Abstract
Motivation: Microarray expression data reveal functionally associated proteins. However, most proteins that are associated are not actually in direct physical contact. Predicting physical interactions directly from microarrays is both a challenging and important task that we addressed by developing a novel machine learning method optimized for this task.
Results: We validated our support vector machine-based method on several independent datasets. At the same levels of accuracy, our method recovered more experimentally observed physical interactions than a conventional correlation-based approach. Pairs predicted by our method to very likely interact were close in the overall network of interaction, suggesting our method as an aid for functional annotation. We applied the method to predict interactions in yeast (Saccharomyces cerevisiae). A Gene Ontology function annotation analysis and literature search revealed several probable and novel predictions worthy of future experimental validation. We therefore hope our new method will improve the annotation of interactions as one component of multi-source integrated systems.
Contact: ts2186@columbia.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
1.1 Protein interactions are crucial to medical biology
Networks of protein–protein interactions provide a framework for the understanding of biological processes and can give insights into the mechanisms of diseases. Interaction networks can assist in designing drugs that modulate specific disease pathways (Ofran et al., 2005; Ryan and Matthews, 2005). The identification of protein–protein interactions is, therefore, of primary importance.
Recent years have seen great advancements in experimental techniques, such as yeast two-hybrid (Y2H) and coimmunoprecipitation (CoIP) that probe protein interactions in a high-throughput fashion (Gavin et al., 2006; Giot et al., 2003; Ho et al., 2002; Ito et al., 2001; Uetz and Pankratz, 2004; Uetz et al., 2000). Y2H focuses on physical interaction between two proteins, while CoIP detects groups of proteins that are part of the same permanent or temporary complex. Most interactions are deposited in databases, such as IntAct (Kerrien et al., 2007), DIP (Salwinski et al., 2004), BIND (Bader et al., 2003) and MIPS (Guldener et al., 2006). In this study, we focus on physical protein–protein interactions.
1.2 Physical interaction versus association
The term ‘protein interaction’ has different meanings. We consider two proteins to interact physically if and only if some of their residues are in contact at some point in time. Assume, protein A activates B at time T1, separates from B at T2 and B regulates C at T3. A and C do not interact by our definition; instead, they are associated. Even for T1 = T2 and the three proteins form a somehow stable complex, by our definition A and C would still not physically interact.
1.3 Expression correlation poorly predicts physical interactions
The Gene Expression Omnibus (GEO) database (Barrett et al., 2005) at the National Center for Biotechnology Information (NCBI) holds >200 000 microarray experiments (February 2008), and this is only one resource (Parkinson et al., 2005; Sherlock et al., 2001). Microarray data has been widely used in elucidating biological mechanisms, specifically in discovering functional modules, pathways (Bar-Joseph et al., 2003; Segal et al., 2003a) and reverse engineering regulatory networks (Hartemink, 2005; Margolin et al., 2006; Segal et al., 2003c).
Microarrays provide noisy measures for the states of a complex biological system. Various types of systematic and stochastic fluctuations contribute to noise during biological sample preparation, hybridization, expression measurement and image processing (Schuchhardt et al., 2000). Another level of noise originates from the fact that each microarray experiment measures a single value for a gene that reflects its activity averaged across many biological processes. This mixing of underlying signals renders the inference of interactions particularly challenging. One approach to filtering systematic noise is the projection technique, which includes methods such as principal component analysis (PCA) and independent component analysis (ICA). They transform high-dimensional input data into lower dimensional components that capture the most important variations in the original data (Alter et al., 2000; Lee and Batzoglou, 2003; Liebermeister, 2002).
Since interacting proteins need to be present at the same time and place to physically contact each other, their expression as measured at the mRNA level by microarrays does not predict protein–protein interactions very well. In fact, many associated proteins showed levels of correlations almost indistinguishable from non-associated ones (Jansen et al., 2002). Associations through permanent protein complexes such as the ribosome and the proteosome are exceptions to this (Jansen et al., 2002).
Despite this limitation, microarray data has been widely combined with other evidence such as sequence homology, function annotations and sequence motifs to predict protein–protein interactions (Jansen et al., 2003; Rhodes et al., 2005). Those attempts did not distinguish between associations and physical interactions, and they all relied on correlations in the microarray data.
Here, we hypothesized that we could squeeze physical interactions out of microarray data. We introduced a novel method that effectively improved the direct inference of physical protein–protein interactions from microarrays, with the ultimate goal of providing a better plug-in for integrated systems (Ben-Hur and Noble, 2005; Jansen et al., 2003; Rhodes et al., 2005). We collected many yeast microarray experiments from GEO and extracted principal components by PCA. Using trusted interaction data from DIP, we applied support vector machines (SVMs) (Vapnik, 1998) to effectively learn from our supervised training data (DIP) which types of correlations reveal physical interactions and which do not. Our method predicted physical protein–protein interactions better than the conventional correlation method, and it discovered meaningful new physical interactions.
