFATC1, or AP-1 genes, such as ATF2 (10, 41, 42).
What is the molecular explanation for the distinct functions of JNK1 and JNK2? Protein sequence analysis of JNK from different species showed that JNK2 is phylogenetically distant from JNK1 and JNK3 (Supplemental Figure 4A). Two subdomains of amino acid residues 218–230 and 363–380 differ significantly between JNK1 and JNK2 (Supplemental Figure 4B). Modeling the structures of JNK1 and JNK2 using a JNK3 crystal structure revealed that these 2 subdomains contain hydrophilic amino acid residues and are surface exposed, implying that these domains may serve specific functions, such as docking sites for substrates (Supplemental Figure 4C). Indeed, it has been shown that these 2 subdomains are responsible for distinctive features of JNK1 and JNK2, such as efficient binding of c-Jun and autophosphorylation (43, 44). Therefore, it is possible that these subdomains might also be involved in the different binding affinities of JNK1 and JNK2 in regulating c-Myc expression.
Our results demonstrate the importance of JNK1 in human HCCs and the potential application of JNK targeting for HCC therapy. Recently, a new strategy using “cocktail” drugs targeting multiple kinases was proposed for cancer treatment (45, 46). It is worth mentioning that abnormal activation of ERK is often observed in human HCC. It has also been shown that systemic administration of sorafenib, an inhibitor of the Raf/ERK pathway and tyrosine receptor kinases, significantly extended survival of HCC patients (47, 48). In light of our findings, a combined inhibition of JNK and ERK activities may profoundly improve HCC therapies.
Methods
Human HCC samples.
Human HCC samples were obtained from the biobank of the Institute of Pathology at the Medical University of Graz (Genome Austria Tissue Bank). All samples were histopathologically evaluated before further analysis. Seven HCC and paired liver samples were used for JNK kinase assay. These tissues were harvested on ice, embedded in OCT immediately, and stored at –80°C before use. An additional 53 formaldehyde-fixed and paraffin-embedded HCC samples were examined for p-JNK staining on a tissue microarray (37 males, 16 females; mean age, 59.5 years). The collection and use of human tissue samples was approved by the Ethical Committee of the Institute of Pathology, Medical University of Graz.
Animals.
All mice used in this study were maintained on a mixed genetic background (C57BL/6; 129/Sv). JNK1-knockout mice (JNK1–/–) were crossed with heterozygous mice with 1 wild-type allele of JNK1 (JNK1+/–) to obtain JNK1+/– and JNK1–/– mice. JNK2+/– and JNK2–/– mice were bred to obtain JNK2+/– and JNK2–/– mice. JNK1–/– mice were crossed with p21–/– to obtain JNK1+/–p21+/–, JNK1–/–p21+/–, JNK1+/–p21–/–, and JNK1–/–p21–/– mice. All animal experiments in this study were approved by the Magistratsabteilung 58, Vienna, Austria.
Chemical-induced liver carcinogenesis protocol.
A single dose of DEN (Sigma-Aldrich) at 100 mg/kg body weight was injected i.p. into 4-week-old male mice. Mouse antibodies specific for O6-Et-dG (EM-21) and O2-Et-dT (EM-4-1) (a gift from M. F. Rajewsky, University of Essen, Essen, Germany) were used to analyze the DNA adducts after DEN treatment. Mice were fed with Pb in a 0.07% diet from the age of 6 weeks until sacrifice. JNK1+/– and JNK1–/– mice were killed 7 months after DEN treatment. JNK2+/–, JNK2–/–, JNK1+/–p21+/–, JNK1–/–p21+/–, JNK1+/–p21–/–, and JNK1–/–p21–/– mice were killed 9 months after DEN treatment. Liver cancer development was compared among offspring from the same breeding.
Liver tumors were identified on H&E-stained sections and quantified as described before (2, 3). In brief, tumor size was quantified using ImageJ software (http://rsbweb.nih.gov/ij/index.html). Tumor mass was calculated as a percentage of total liver size for each section. Tumor number was calculated as the total number of tumors in relation to the total liver size of the section. Tumor size was calculated as the average size of all tumors in the section.
D-JNKI1 peptide treatment.
