Abstract
To be diagnostically useful, structural MRI must reliably distinguish Alzheimer’s disease (AD) from normal aging in individual scans. Recent advances in statistical learning theory have led to the application of support vector machines to MRI for detection of a variety of disease states. The aims of this study were to assess how successfully support vector machines assigned individual diagnoses and to determine whether data-sets combined from multiple scanners and different centres could be used to obtain effective classification of scans. We used linear support vector machines to classify the grey matter segment of T1-weighted MR scans from pathologically proven AD patients and cognitively normal elderly individuals obtained from two centres with different scanning equipment. Because the clinical diagnosis of mild AD is difficult we also tested the ability of support vector machines to differentiate control scans from patients without post-mortem confirmation. Finally we sought to use these methods to differentiate scans between patients suffering from AD from those with frontotemporal lobar degeneration. Up to 96% of pathologically verified AD patients were correctly classified using whole brain images. Data from different centres were successfully combined achieving comparable results from the separate analyses. Importantly, data from one centre could be used to train a support vector machine to accurately differentiate AD and normal ageing scans obtained from another centre with different subjects and different scanner equipment. Patients with mild, clinically probable AD and age/sex matched controls were correctly separated in 89% of cases which is compatible with published diagnosis rates in the best clinical centres. This method correctly assigned 89% of patients with post-mortem confirmed diagnosis of either AD or frontotemporal lobar degeneration to their respective group. Our study leads to three conclusions: Firstly, support vector machines successfully separate patients with AD from healthy aging subjects. Secondly, they perform well in the differential diagnosis of two different forms of dementia. Thirdly, the method is robust and can be generalized across different centres. This suggests an important role for computer based diagnostic image analysis for clinical practice.
Keywords: Alzheimer’s, frontotemporal lobar degeneration, linear support vectors, classification
Introduction
Alzheimer’s disease (AD), the commonest cause of dementia, is a neurodegenerative disorder. Definitive diagnosis can only be made with histopathological confirmation of amyloid plaques and neurofibrillary tangles, usually at autopsy. Early detection of AD is seen as important because treatment may be most efficacious if introduced as early as possible. In practice, a diagnosis is largely based on clinical history and examination supported by neuropsychological evidence of the pattern of cognitive impairment (Blennow et al., 2006). However, the reality is that only about half of those with probable dementia are actually recognized in the primary care setting (Valcour et al., 2000; Solomon and Murphy, 2005). When time-consuming criteria such as those published by the National Institute of Neurological and Communicative Disorders and Stroke/Alzheimer’s disease and related Disorders Association (NINCDS-ADRDA) (McKhann et al., 1984) are used, the diagnostic accuracy is improved. Based on a review of 13 studies using neuropathological confirmation, a probable AD diagnosis using NINCDS-ADRDA or the Diagnostic and Statistical Manual, 3rd edition, revised (DSM-IIIR) (American Psychiatric Association, 1987) criteria has an average sensitivity of 81% (range 49-100%) and specificity of 70% (range 47-100%) when patients are followed to autopsy (Knopman et al., 2001).
Historically, brain imaging, e.g. MRI, has largely been used to rule out alternative causes of dementia. This approach is consistent with established diagnostic consensus criteria such as those published by the NINCDS-ADRDA (McKhann et al., 1984). More recently, there has been a realization that MRI may add positive predictive value to a diagnosis of Alzheimer’s disease (Fox and Schott, 2004). Several studies demonstrate that using MRI to evaluate atrophy of temporal lobe structures can contribute to diagnostic accuracy (Barnes et al., 2004; Wahlund et al., 2005), but these findings have yet to be applied to routine clinical radiological practice, let alone the general practice setting (Wahlund et al., 2005). Manual measurements of these structures on MR images (Jack et al., 1992) are time-consuming and do not capture the full pattern of atrophy. The few studies of temporal lobe structures which utilize ante-mortem clinical data and structural MR in subjects where there is histopathological verification of AD also show hippocampal volume to be a sensitive marker for pathological AD stage (Gosche et al., 2002; Jack et al., 2002; Csernansky et al., 2004). If clinical MR scans are to be useful in the diagnosis of dementia, non-expert dependent, automated methods are needed that perform equally well or better than those seen in clinical practice so far. Furthermore, a method that has the capacity to utilize information from the whole brain will be more likely to distinguish among the dementias than techniques that rely solely on small regions, e.g. the medial temporal lobe, since hippocampal atrophy is present in other forms of dementia as well as AD (Chan et al., 2001; Jack et al., 2002).