2 MATERIALS AND METHODS
2.1 Microarray data
We used microarray data as a proxy for protein expression and downloaded 349 yeast microarray experiments (Affymetrix S98 chipset, GPL 90 GEO platform) from GEO. Expression values were log2 transformed and quantile-normalized to render measurements from different sources and conditions more comparable. Missing expression values were filled in using k-nearest-neighbor imputation (Troyanskaya et al., 2001). Affymetrix probe identifiers were converted to SWISS-PROT identifiers (Boeckmann et al., 2003); data without corresponding identifiers were discarded. When multiple probes corresponded to the same SWISS-PROT identifier, we averaged over all probe intensities. The 349 experiments covered a total of 5823 unique proteins.
2.2 Protein–protein interaction data
We downloaded the core yeast dataset from DIP (Deane et al., 2002; Salwinski et al., 2004) as our set of trusted interaction network. The set/network consisted of 5299 interactions between 2312 proteins. DIP considers these interactions to be of high quality; they mostly originated from Y2H or detailed experiments. These interactions constituted the body of all positives. Since current databases do not document negatives, we generated 5299 non-interactions by randomly pairing the 2312 proteins and excluding those known to interact (i.e. annotated in DIP). Our solution provides a more conservative estimate of accuracy than common approaches that pair proteins from different compartments (Ben-Hur and Noble, 2006; Jansen and Gerstein, 2004; Jansen et al., 2003).
2.3 Noise removal and feature extraction: expression modes
PCA and ICA are statistical techniques for revealing hidden factors that underlie sets of random variables, measurements or signals. It has been demonstrated that by processing microarray data through PCA or ICA, proteins with extremely high or low activity in a principal component are usually involved in related biological processes (Lee and Batzoglou, 2003). Mathematically, the transformation of microarray data into principal components is:
| (1) |
where X is a 349×5823 matrix containing the original microarray expression values, P is a 349×349 matrix discovered by PCA or ICA representing the important directions of variation in the microarray data and Y is a 349×5823 matrix of principal components containing the relative protein activity along these directions. The rows of Y are by convention sorted by their importance (i.e. corresponding eigenvalues). We refer to each row of Y as an expression mode and use the top n to represent proteins. We applied PCA to our microarray dataset without using any knowledge of protein function. As expected (Lee and Batzoglou, 2003), we found proteins with highly activated or repressed activity in an expression mode to usually have coherent biological roles (Supplementary Material). In our context, PCA slightly outperformed ICA (T.T. Soong and B. Rost, unpublished data). For simplicity, we only present PCA results here.
2.4 Input features
We used the expression modes to represent individual proteins: each protein i is a vector mi of n real values taken from the top n expression modes as obtained via PCA (i.e. Y1:n,i). We then applied an idea from the prediction of intra-chain residue contacts (Punta and Rost, 2005): a pair of proteins A and B was represented by concatenating the expression modes mA and mB. We also included the Pearson correlation rAB to reflect the information captured by the single ‘expression component’ used conventionally when inferring interactions from microarrays (Jansen et al., 2003; Rhodes et al., 2005). The input features FAB for a protein pair A–B thus became:
| (2) |
where ⊕ is the concatenation operator. To maintain symmetry (A–B identical to B–A) we trained on both FAB and FBA. To infer unknown interactions, we averaged the scores of A–B and B–A.
2.5 Using machine learning to improve prediction
The naïve Bayes algorithm in Jansen et al. (2003) and Rhodes et al. (2005) integrates many types of evidence such as microarray expression and function annotation. Given n types of evidence E1,…,En, whether two proteins interact (posterior odds) depends on how each evidence Ei supports the interaction (likelihood ratioi), and our knowledge of how often proteins interact by chance (prior odds):
 |
(3) |
where each evidence Ei multiplicatively contributes likelihood ratioi to posterior odds. When we use microarray data as evidence E1, the corresponding likelihood ratio1 becomes:
| (4) |
where r is the Pearson correlation between two proteins’ microarray expression. Improving this microarray component could thus directly add to the performance of the integrative system.
Here, we used the SVM to improve this microarray component. The SVM is a machine learning method based on statistical learning theory. It projects the input data into a higher dimensional space and finds a hyperplane that best separates the data. The SVM maximizes the shortest distance from the data points to the hyperplane to minimize generalization error. SVMs have been used extensively in computational biology (Liu et al., 2006; Melvin et al., 2007; Nair and Rost, 2005). We used the LIBSVM (Chang and Lin, 2001) with Gaussian RBF and default parameters. An SVM score is reported for every protein pair [Equation (S2), Supplementary Material].
We implemented the correlation-based module [Equation (4)] as the baseline for comparison. For simplicity, we refer to it as the ‘Bayesian model’ and the likelihood ratio as the ‘Bayes score’. We used Gaussian kernel density estimation to calculate the likelihoods for continuous levels of r. Note that the prior odds do not affect the Bayes score calculation [Equation (4)] and the classifier comparison.
2.6 Cross-validation
We performed standard 10-fold cross-validation experiments where the positives and negatives were randomly split into 10 subsets of equal size; nine subsets were used for training, and one for testing. We cycled through the sets such that each example was used for testing exactly once. For the SVM, to account for noise in the data, we used the default parameters (e.g. cost and class weights) without further optimizing them by a grid search approach on the training data (i.e. cross-training). All reported levels of performance are valid for the test sets and reflect the expected performance for protein pairs never encountered before.