D-JNKI1 peptide of D amino acid DQSRPVQPFLNLTTPRKPR-PP-RRRQRRKKRG and TAT control peptide PP-RRRQRRKKRG were synthesized by the Protein Chemistry Department at the Research Institute of Molecular Pathology (IMP) (49). Specific inhibition of JNK activation by D-JNKI1 has been shown previously (3, 49). Pilot experiments showed that p-JNK levels were reduced in mouse liver tumors even 2 days after 5 mg/kg D-JNKI1 treatment; on the other hand, NMDA-induced p-JNK levels were not decreased in D-JNKI1–treated cortical neuronal cultures (49), which may be explained by different stimuli and cell types. Wild-type mice were administered D-JNKI1 or TAT control peptide 5 months after DEN injection. D-JNKI1 or TAT peptides were injected at 5 mg/kg body weight i.p. twice a week for 3 months.
Xenograft liver cancer model.
Human Huh7 cells were cultured in DMEM with 10% FCS at 37°C. Cells that grew to 80% confluent were trypsinized and resuspended in DMEM medium. 2 × 106 cells in 200 μl DMEM were subcutaneously injected into the flank sides of nude mice (Foxn1nu/nu, MF1 background) at 8–12 weeks of age. Two days after implantation of tumor cells, mice were injected i.p. with D-JNKI1 or TAT twice a week for 4 weeks at 5 mg/kg body weight. We measured the lengths (L) and widths (W) of tumors twice a week by caliper and calculated the tumor sizes as: V = L × W × W/2, with V indicating volume.
Partial hepatectomy.
Partial hepatectomy was described elsewhere (24, 27). Mice 3 to 5 months old were used for experiments. Surgeries were performed between 8 and 12 am. Mice were anesthetized by avertin treatment. The abdominal cavity was opened below the rib cage, and the large left lateral and median lobes were removed. Animals were recovered on a 37°C heating block for 1–2 hours after surgery.
Hydrodynamics-based transfection of mouse hepatocytes.
The human c-Myc gene was inserted in the pIRES2-AcGFP vector to generate the p-cMyc-IRES-GFP plasmid. Mice between 3 and 5 months old were used. 30 μg of plasmid in 2 ml Krebs buffer was injected into each mouse through the tail vein within 5 seconds. Two days after injection, transfection of hepatocytes was monitored by GFP expression using liver cryosections. Mice were subjected to partial hepatectomy 2 days after injection.
Immunohistochemistry.
Immunohistochemistry stainings for Ki67 (Novocastra; Leica Microsystems) and vWF (Dako) were performed using Discovery XT system (Ventana Medical Systems Inc.). Immunostaining for p-JNK (Promega) and p21 (Santa Cruz Biotechnology Inc.) was performed using standard protocols with citrate buffer (pH 6.0) pretreatment. The in situ cell death detection kit (ApoAlert; Clontech) was used for TUNEL assay.
RNAi-mediated gene knockdown.
Lentivirus vectors expressing shRNAs against human JNK1 (TRCN0000001055-57, TRCN0000010580-81), JNK2 (TRCN0000001012-16), and scramble shRNA (SHC002, CAACAAGATGAAGAGCACCAA) were obtained from Sigma-Aldrich. Scramble shRNA, which does not target any human or mouse genes, was used as control.
Huh7 cells were infected with lentivirus with polybrene for 2 days and then selected for puromycin resistance for 5 days. The first 3 passages of Huh7 cells with shRNA knockdown were used for experiments. TRCN0000001055 and TRCN0000001081 showed similar efficiency of JNK1 knockdown and proliferation inhibition in Huh7 cells. TRCN0000001012–TRCN0000001015 showed similar JNK2 knockdown efficiency, and none of the shRNAs inhibited Huh7 proliferation. Results of TRCN0000001081 shRNA (GACTCAGAACACAACAAACTT) against JNK1 and TRCN0000001015 shRNA (GGGATTGTTTGTGCTGCATT) against JNK2 shRNA are shown (Figure 1, D and E, and Figure 6, C and D).
For p21 and c-Myc knockdown, ON-TARGETplus SMARTpool siRNAs against human p21 or c-Myc were purchased from Dharmacon. siRNAs were transfected to the Huh7 cells using Lipofectamine reagent (Invitrogen). Two days after transfection, cells were replated and proliferation was measured in the following 3 days.
ChIP assay.
ChIP was performed as described elsewhere (29). In brief, DEN-induced mouse liver tumors were homogenized and cross-linked in 1% formaldehyde. Lysates were then sonicated to yield DNA fragments around 500–1000 bp. DNAs were coimmunoprecipitated by c-Myc or p53 antibodies or rabbit IgG at 4°C overnight. p21 promoter (–216 to –108) was amplified by qRT-PCR using the primer pairs listed in Supplemental Table 1.
Western blot and JNK kinase assays.