There has been recent interest in machine-learning techniques such as support vector machines (SVMs) to categorize individual structural or functional brain images by differentiation of images from two groups (e.g. male/female or patient/control) (Lao et al., 2004; Fan et al., 2005; Mourao-Miranda et al., 2005; Kawasaki et al., 2007). Machine learning based pattern recognition techniques are multivariate and take into account specific inter-regional dependencies characteristic of different distributed pathologies, using such information to help categorize scans (Lao et al., 2004; Fan et al., 2005). SVMs are trained using a specific algorithm on well-characterized data (e.g. AD or normality). New scans can be tested against trained sets and in turn categorized as members of a particular clinical group (e.g. AD). Such categorization methods potentially satisfy the requirements of a diagnostic tool. The feasibility of such an approach using MR scans has recently been shown by automatic gender-based classification (Lao et al., 2004) and by detection of a variety of diseases (Fan et al., 2005; Kawasaki et al., 2007). In AD, automatic image classification has previously been used in functional imaging and cortical thickness measurements (deFigueiredo et al., 1995; Herholz et al., 2002; Lerch et al., 2006) to differentiate scans from patients with dementia and controls. More recently, pattern recognition methods applied to structural MRI were reported for the separation of mild cognitive impairment (MCI) from cognitively normal individuals (Davatzikos et al., 2006; Teipel et al., 2007).
To date, the application of SVM to structural MR scans for the purpose of AD diagnosis has not been demonstrated using pathologically confirmed cases for training data, nor to differentiate different forms of dementia. We applied SVM classification to examine various sets of MR brain scans from AD patients and elderly normal persons. In the first set, AD patients were largely from a community-based setting and all AD diagnoses were confirmed with neuropathology. The second set consisted of neuropathologically confirmed AD patients and controls from a quaternary referral centre allowing us to test how well results can be generalized and data-sets combined from different centres and scanners. The third set consisted of probable AD patients limited to 80 years of age or younger with Mini Mental State Examination (MMSE) scores ≥20 and age/sex matched cognitively normal controls. Finally, a dataset of subjects with neuropathologically proven frontotemporal lobar degeneration (FTLD) having comparable MMSE scores with the first two groups were included to explore whether SVMs can further distinguish between AD and FTLD. FTLD is characterized by frontal and temporal lobe atrophy with corresponding cognitive and behavioural deficits. Nonetheless, FTLD is sometimes difficult to distinguish from AD clinically. Although pathologically heterogeneous, FTLD can be neuropathologically separated from AD (McKhann et al., 2001; Cairns et al., 2007).
Materials and Methods
Subjects
Group I consisted of 20 patients and 20, age and gender matched cognitively normal controls from a community and referral based sample in Rochester, Minnesota, USA (see Table 1 for details). Cases had an ante-mortem MRI and an autopsy as part of a long-term research program in which the AD was confirmed neuropathologically. Neuropathological diagnosis was made according to criteria formulated by a working group of the National Institute on Aging and the Reagan Institute of the Alzheimer’s Association (NIA-RIA, 1997). Subjects were excluded from analysis if their scans revealed gross structural abnormalities other than atrophy. Diagnostic assignation was based on the combined results of medical history, clinical examination, psychometry and neuropathology. Criteria for the diagnosis of normal cognition were as follows: (i) independently functioning community membership, with (ii) no active neurological or psychiatric disorder, (iii) no psychoactive medication, (iv) a normal neurological examination, (v) no ongoing medical problem and (vi) no associated treatment that might interfere with cognitive function (Jack et al., 2004). Enrolled controls had an MMSE score ≥27 and a delayed paragraph recall score (Wechsel, 1987) >10 for those with 16 or more years of education, >6 for those with 8-15 years of education, and >4 for those with 7 or fewer years of education.