2.7 Performance measures
We assessed performance through the receiver operating characteristic (ROC) curve and the area under the ROC curve (AUC). The true positive rate (TPR) and the false positive rate (FPR) were compiled as follows:
| (5) |
where TP is the number of correctly inferred interactions, P the total number of all observed interactions, FP the number of incorrectly inferred interactions and N the total number of all non-interactions. For each classifier, we tried different threshold values above which protein pairs were classified to interact, thereby yielding a complete ROC curve. The calculation was performed using the ROCR program (Sing et al., 2005). Results were reported over all protein pairs in all 10 cross-validation test sets.
3 RESULTS
3.1 SVM gets physical protein–protein interactions from microarrays
We trained our SVMs with different numbers of expression modes and evaluated the performance using 10-fold cross-validation. We compared the SVMs to the conventional correlation method, which we had implemented as a Bayesian model [Equation (4)] and evaluated using the same data and cross-validation procedure.
The Bayesian model alone inferred physical interactions from microarrays slightly better than random (Fig. 1A, green line versus diagonal). The SVM with only 20 expression modes (blue) already improved significantly over the Bayesian model (green) using all 349 microarrays. Increasing the number of expression modes used as SVM input improved performance until saturation at ∼150 expression modes (Table 1). The improvement originated from two sources: the SVM and the expression mode extraction. A small number of expression modes improved the performance over using all microarray data (e.g. SVM20>SVMALLMA), and the performance continued to increase when we incorporated more expression modes (e.g. SVM150>SVM50>SVM20, see Fig. 1, Table 1).
Fig. 1.
ROC curves for inferring physical interactions. (A) The green line marks the baseline Bayesian classifier trained on all 349 microarrays. The other lines represent the SVMs, e.g. SVM20 used 20 expression modes (blue); SVMALLMA (red) used all 349 original microarrays as input. In addition to the expression modes or original microarray data, all SVMs used the correlation information (see Section 2). Optimal predictions are close to the top left, random predictions close to the diagonal (dotted gray line). (B) A close-up of the ROC curves for the most confident predictions (FPR <0.01, i.e. ∼50 false positives). For clarity, we only show the curves for SVM150, SVMALLMA and the Bayesian model. Our best SVM model SVM150 consistently outperforms SVMALLMA and Bayes at all confidence levels.
Table 1.
AUC for inferring interactionsa
| Classifier | AUC (all) | AUC (FPR <0.1) | AUC (FPR <0.01) |
|---|---|---|---|
| SVM20 | 0.748 | 0.241 | 0.052 |
| SVM50 | 0.765 | 0.277 | 0.063 |
| SVM100 | 0.768 | 0.290 | 0.067 |
| SVM150 | 0.766 | 0.289 | 0.079 |
| SVM200 | 0.766 | 0.286 | 0.076 |
| SVM250 | 0.758 | 0.278 | 0.074 |
| SVMALLMA | 0.719 | 0.220 | 0.047 |
| Bayesian model | 0.630 | 0.157 | 0.039 |
aComparison of performance through AUC based on 10-fold cross-validation: AUC (all): full ROC curve, AUC (FPR <0.1): area for high accuracy (FDR <0.1), AUC (FPR <0.01): area for highest accuracy (FDR <0.01).
High improvements in AUC may be meaningless if we failed to identify at least some interactions without mistakes. Closer inspection of the low error region revealed that the SVM method clearly recovered more true interactions in this realm than the Bayesian model (Fig. 1B, FPR <0.01, i.e. ∼50 false positives).
3.2 SVM scores partially reflected network distance
We hypothesized that the SVM might have implicitly learned important information not explicitly used for training, in particular, that the SVM score might reflect biological relations such as network distance. We defined the network distance between two proteins as the number of interactions needed for one protein to pass information to the other (e.g. A binds B, B binds C; A–C have a distance of 2). If our assumption is correct, scores will be highest for physically interacting proteins, lower for pairs associated through one intermediate and much lower for pairs far apart in the network.
To examine the relationship between microarray-derived scores and network distances, we trained the SVM and Bayesian classifiers with our trusted interactions (see Section 2) and inferred two sets of interaction scores for all remaining pairs in the DIP network. The first set is from our final SVM (using 150 expression modes); the other is from the Bayesian model. Technically, this provided two sets of scores for all remaining 2 660 918 protein pairs. We calculated the network distances in DIP and compared them to our interaction scores (Fig. 2A and B).
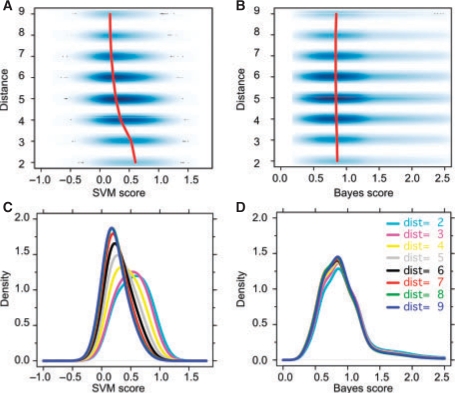
Fig. 2.