Antibodies against ERK, p-ERK, p-AKT, AKT, p-PTEN, p-GSK3β, p-Raf, p-PDK1 (Cell Signaling Technology), cyclin D1, c-Jun, Cdc2, Cdk2, p21 (BD Transduction Laboratories), cyclin A, cyclin E, c-Myc (Santa Cruz Biotechnology Inc.), and p53 (Novocastra) were used for Western blot. Western blots were quantified using ImageQuant TL v2003.03 (GE Healthcare).
For total JNK kinase assay, a JNK kinase assay kit was used (9810; Cell Signaling Technology). In brief, protein extracts were incubated with GST–c-Jun–conjugated (GST, glutathione S-transferase) beads overnight at 4°C. ATP was added, and kinase reaction was carried out at 30°C for 30 minutes. p–GST–c-Jun protein was separated by SDS-PAGE gel and detected by a p–c-Jun antibody from the kit.
For the JNK1 kinase assay, we immunoprecipitated JNK1 by antibody specifically against JNK1 (sc-474; Santa Cruz Biotechnology Inc.) and followed this with radioactive kinase assay using GST-fused c-Jun (Cell Signaling Technology) as substrate. For the JNK2 kinase assay, several antibodies against JNK2 from various companies were tested for immunoprecipitate-based JNK2 kinase assay. However, all of the tested antibodies showed nonspecific binding to JNK1 to some extent. Therefore, a p-JNK2–specific ELISA kit was used to measure p-JNK2 levels (DYC2236-2; R&D Systems). The specificities of JNK1 kinase assay and p-JNK2 ELISA kit are shown in Supplemental Figure 1, A and B.
Statistics.
Two-tailed Student’s t test was applied for calculating statistical probability in this study. Statistical significance was set as P < 0.05.
Supplementary Material
Acknowledgments
We are grateful to V. Komnenovic (IMP) and I. Kufferath (Medical University of Graz) for histological analyses; H. Denk (Medical University of Graz) for expert advice in human HCC; H. Zöbel (ORIDIS Biomed) for production of tissue microarrays; M. Richter (IMP) for lentivirus shRNA preparation methods; A. Schleiffer (IMP) for bioinformatic analysis of JNK sequences and structures; O. Sansom (Beatson Institute for Cancer Research) for immunohistochemical staining of p21; C. Bonny for D-JNKI1 peptide samples and discussion of the D-JNKI1 experiments; and M. Eilers (Institute of Molecular Biology and Tumor Research, Marburg) for c-Myc plasmids. We thank R. Eferl, M. Eilers, L. Gresh, J. Guinea-Viniegra, P. Hasselblatt, A. Nebreda, K. Sabapathy, M. Sibilia, and especially, L. Bakiri for critical reading of the manuscript. The IMP is funded by Boehringer Ingelheim. The biobank of the Institute of Pathology was supported by the Austrian Genome Program. L. Hui was supported by an EMBO long-term fellowship and a Marie Curie individual fellowship.
Footnotes
Nonstandard abbreviations used: CDK, cyclin-dependent kinase; DEN, diethylnitrosamine; GST, glutathione S-transferase; HCC, hepatocellular carcinoma; IRES, internal ribosome entry site; O6-Et-dG, O6-ethyldeoxyguanosine; O2-Et-dT, O2-ethyldeoxythymidine; p, phosphorylated; Pb, phenobarbital; qRT-PCR, quantitative RT-PCR.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:3943–3953 (2008). doi:10.1172/JCI37156
Ewa Stepniak’s present address is: Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
References
- 1.Farazi P.A., DePinho R.A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 2.Eferl R., et al. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112:181–192. doi: 10.1016/S0092-8674(03)00042-4. [DOI] [PubMed] [Google Scholar]
- 3.Hui L., et al. p38alpha suppresses normal and cancer cell proliferation by antagonizing the JNK-c-Jun pathway. Nat. Genet. 2007;39:741–749. doi: 10.1038/ng2033. [DOI] [PubMed] [Google Scholar]
- 4.Maeda S., Kamata H., Luo J.L., Leffert H., Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Sakurai T., Maeda S., Chang L., Karin M. Loss of hepatic NF-kappa B activity enhances chemical hepatocarcinogenesis through sustained c-Jun N-terminal kinase 1 activation. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10544–10551. doi: 10.1073/pnas.0603499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakurai T., et al. Hepatocyte necrosis induced by oxidative stress and IL-1 alpha release mediate carcinogen-induced compensatory proliferation and liver tumorigenesis. Cancer Cell. 2008;14:156–165. doi: 10.1016/j.ccr.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weston C.R., Davis R.J. The JNK signal transduction pathway. Curr. Opin. Cell Biol. 2007;19:142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Hochedlinger K., Wagner E.F., Sabapathy K. Differential effects of JNK1 and JNK2 on signal specific induction of apoptosis. Oncogene. 2002;21:2441–2445. doi: 10.1038/sj.onc.1205348. [DOI] [PubMed] [Google Scholar]