Table 1. Demographic information on groups I, II, and IV with post-mortem confirmation of AD obtained at different centres.
| Group I |
Group II |
Group III |
Group IV |
|||||
|---|---|---|---|---|---|---|---|---|
| Group (n) | AD (20) | controls (20) | AD (14) | controls (14) | AD (33) | Controls (57) | AD (18) | FTLD (19) |
| Sex (F/M) | 11/9 | 10/10 | 5/9 | 5/9 | 10/23 | 16/41 | 6/12 | 8/11 |
| Age at MRI-scan (mean, range) | 81.0 (51-102) | 79.5 (55-91) | 65.0 (53-85) | 63.0 (51-81) | 73.1 (61-80) | 71.9 (61-80) | 66.0** (53-85) | 61.7** (46-73) |
| MMSE - score (mean, range) | 16.7 (7-29) | 29.0 (27-30) | 16.1* (10-20) | 29.2 (28-30) | 23.5 (20-28) | 29.1 (27-30) | 16.2* (5-29) | 18.0 (0-26) |
| Years from MRI-scan to death (mean, range) | 1.7 (0.2-3.4) | NA | 3.6 (0.3-7.2) | NA | NA | NA | 3.5 (0.3-7.2) | 5.8 (1.3-11.0) |
AD = Alzheimer’s disease; FTLD = frontotemporal lobar degeneration; MMSE=Folstein Mini Mental State Examination;
= MMSE scores obtained around the time of scanning only available from 12 subjects;
P = 0.1.
Group II consisted of 14 patients and 14 age and gender matched cognitively normal controls from the Dementia Research Centre, University College London (see Table 1 for details). The patients all fulfilled NINCDS-ADRDA criteria for ‘definite AD’ in that the clinical diagnosis of AD was confirmed histopathologically either from cerebral biopsy or at autopsy (McKhann et al., 1984) according to CERAD (Mirra et al., 1991) and NIA-RIA criteria (NIA-RIA, 1997). They tended to be younger than AD patients from group I. No strong family history was present in any of the subjects. Controls were determined to be cognitively normal either by subsequent clinical exam in follow-up or through pathological confirmation.
Group III consisted of 33 patients with probable mild AD and 57 age and gender matched cognitively normal controls from a community and referral based sample in Rochester, Minnesota, USA. The diagnosis of probable AD was made according to the DSM-III-R (American Psychiatric Association, 1987) and NINCDS-ADRDA criteria for AD (McKhann et al., 1984). Subjects were excluded from analysis if their scan revealed gross structural abnormalities other than atrophy. Diagnostic assignation was based on the combined results of medical history, clinical examination and psychometry. Controls were selected using the same criteria as outlined for group I and age/sex matched to AD patients. Patients with MMSE scores from 20-30 were considered in the mild stage of AD (Morris et al., 1989; Wolfson et al., 2002; Perneczky et al., 2006). In an attempt to restrict the group to typical patients from this largely community based sample for which diagnosis is both more critical and difficult, patients were included if they were 80 years old or younger with MMSE scores ≥20 at the time of scanning (see Table 1).
To test the ability to differentiate different forms of dementia we included an additional group of 19 subjects with pathologically confirmed FTLD (group IV, see Table 1). All the patients were diagnosed during life into one of the three FTLD subtypes according to consensus criteria (Neary et al., 1998): 9 patients had behavioural variant FTLD, 8 had semantic dementia and 2 had progressive non-fluent aphasia. In total there were 8 patients with tau-positive pathology and 11 patients with ubiquitin-positive, tau-negative pathology, diagnosed according to consensus pathological criteria (McKhann et al., 2001): behavioural variant FTLD (5 tau-positive, 4 ubiquitin-positive), semantic dementia (2 taupositive, 6 ubiquitin-positive), progressive non-fluent aphasia (1 tau-positive, 1 ubiquitin-positive). Patients in this group tended to be younger than AD-group II but not significantly so (P = 0.1).