The SVM captures network distance as shown by the relation between interaction score and network distance for the SVM (A+C) and for the Bayesian model (B+D). For clarity, we divided all protein pairs into eight DIP-distance groups (dist = 2, …, 9, see Section 2); hue (A+B) is proportional to data density; red lines trace the peaks of the score distributions. (C) SVM: the scores of closely associated proteins (e.g. dist = 2, cyan) are considerably higher than those of distantly associated proteins (e.g. dist = 9, blue). On average the SVM score is indicative of the network distance. (D) Bayesian model: the scores of closely associated proteins (e.g. dist = 2, cyan) mostly overlapped with the scores of proteins of all other distances.
The distances d between proteins in the DIP network ranged between 2 and 13, with an average of 5.2. We further plotted the score distributions with respect to distance for visual clarity (Fig. 2C and D). SVM scores and network distance were somehow correlated, i.e. the higher the score, the closer the proteins in the network and vice versa (e.g. cyan, d = 2 versus blue, d = 9). The scores for the Bayesian model on the other hand overlapped almost completely, although there were slightly more low scores for distant protein pairs (cyan lower than blue). The relationship between distance and score was much stronger (P ≪ 0.05) for the SVM (Spearman r =−0.29) than for the Bayesian model (Spearman r = −0.04) as verified using (Cohen et al., 2003).
3.3 Performance on independent datasets
In addition to comparing the performance by cross-validation (Fig. 1, Table 1), we also evaluated our methods on two independent datasets: (i) 29 133 interactions from IntAct, and (ii) 68 755 interactions from known protein complexes in MIPS. A better method should assign higher interaction scores to important interactions. Using the SVM and Bayesian classifiers trained on the previous datasets (see Section 2), we now scored the interactions in IntAct and MIPS. Since the SVM and the Bayesian scores differed in their absolute scale, we converted raw interaction scores into estimated confidence levels (accuracies). In contrast to calculations of the AUC [Equation (5)], the estimation of accuracy (TF/TP+FP) depends on the relative numbers of interactions and non-interactions. This opens up the question of how many interactions exist in yeast: if the numbers of interactions and non-interactions were similar (positives:negatives ≈1:1), a random predictor would achieve ∼50% accuracy. We do not know the true numbers, but it has been suggested that most proteins do not interact with each other (Bader and Hogue, 2002; Kumar and Snyder, 2002). As some publications nevertheless use the 1:1 ratio to evaluate accuracy, we estimated the interaction score versus accuracy relation in two extreme scenarios: 1:1 and 1:284 (Fig. S1 and Fig. S2, Supplementary Material). We ranked the interactions in each database by SVM or Bayesian score and looked at the minimum confidence (accuracy) corresponding to the strongest n retrieved interactions (Fig. 3).
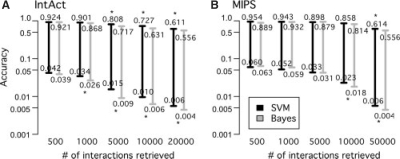
Fig. 3.
Performance on independent datasets. We tested the accuracy-coverage (or precision-recall) performance on unseen interactions in two independent datasets: (A) IntAct and (B) MIPS. The SVM (black) in general outperformed the Bayesian model (gray), classifying real IntAct interactions as more likely to occur. The SVM and the Bayesian model mostly performed equally well for the MIPS interactions of protein complexes. The error bars indicate assigned accuracies estimated using different positive:negative ratios (top = 1:1, bottom = 1:284; Supplementary Material). Asterisks indicate statistical significance (P <0.05; t-test).
For the IntAct dataset, the interactions were mostly classified as more likely to occur by the SVM (Fig. 3A). For the MIPS dataset, the strongest 5000 pairs were rated similarly by the SVM and the Bayesian model (P >0.05; Fig. 2B). This result might be due to the large variation in high-accuracy score estimates (Figures S1 and S2, Supplementary Material). More likely, however, this result confirmed our hypothesis that the SVM-based method improves for transient physical interactions, while the correlation-based method already captures very stable complexes that are over-represented in the MIPS dataset.
The experiments on independent datasets also demonstrated the challenge of identifying the drops (new interactions) in the ocean (non-interactions): despite the improved performance (50-fold increase over random), the SVM still was bound to <20% accuracy on a genomic scale (Fig. S2, Supplementary Material).
3.4 SVM explored different aspects of protein interaction
For all 2312 proteins in the core DIP network, we used the SVM (trained on all previous trusted data; Section 2) to identify interactions not annotated in DIP. We compared our predictions to BioGRID (Breitkreutz et al., 2008). BioGRID contains high-throughput as well as literature-derived data and comprehensively catalogs several aspects of protein interaction and association (e.g. affinity capture, two-hybrid and synthetic lethality). The SVM shows more confirmed predictions than the Bayesian method in most of these categories (Fig. S4, Supplementay Material). Furthermore, when summing over all categories that are more likely to capture physical interactions than associations (Supplementary Material), the SVM outperformed the Bayesian model (Fig. 4A). The Bayesian method had many more predictions confirmed by Affinity Capture-MS, a method that detects whether proteins belong to the same protein complex where most data come from high-throughput experiments (Fig. 4B). This might be explained by the observation that proteins in stable protein complexes have highly correlated microarray expression (Jansen et al., 2002). However, the SVM fared equally well by small-scale Affinity Capture-Western experiments (Fig. S4C, Supplementary Material) that also detect protein complexes. Thus, the discrepancy could also be due to the technical differences between high-throughput and small-scale affinity capture experiments. Overall, the SVM has significantly more predictions confirmed by BioGRID than expected by chance (Fig. S4, Supplementary Material).