- 9.Wada T., et al. Antagonistic control of cell fates by JNK and p38-MAPK signaling. Cell Death Differ. 2008;15:89–93. doi: 10.1038/sj.cdd.4402222. [DOI] [PubMed] [Google Scholar]
- 10.Sabapathy K., et al. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol. Cell. 2004;15:713–725. doi: 10.1016/j.molcel.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 11.Jaeschke A., et al. JNK2 is a positive regulator of the cJun transcription factor. Mol. Cell. 2006;23:899–911. doi: 10.1016/j.molcel.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 12.Papa S., et al. Gadd45beta promotes hepatocyte survival during liver regeneration in mice by modulating JNK signaling. J. Clin. Invest. 2008;118:1911–1923. doi: 10.1172/JCI33913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwabe R.F., et al. c-Jun-N-terminal kinase drives cyclin D1 expression and proliferation during liver regeneration. Hepatology. 2003;37:824–832. doi: 10.1053/jhep.2003.50135. [DOI] [PubMed] [Google Scholar]
- 14.Bain J., McLauchlan H., Elliott M., Cohen P. The specificities of protein kinase inhibitors: an update. Biochem. J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.She Q.B., Chen N., Bode A.M., Flavell R.A., Dong Z. Deficiency of c-Jun-NH(2)-terminal kinase-1 in mice enhances skin tumor development by 12-O-tetradecanoylphorbol-13-acetate. Cancer Res. 2002;62:1343–1348. [PubMed] [Google Scholar]
- 16.Tong C., et al. c-Jun NH2-terminal kinase 1 plays a critical role in intestinal homeostasis and tumor suppression. Am. J. Pathol. 2007;171:297–303. doi: 10.2353/ajpath.2007.061036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasselblatt P., Gresh L., Kudo H., Guinea-Viniegra J., Wagner E.F.2008The role of the transcription factor AP-1 in colitis-associated and beta-catenin-dependent intestinal tumorigenesis in mice. Oncogene. 276102–6109. . doi: 10.1038/onc.2008.211 [DOI] [PubMed] [Google Scholar]
- 18.Shibata W., et al. c-Jun NH2-terminal kinase 1 is a critical regulator for the development of gastric cancer in mice. Cancer Res. 2008;68:5031–5039. doi: 10.1158/0008-5472.CAN-07-6332. [DOI] [PubMed] [Google Scholar]
- 19.Chen N., et al. Suppression of skin tumorigenesis in c-Jun NH(2)-terminal kinase-2-deficient mice. Cancer Res. 2001;61:3908–3912. [PubMed] [Google Scholar]
- 20.Fausto N., Campbell J.S., Riehle K.J. Liver regeneration. Hepatology. 2006;43:S45–53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 21.Lin W.W., Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J. Clin. Invest. 2007;117:1175–1183. doi: 10.1172/JCI31537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Bergami P., et al. Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell. 2007;11:447–460. doi: 10.1016/j.ccr.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vivanco I., et al. Identification of the JNK signaling pathway as a functional target of the tumor suppressor PTEN. Cancer Cell. 2007;11:555–569. doi: 10.1016/j.ccr.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 24.Behrens A., et al. Impaired postnatal hepatocyte proliferation and liver regeneration in mice lacking c-jun in the liver. EMBO J. 2002;21:1782–1790. doi: 10.1093/emboj/21.7.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuzaki K., et al. Chronic inflammation associated with hepatitis C virus infection perturbs hepatic transforming growth factor beta signaling, promoting cirrhosis and hepatocellular carcinoma. Hepatology. 2007;46:48–57. doi: 10.1002/hep.21672. [DOI] [PubMed] [Google Scholar]
- 26.Wu H., et al. Targeted in vivo expression of the cyclin-dependent kinase inhibitor p21 halts hepatocyte cell-cycle progression, postnatal liver development and regeneration. Genes Dev. 1996;10:245–260. doi: 10.1101/gad.10.3.245. [DOI] [PubMed] [Google Scholar]
- 27.Stepniak E., et al. c-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev. 2006;20:2306–2314. doi: 10.1101/gad.390506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.el-Deiry W.S., et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-P. [DOI] [PubMed] [Google Scholar]