Differentiation of FTLD from AD on clinical or neuropsychological grounds can sometimes be difficult and, in particular, the MMSE is rarely helpful.
Consent was obtained according to the Declaration of Helsinki, and the study was approved by the Mayo Clinic Institutional Review Board and the Local Research Ethics Committee in London. All subjects gave written informed consent.
MR imaging
For groups I and III, MR scans were collected over a period of about 10 years with a total of 13 different scanners. Several software updates occurred at different times for different scanners. However, a closely followed quality control program ensured uniformity over time. All scanners were monitored with daily phantom quality checks which calibrated the gradients to within +/- 1 mm over a 200 mm volume centered on the iso-center, monitored signal to noise and radio frequency transmit gain. All scans were done on the same platform, General Electric Signa 1.5T scanners (T1-weighted image parameters: TR = 23 to 27 ms, TE = 6 to 10 ms, flip angle 25° or 45°, voxel size 0.86 mm × 0.86 mm × 1.6 mm or 0.94 mm 0.94 mm × 1.6 mm, matrix dimensions 256 × 192). The major hardware elements (body resonance module, gradient coil and birdcage head transmit-receive volume coil) were unchanged throughout time and across all scanners. Importantly, there was no evidence that the effect of the different scanners or upgrades interacted with the effect of disease (Stonnington et al., 2007).
For Groups II and IV, data was acquired from three different 1.5 T scanners. Image parameters were TR = 35 or 15, TE = 5 or 5.4 or 7, flip angle 35° or 15°. Scanners and scanning parameters were balanced across groups and within groups as well as between AD patients from group II and FTLD patients (group IV). This was ensured by excluding four AD subjects from group II when compared to their control group. Because the mix of scanners used was different for normal elderly controls and FTLD subjects, the same four AD subjects were included for comparison between AD and FTLD subjects of group IV to maintain an equal balance of scanners between groups.
Image processing
Images were visually inspected for artefacts or structural abnormalities unrelated to AD or FTLD. Images were firstly segmented into grey matter (GM), white matter and cerebro-spinal fluid using SPM5 (Wellcome Trust Centre for Neuroimaging, Institute of Neurology, UCL, London UK—http://www.fil.ion.ucl.ac.uk/spm). Then, GM segments were further normalized to the population templates generated from all the images in each group and the combined images of groups I and II as well as of patients from group II and IV using an in-house implementation of a diffeomorphic registration algorithm (Ashburner, 2007). This non-linear warping technique minimizes structural variation between subjects. A separate ‘modulation’ step (Ashburner and Friston, 2000) was used to ensure that the overall amount of each tissue class remained constant after normalization. No spatial smoothing was performed.
Support vector classification
A support vector machine (SVM) is an example of a supervised, multivariate classification method. SVMs are supervised in the sense that they include a training step to learn about differences between groups to be classified. The method has previously been applied to neuroimaging data (Lao et al., 2004; Fan et al., 2005; Mourao-Miranda et al., 2005; Kawasaki et al., 2007). Data for this method need not satisfy assumptions of Random field theory, making additional smoothing unnecessary.
Here we describe an SVM intuitively to help readers understand the concept without recourse to technical detail. In the context of machine learning, individual MR images are treated as points located in a high dimensional space. Figure 1 illustrates this procedure in an imaginary 2D space: in this example the two groups to be classified (A and B) are represented by circles and squares. It can be seen that the groups cannot be separated on the basis of values along 1D only and that only a combination of the two leads to adequate separation. The space used for classifying image data is of much higher dimension; the total number of dimensions is determined by the numbers of voxels in each MR image.
Fig. 1.
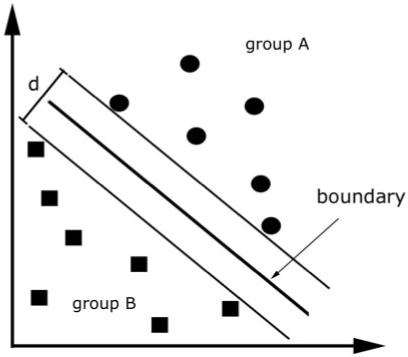
Illustration of the concept used in support vector machines. The algorithm tries to find a boundary that maximizes the distance (d) between groups. The figure reduces the problem to two groups and two dimensions for the purpose of illustration only.