Fig. 4.
Predicted interactions confirmed by BioGRID. We show the numbers of confirmed SVM (black) and Bayesian (gray) predictions. (A) The SVM predicted more interactions in all categories that tend to capture physical interactions rather than associations. (B) The Bayesian method on the other hand predicted more associations through stable protein complexes as discovered by high-throughput affinity capture experiments.
3.5 Prediction annotations suggested potential interactions
In yet another validation, we carefully inspected the Gene Ontology (GO) (Ashburner et al., 2000) annotations of the most confidently predicted protein pairs. Interacting proteins often perform similar biological roles (Jansen et al., 2003; Rhodes et al., 2005). Since our methods did not use any information about protein function, similar annotations between a predicted protein pair would indicate their interaction as biologically plausible.
We quantified the similarity between GO annotations according to a previous suggestion (Lord et al., 2003). We identified a minimum GO score (5.6) above which two proteins are most likely to interact (Table S2, Supplementary Material). The GO scores suggested many of the top SVM predictions to be biologically plausible. For example, 82 of the top 1000 predictions had GO scores >5.6, while only 15.8±4.5 high scoring pairs were expected among an equal number of non-interacting proteins. The GO scores of our top 1000 predictions were also significantly higher than those of 1000 random pairs (P ≪ 0.05, Mann–Whitney test).
Predictions with high GO scores include: elo3_yeast (Sur4p, YLR372W) and elo2_yeast (Fen1p, YCR034W) with a GO score of 8.95. These two proteins are required in the formation of long-chain fatty acids as identified through synthetic lethal experiments (Oh et al., 1997). The two transmembrane proteins catalyze specific products in the condensation of long-chain fatty acids (Dickson et al., 2006). An interaction prediction would suggest a tandem reaction process or a possible interaction within lipid micro-domains or rafts, a type of unexpected prediction that can minimize the experimental limitations of identifying interactions among transmembrane proteins. A GO score of 6.5 is attributed to the predicted pair of pob3_yeast (YMLO6W) and ctk3_yeast (YML11W), two proteins involved in chromatin modulated transcription functions, suggesting a possible role in regulation of FACT via the Ctk kinase complex (Singer and Johnston, 2004; Wood et al., 2007). Two ER-Golgi retrograde transport proteins copb2_yeast (Sec27p, YGL137W) and gcs1_yeast (YDL226C) have a GO score of 7.1 and have been implicated through E-MAP experiments (Schuldiner et al., 2005). As one of the proteins of the COP1 coatomer involved in retrograde transport of proteins from the Golgi to the ER, Sec27p is known to bind the di-lysine motif critical to this function. The Gcs1p protein contains the di-lysine motif and also acts as a mediator in the secretory pathway thereby suggesting a plausible interaction between the two proteins.
In addition to GO annotations, we manually searched the literature for some of the strong predictions and discovered several interesting cases worthy of further investigation. For instance, we predicted an interaction between the mRNA binding proteins mex67_yeast (YPL169C) and pub1_yeast (YNL016W). The two proteins share a common interaction partner, Npl3p (YDR432W) (Deka et al., 2008). Npl3p interacts with Mex67p in vitro and is associated in vivo with Mex67p-mRNA (Gilbert and Guthrie, 2004). The Pub1p and Npl3p interaction was observed in a large-scale TAP-MS study of the yeast proteome (Gavin et al., 2006). Pub1p resides in both the nucleus and cytoplasm and is involved in the regulation of mRNA decay and other post-transcriptional processes (Duttagupta et al., 2005). Mex67p is involved in exporting RNA out of the cell through the nuclear pore complex and has been partnered with various accessory proteins within mRNPs (Stewart, 2007). Homology transfer (Mika and Rost, 2006) did not reveal this pair; the prediction that Mex67p and Pub1p interact is therefore novel and awaits experimental verification.
Other interesting predictions include the interaction between ypt1_yeast (YFL038C) and vac8_yeast (YEL013W). Vac8p, a vacuole membrane protein involved in nucleus–vacuole junction formation (Kvam and Goldfarb, 2006), may also be involved with the Golgi-targeting GTPase Ypt1p in Golgi-vesicle targeting (Matern et al., 2000). We also predict the ER to Golgi transport p24 membrane protein (erv25_yeast, YML012W) (Belden and Barlowe, 2001) having a possible interaction with ypt1_yeast, implicating the Erv25p cytoplasmic tail.
We further explored interaction predictions in the yeast cell-cycle pathway. We compared our predictions to known interactions from BioGRID for all known yeast cell-cycle proteins (Wrzeszczynski and Rost, 2004) and also separately to those found in the current KEGG database release 45.0 (Kanehisa et al., 2008). In our top 1000 predictions, we found 213 new interactions for 15 KEGG cell-cycle proteins and 176 new interactions for 20 proteins from our cell-cycle dataset. The predicted interactions as well as their GO annotations and scores are available online at http://rostlab.org/svmppi.