- 29.Herold S., et al. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell. 2002;10:509–521. doi: 10.1016/S1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 30.Seoane J., Le H.V., Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 31.Oskarsson T., et al. Skin epidermis lacking the c-Myc gene is resistant to Ras-driven tumorigenesis but can reacquire sensitivity upon additional loss of the p21Cip1 gene. Genes Dev. 2006;20:2024–2029. doi: 10.1101/gad.381206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabapathy K., Wagner E.F. JNK2: a negative regulator of cellular proliferation. Cell Cycle. 2004;3:1520–1523. doi: 10.4161/cc.3.12.1315. [DOI] [PubMed] [Google Scholar]
- 33.Sanna M.G., Duckett C.S., Richter B.W., Thompson C.B., Ulevitch R.J. Selective activation of JNK1 is necessary for the anti-apoptotic activity of hILP. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6015–6020. doi: 10.1073/pnas.95.11.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.David J.P., Sabapathy K., Hoffmann O., Idarraga M.H., Wagner E.F. JNK1 modulates osteoclastogenesis through both c-Jun phosphorylation-dependent and -independent mechanisms. . J. Cell. Sci. 2002;115:4317–4325. doi: 10.1242/jcs.00082. [DOI] [PubMed] [Google Scholar]
- 35.Mizukami J., et al. Receptor activator of NF-kappaB ligand (RANKL) activates TAK1 mitogen-activated protein kinase kinase kinase through a signaling complex containing RANK, TAB2, and TRAF6. Mol. Cell. Biol. 2002;22:992–1000. doi: 10.1128/MCB.22.4.992-1000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasselblatt P., Rath M., Komnenovic V., Zatloukal K., Wagner E.F. Hepatocyte survival in acute hepatitis is due to c-Jun/AP-1-dependent expression of inducible nitric oxide synthase. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17105–17110. doi: 10.1073/pnas.0706272104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuntzen C., et al. Inhibition of c-Jun-N-terminal-kinase sensitizes tumor cells to CD95-induced apoptosis and induces G2/M cell cycle arrest. Cancer Res. 2005;65:6780–6788. doi: 10.1158/0008-5472.CAN-04-2618. [DOI] [PubMed] [Google Scholar]
- 38.Fan Y., et al. c-Jun NH2-terminal kinase decreases ubiquitination and promotes stabilization of p21(WAF1/CIP1) in K562 cell. Biochem. Biophys. Res. Commun. 2007;355:263–268. doi: 10.1016/j.bbrc.2007.01.146. [DOI] [PubMed] [Google Scholar]
- 39.Kim G.Y., et al. The stress-activated protein kinases p38 alpha and JNK1 stabilize p21(Cip1) by phosphorylation. J. Biol. Chem. 2002;277:29792–29802. doi: 10.1074/jbc.M201299200. [DOI] [PubMed] [Google Scholar]
- 40.Noguchi K., Kokubu A., Kitanaka C., Ichijo H., Kuchino Y. ASK1-signaling promotes c-Myc protein stability during apoptosis. Biochem. Biophys. Res. Commun. 2001;281:1313–1320. doi: 10.1006/bbrc.2001.4498. [DOI] [PubMed] [Google Scholar]
- 41.Bowman T., et al. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc. Natl. Acad. Sci. U. S. A. 2001;98:7319–7324. doi: 10.1073/pnas.131568898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchholz M., et al. Overexpression of c-myc in pancreatic cancer caused by ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway. EMBO J. 2006;25:3714–3724. doi: 10.1038/sj.emboj.7601246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kallunki T., et al. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 44.Cui J., et al. Identification of a specific domain responsible for JNK2alpha2 autophosphorylation. J. Biol. Chem. 2005;280:9913–9920. doi: 10.1074/jbc.M412165200. [DOI] [PubMed] [Google Scholar]
- 45.Engelman J.A., et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 46.Stommel J.M., et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 47.Abou-Alfa G.K., et al. Phase II study of sorafenib in patients with advanced hepatocellular carcinoma. J. Clin. Oncol. 2006;24:4293–4300. doi: 10.1200/JCO.2005.01.3441. [DOI] [PubMed] [Google Scholar]
- 48.Llovet J.M., et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 49.Borsello T., et al. A peptide inhibitor of c-Jun N-terminal kinase protects against excitotoxicity and cerebral ischemia. Nat. Med. 2003;9:1180–1186. doi: 10.1038/nm911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.