Abstract
Summary: MonkeySNP is a web-based resource created by the Genetic Resource and Informatics Program at the Oregon National Primate Research Center to facilitate access to non-human primate (NHP) single nucleotide polymorphisms (SNP) data. MonkeySNP is a mirror of the NCBI dbSNP database and contains additional NHP subpopulation genotype data and visual genotype displays to support SNP review and selection.
Availability: http://monkeysnp.ohsu.edu/snp/
Contact: dubayc@ohsu.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Efforts to identify and characterize SNPs in rhesus macaques (Macaca mulatta) have uncovered the potential value of these variants for use in research (Ferguson et al., 2007; Hernandez et al., 2007; Malhi et al., 2007). In addition, comparisons of SNPs in Indian-origin rhesus and Chinese-origin rhesus, the rhesus populations used in US biomedical research, identified them as genetically divergent. The level of heterogeneity observed in non-human primates make SNPs ideally suited for use in both breeding management (e.g. ancestry and lineage) and research (e.g. association studies) (Ferguson et al., 2007; Malhi et al., 2007).
A central public repository for SNPs exists at the National Center for Biotechnology Information: http://www.ncbi.nlm.nih.gov/SNP. dbSNP in its current build (129) contains over 16 million validated SNPs from 44 organisms. One of the organisms represented in dbSNP is the rhesus macaque, an important animal model widely used for basic and translational research due to its genetic and physiological similarity to humans (Gibbs et al., 2007). There are currently 789 SNPs reported for the rhesus macaques in dbSNP (at species-level build 128).
The dbSNP database schema provides high-level population and taxonomy identifiers, subpopulation and individual genotypes to be defined by the submitter; however none of the rhesus submissions to date in dbSNP contain genotypic data. Allele frequency data are available through dbSNP, though it can be difficult to extract for comparisons between subpopulations.
We have developed a web portal called MonkeySNP, to facilitate access to NHP dbSNP information. MonkeySNP data are organized by species, as well as by subpopulation, and are augmented with genotypes submitted by the SNP discoverer. Allele frequency data are listed by population, to inform SNP selection for use in studies or to facilitate identification of subpopulation-specific alleles. MonkeySNP calculates and displays general population frequencies of alleles, as well as subpopulation allele frequencies from genotypes and displays them in a manner that makes identification and comparison of SNPs intuitive.
dbSNP is an invaluable central repository that maintains general SNP information for the scientific community. However, it does not easily provide subpopulation or subpopulation reports, nor does it produce them in a visual or intuitive manner. In order to bridge this gap, monkeySNP was created based on SNP researchers’ requests.
2 FEATURES AND FUNCTIONALITIES
The dbSNP database schema forms the basis for the MonkeySNP schema. SNP data for the taxonomy identifier Macaca mulatta are downloaded from the NCBI dbSNP server whenever they release a new build. dbSNP-submitted genotype data that is not in the public build are added locally, if available, between build releases, so all data have dbSNP provenance. We have augmented the dbSNP schema with additional tables to facilitate calculation and display of allele frequencies for subpopulations.
The monkeySNP website home page consists of a Search, Summaries, Registration and Contact Us tabs. The Search tab brings the user to a search page where the user can search for a SNP by GenBank gene name, chromosome or dbSNP ID (numeric portion of dbSNP ‘rs’ number). The Summaries tab brings the user to a summary page of all SNPs within the database. A user can sign up for a newsletter giving updates on changes to our website via the Registration tab. The Contact Us tab allows the user to send us any comments or suggestions.
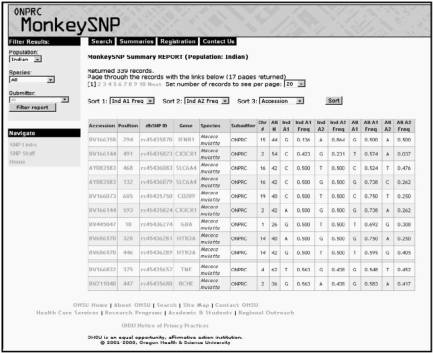
The SNP information page lists the SNPs with GenBank Accession Number, Position, dbSNP ID, Gene, Species, Submitter, chromosome number, N (number of chromosomes genotyped), genotypes and frequencies (Fig. 1). Results can be sorted on up to three categories. Furthermore, the results can be filtered by population, species and by submitter.
Fig. 1.
Results page after clicking on Summaries tab with filter set for Indian population. List was sorted on Indian frequency allele 1, Indian frequency allele 2 and accession number.
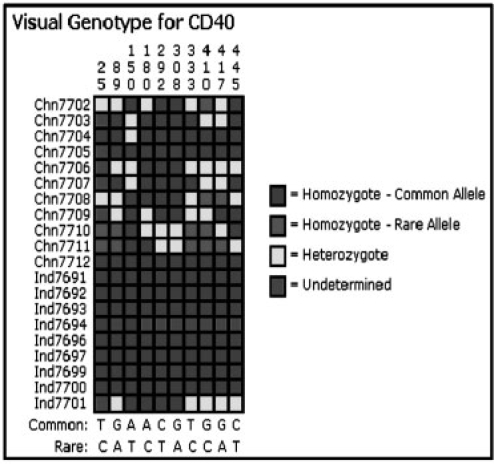
The results table also contains hyperlinks to more information on the SNP. The ‘Accession Number’ is linked to the GenBank page for the SNP reference. The ‘Position’ links to SNP details which include subpopulation, total number of chromosomes observed (N), allele frequencies in the subpopulation and flanking DNA sequence 5′ to 3′. The ‘dbSNP ID’ hyperlinks to the associated dbSNP webpage. The ‘Gene’ links to a visual genotype (VG) diagram (Fig. 2). The VG displays individual genotypes, positions, as well as the common and rare alleles for all SNPs in a gene. The alleles are color coded according to homozygosity or heterozygosity. At a glance, population-specific information can be gleaned from the color coding.
Fig. 2.
VG diagram of CD40 gene. Differences in genotype frequency become apparent between subpopulations.
In summary, the goal of this website is to allow the user to easily identify SNPs that are subpopulation specific through frequency information displayed numerically and visually.
This resource will continue to expand and change according to the scientific community's needs and feedback. We are currently working on enhancements to this website. One in-process enhancement is to make all results downloadable as a Microsoft Excel spreadsheet. We are also developing tools to aid researchers in making submissions of subpopulation and genotype data directly to dbSNP easier. Comments on the site are welcome.
Funding
National Institutes of Health/National Center for Research Resources (RR00163) for the operation of the Oregon National Primate Research Center.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Ferguson B, et al. Single nucleotide polymorphisms (SNPs) distinguish Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta) BMC Genomics. 2007;8:43. doi: 10.1186/1471-2164-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Hernandez RD, et al. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science. 2007;316:240–243. doi: 10.1126/science.1140462. [DOI] [PubMed] [Google Scholar]
- Malhi RS, et al. MamuSNP: a resource for rhesus macaque (Macaca mulatta) genomics. PLoS. 2007;5:e438. doi: 10.1371/journal.pone.0000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DG. Genetic characterization of Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta) Comp. Med. 2005;55:227–230. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.