4 DISCUSSION AND CONCLUSIONS
4.1 Better inference of physical interactions
We demonstrated that proper preprocessing and machine learning improve the inference of direct physical protein–protein interactions from microarrays. Our method began by removing systematic noise using PCA, thereby implicitly reconstructing the underlying biological processes (expression modes) that reflect protein activity more distinctly than the original expression data. The SVM employed the expression modes and outperformed the conventional Bayesian correlation method in predicting interactions; this was true both for our original 10-fold cross-validation experiment and all subsequent independent datasets (Figures 1, 3 and 4A, Table 1). Our method found several interesting predictions of biological significance.
4.2 SVM provides new measure for protein function annotation
Besides being more accurate in predicting interactions, the SVM model also provided a good measure of microarray coexpression and reflected the relative distance between proteins in the interaction network (Fig. 2). The SVM's ability to implicitly capture network distances may constitute an important improvement over the Bayesian model: in reconstructing the network of all interactions, the cost of mistaking distantly associated proteins for interacting ones is much higher than mistaking closely associated proteins for interacting ones. In a system of communicating entities, information degrades when transmitted from the source to the receiver through intermediates (Shannon, 1948). In the interaction network, the mutual information between directly interacting proteins is therefore higher than between proteins that communicate through intermediates. Although the SVM is not explicitly taught to learn network distances, the information embedded in the network effectively allows such a relationship to be learned.
New methods for functional annotation increasingly use global information from the interaction network. These methods annotate a protein based on the functions of its immediate interaction partners or corresponding modules (Bader and Hogue, 2003; Letovsky and Kasif, 2003; Rost et al., 2003; Schwikowski et al., 2000; Segal et al., 2003b; Sharan et al., 2007). Since the SVM model can easily avoid falsely connecting functionally dissimilar proteins, the resultant interactions are expected to be more functionally coherent and can further improve protein annotation.
4.3 Limitations and extensions
One limitation of our approach is in data quality. Interactions used for training and microarrays used for input need to be clean. Current high-throughput technologies remain error prone and may be far from complete. Improvements in experimental data will improve our approach.
Microarrays measure mRNA levels rather than protein abundance in the cell. Microarray expression is correlated with protein abundance (Ghaemmaghami et al., 2003), but not enough to predict protein abundance from mRNA levels. Since the expression of interacting proteins has been shown to co-evolve in multiple organisms (Bhardwaj and Lu, 2005; Fraser et al., 2004), an approach based on co-evolution might augment our predictions.
A formidable challenge to our method as well as to any interaction prediction method is the ocean of false positives: of all the ∼18 million possible protein pairs in yeast a tiny fraction interact in vivo. Even tiny false positive rates yield huge numbers of false positives when trying to predict the entire interactome. Although we have demonstrated an improvement over the conventional correlation method and shown many biologically plausible predictions, the large number of non-interacting pairs still prevents us from making predictions without giving false positives.
4.4 Future work
As demonstrated by the examples that we looked at carefully, predictions with similar annotations of function are likely to be true interactions. Besides microarrays, there are many other data sources that provide information about protein interactions (Liu and Rost, 2004; Lu et al., 2002; Pavlidis et al., 2002; Pazos and Valencia, 2001; Pellegrini et al., 1999; Rzhetsky et al., 2004; Sprinzak and Margalit, 2001). Here, we have achieved the goal of improving the prediction of physical interactions based only on microarray data, now it is time to benefit from this improvement by integrating other sources. With such an integrated system, protein interactions could be combined with other levels of cellular networks (e.g. transcriptional regulatory and signaling networks) along with temporal and spatial data to shed light on the phenotypes and dynamic behavior of cells (de Lichtenberg et al., 2005; Han et al., 2004; Qi and Ge, 2006) and help understand disease pathways. A particular advantage of our new module is that it captures interactions between types of proteins that may not be contained in other experimental data.
Funding
National Institutes of Health (U54-GM074958-01 from the Protein Structure Initiative (PSI) of the NIGMS to T.T.S., K.W. and B.R., U54-TM072980); National Library of Medicine (R01-LM07329 to T.T.S., K.W. and B.R.).
Conflict of Interest: none declared.
Supplementary Material
ACKNOWLEDGEMENTS
Thanks to Yanay Ofran (Bar Eilan University), Rajesh Nair (Columbia), Avner Schlessinger (Columbia), Marco Punta (Columbia) and Jinfeng Liu (Genentech) for valuable discussions. Last but not least, thanks to all those who deposit their experimental data in public databases, and to those who maintain these databases.
REFERENCES
- Alter O, et al. Singular value decomposition for genome-wide expression data processing and modeling. Proc. Natl Acad. Sci.USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. Analyzing yeast protein-protein interaction data obtained from different sources. Nat. Biotechnol. 2002;20:991–997. doi: 10.1038/nbt1002-991. [DOI] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, et al. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Joseph Z, et al. Computational discovery of gene modules and regulatory networks. Nat. Biotechnol. 2003;21:1337–1342. doi: 10.1038/nbt890. [DOI] [PubMed] [Google Scholar]
- Barrett T, et al. NCBI GEO: mining millions of expression profiles-database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Barlowe C. Deletion of yeast p24 genes activates the unfolded protein response. Mol. Biol. Cell. 2001;12:957–969. doi: 10.1091/mbc.12.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Hur A, Noble WS. Kernel methods for predicting protein-protein interactions. Bioinformatics. 2005;21(Suppl. 1):i38–i46. doi: 10.1093/bioinformatics/bti1016. [DOI] [PubMed] [Google Scholar]
- Ben-Hur A, Noble WS. Choosing negative examples for the prediction of protein-protein interactions. BMC Bioinformatics. 2006;7(Suppl. 1):S2. doi: 10.1186/1471-2105-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj N, Lu H. Correlation between gene expression profiles and protein-protein interactions within and across genomes. Bioinformatics. 2005;21:2730–2738. doi: 10.1093/bioinformatics/bti398. [DOI] [PubMed] [Google Scholar]
- Boeckmann B, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz BJ, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-C, Lin C.-J. LIBSVM: a library for support vector machines. 2001. [Google Scholar]
- Cohen J, et al. Applied Multiple Regression/Correlation Analysis for the Behavioral Sciences. Mahwah, N.J: L. Erlbaum Associates; 2003. [Google Scholar]
- de Lichtenberg U, et al. Dynamic complex formation during the yeast cell cycle. Science. 2005;307:724–727. doi: 10.1126/science.1105103. [DOI] [PubMed] [Google Scholar]
- Deane CM, et al. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell. Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- Deka P, et al. Structure of the yeast SR protein Npl3 and interaction with mRNA 3′-end processing signals. J. Mol. Biol. 2008;375:136–150. doi: 10.1016/j.jmb.2007.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC, et al. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog. Lipid Res. 2006;45:447–465. doi: 10.1016/j.plipres.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Duttagupta R, et al. Global analysis of Pub1p targets reveals a coordinate control of gene expression through modulation of binding and stability. Mol. Cell. Biol. 2005;25:5499–5513. doi: 10.1128/MCB.25.13.5499-5513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, et al. Coevolution of gene expression among interacting proteins. Proc. Natl Acad. Sci. USA. 2004;101:9033–9038. doi: 10.1073/pnas.0402591101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol. Cell. 2004;13:201–212. doi: 10.1016/s1097-2765(04)00030-9. [DOI] [PubMed] [Google Scholar]
- Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Guldener U, et al. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JD, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- Hartemink AJ. Reverse engineering gene regulatory networks. Nat. Biotechnol. 2005;23:554–555. doi: 10.1038/nbt0505-554. [DOI] [PubMed] [Google Scholar]
- Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Gerstein M. Analyzing protein function on a genomic scale: the importance of gold-standard positives and negatives for network prediction. Curr. Opin. Microbiol. 2004;7:535–545. doi: 10.1016/j.mib.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Jansen R, et al. Relating whole-genome expression data with protein-protein interactions. Genome Res. 2002;12:37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, et al. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S, et al. IntAct-open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Snyder M. Protein complexes take the bait. Nature. 2002;415:123–124. doi: 10.1038/415123a. [DOI] [PubMed] [Google Scholar]
- Kvam E, Goldfarb DS. Nucleus-vacuole junctions in yeast: anatomy of a membrane contact site. Biochem. Soc. Trans. 2006;34:340–342. doi: 10.1042/BST0340340. [DOI] [PubMed] [Google Scholar]
- Lee SI, Batzoglou S. Application of independent component analysis to microarrays. Genome Biol. 2003;4:R76. doi: 10.1186/gb-2003-4-11-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letovsky S, Kasif S. Predicting protein function from protein/protein interaction data: a probabilistic approach. Bioinformatics. 2003;19(Suppl. 1):i197–i204. doi: 10.1093/bioinformatics/btg1026. [DOI] [PubMed] [Google Scholar]
- Liebermeister W. Linear modes of gene expression determined by independent component analysis. Bioinformatics. 2002;18:51–60. doi: 10.1093/bioinformatics/18.1.51. [DOI] [PubMed] [Google Scholar]
- Liu J, Rost B. CHOP proteins into structural domains. Proteins Struct. Funct. Bioinform. 2004;55:678–688. doi: 10.1002/prot.20095. [DOI] [PubMed] [Google Scholar]
- Liu J, et al. Distinguishing protein-coding from non-coding RNAs through support vector machines. PLoS Genetics. 2006;2:e29. doi: 10.1371/journal.pgen.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord PW, et al. Investigating semantic similarity measures across the Gene Ontology: the relationship between sequence and annotation. Bioinformatics. 2003;19:1275–1283. doi: 10.1093/bioinformatics/btg153. [DOI] [PubMed] [Google Scholar]
- Lu L, et al. MULTIPROSPECTOR: an algorithm for the prediction of protein-protein interactions by multimeric threading. Proteins. 2002;49:350–364. doi: 10.1002/prot.10222. [DOI] [PubMed] [Google Scholar]
- Margolin AA, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl. 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matern H, et al. A novel Golgi membrane protein is part of a GTPase-binding protein complex involved in vesicle targeting. EMBO J. 2000;19:4485–4492. doi: 10.1093/emboj/19.17.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melvin I, et al. SVM-Fold: a tool for discriminative multi-class protein fold and superfamily recognition. BMC Bioinformatics. 2007;8(Suppl. 4):S2. doi: 10.1186/1471-2105-8-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika S, Rost B. Protein-protein interactions more conserved within species than across species. PLoS Comput. Biol. 2006;2:e79. doi: 10.1371/journal.pcbi.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair R, Rost B. Mimicking cellular sorting improves prediction of subcellular localization. J. Mol. Biol. 2005;348:85–100. doi: 10.1016/j.jmb.2005.02.025. [DOI] [PubMed] [Google Scholar]
- Ofran Y, et al. Beyond annotation transfer by homology: novel protein-function prediction methods to assist drug discovery. Drug Discov. Today. 2005;10:1475–1482. doi: 10.1016/S1359-6446(05)03621-4. [DOI] [PubMed] [Google Scholar]
- Oh CS, et al. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- Parkinson H, et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2005;33:D553–D555. doi: 10.1093/nar/gki056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, et al. Learning gene functional classifications from multiple data types. J. Comput. Biol. 2002;9:401–411. doi: 10.1089/10665270252935539. [DOI] [PubMed] [Google Scholar]
- Pazos F, Valencia A. Similarity of phylogenetic trees as indicator of protein-protein interaction. Protein Eng. 2001;14:609–614. doi: 10.1093/protein/14.9.609. [DOI] [PubMed] [Google Scholar]
- Pellegrini M, et al. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Rost B. PROFcon: novel prediction of long-range contacts. Bioinformatics. 2005;21:2960–2968. doi: 10.1093/bioinformatics/bti454. [DOI] [PubMed] [Google Scholar]
- Qi Y, Ge H. Modularity and dynamics of cellular networks. PLoS Comput. Biol. 2006;2:e174. doi: 10.1371/journal.pcbi.0020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, et al. Probabilistic model of the human protein-protein interaction network. Nat. Biotechnol. 2005;23:951–959. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- Rost B, et al. Automatic prediction of protein function. Cell. Mol. Life Sci. 2003;60:2637–2650. doi: 10.1007/s00018-003-3114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DP, Matthews JM. Protein-protein interactions in human disease. Curr. Opin. Struct. Biol. 2005;15:441–446. doi: 10.1016/j.sbi.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Rzhetsky A, et al. GeneWays: a system for extracting, analyzing, visualizing, and integrating molecular pathway data. J. Biomed. Inform. 2004;37:43–53. doi: 10.1016/j.jbi.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Salwinski L, et al. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchhardt J, et al. Normalization strategies for cDNA microarrays. Nucleic Acids Research. 2000;28:e47. doi: 10.1093/nar/28.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Schwikowski B, et al. A network of protein-protein interactions in yeast. Nat. Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- Segal E, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 2003a;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- Segal E, et al. Discovering molecular pathways from protein interaction and gene expression data. Bioinformatics. 2003b;19(Suppl. 1):i264–i271. doi: 10.1093/bioinformatics/btg1037. [DOI] [PubMed] [Google Scholar]
- Segal E, et al. Genome-wide discovery of transcriptional modules from DNA sequence and gene expression. Bioinformatics. 2003c;19(Suppl. 1):i273–i282. doi: 10.1093/bioinformatics/btg1038. [DOI] [PubMed] [Google Scholar]
- Shannon CE. A mathematical theory of communication. Bell Sys. Tech. J. 1948;27:379–423. 623–656. [Google Scholar]
- Sharan R, et al. Network-based prediction of protein function. Mol. Syst. Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlock G, et al. The Stanford Microarray Database. Nucleic Acids Res. 2001;29:152–155. doi: 10.1093/nar/29.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T, et al. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- Singer RA, Johnston GC. The FACT chromatin modulator: genetic and structure/function relationships. Biochem. Cell Biol. 2004;82:419–427. doi: 10.1139/o04-050. [DOI] [PubMed] [Google Scholar]
- Sprinzak E, Margalit H. Correlated sequence-signatures as markers of protein-protein interaction. J. Mol. Biol. 2001;311:681–692. doi: 10.1006/jmbi.2001.4920. [DOI] [PubMed] [Google Scholar]
- Stewart M. Ratcheting mRNA out of the nucleus. Mol. Cell. 2007;25:327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Troyanskaya O, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Uetz P, Pankratz MJ. Protein interaction maps on the fly. Nat. Biotechnol. 2004;22:43–44. doi: 10.1038/nbt0104-43. [DOI] [PubMed] [Google Scholar]
- Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Vapnik VN. Statistical Learning Theory. New York: Wiley; 1998. [Google Scholar]
- Wood A, et al. Ctk complex-mediated regulation of histone methylation by COMPASS. Mol. Cell. Biol. 2007;27:709–720. doi: 10.1128/MCB.01627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzeszczynski KO, Rost B. Cataloging proteins in cell cycle control. Methods Mol. Biol. 2004;241:219–233. doi: 10.1385/1-59259-646-0:219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.