In practical terms, a linear kernel matrix is created from normalized grey matter segmented images. To this end, each MRI scan undergoes a pair wise multiplication with all other scans. Each element in the kernel matrix is therefore a dot product of two images. Intuitively, the kernel matrix can be viewed as a similarity measure among subjects belonging to a characterized group. The voxels are effectively treated as coordinates of a high dimensional space and their location is determined by the intensity value at each voxel. The images do not span the whole high dimensional space, but rather cluster in subspaces containing images that are very similar. This is one reason why image normalization into a standard space is an important preprocessing step. Good normalization will tighten clustering and reduce dimensionality.
The use of an SVM for image classification is an example of a linear discrimination. In the basic model it is a binary classifier, which means it divides the space into which the MR images are distributed into two classes by identifying a separating hyperplane. In a simple 2D space, the boundary is represented by a line, but is called a hyperplane in higher dimensional space. Fisher’s linear discriminate analysis or linear perceptrons can both identify linear discriminant hyperplanes. However, the motivation behind using an SVM is that it uses the principle of ‘structural risk minimization’, which aims to find a hyperplane that maximizes the distance between training classes (see Fig. 1). Intuitively, it can be seen that the optimal separating hyperplane (OSH) produced by an SVM is defined by those voxels that are closest to the separating boundary between them, i.e. the voxels that are most ambiguous. These voxels are called the ‘support vectors’. Voxels that are further away from a separating boundary are distinctively different, hence are not used to calculate the OSH. This fact suggests that adding more images to a training set will have little effect on an OSH if they are distant from it.
After training, an OSH contains learned differences between classes—in our case, AD and control images. That information is then used to assign any new image to its appropriate class (leaveone-out method). This procedure iteratively leaves successive images out of training for subsequent class assignation until each has been used in this way. This validation procedure ensures that a trained SVM can generalize and be used on scans that have never been presented to the SVM algorithm previously.
In addition to identification of an OSH we determined what voxels contributed most to classification and their distribution in the brain. During training an SVM assigns a specific weight to every scan reflecting its importance to group separation.
This weight is multiplied by a label vector indicating which group the scan belongs to (e.g. −1 for AD and +1 for controls). Each scan is then multiplied by the product of weight and label and summed, resulting in a value for each voxel indicating its importance for group discrimination.
For technical information on SVMs, interested readers are referred to the following textbooks (Vapnik, 1998; Bishop, 2006).
In order to test whether scan data from different centres can be aggregated to provide generic SVMs we investigated using one pathologically confirmed image set for training and another for testing. We also combined data-sets from groups I and II with the leave-one-out method described above. The aim of this procedure was to show generalizability, of great importance to the introduction of these methods into clinical radiological practice.
Mild AD affects the hippocampus and immediately adjacent cortex primarily (Braak and Braak, 1991). For classification of group III we therefore performed a standard whole-brain analysis and then also attempted classification using data from a hippocampus centred volume of interest (VOI: dimensions 12, 16 and 12 mm in x,y,z directions, respectively equivalent to x, y, z= −17, −8, −18 and 16, −9, −18 in MNI-space) (Hirata et al., 2005). We created a separate kernel matrix from the whole brain and from the brain within the VOI from group III. These kernels were used separately and in combination.
In order to test how well SVMs can differentiate different forms of dementia we created a separate kernel from subjects with pathologically confirmed FTLD and confirmed AD-patients, both acquired with the same scanning hardware and sequence.
Results
Confirmed AD-patients versus controls
Subjects from group I were correctly assigned to the appropriate diagnostic category in 95.0% of trials with the leave-one out method using whole brain images (sensitivity 95.0%, specificity 95.0%). One 89 year old AD patient with an MMSE of 29 and one 86 year old control were misclassified. Fig. 2 displays regions that were most influential in making the binary classification between AD and normal when the grey matter of the whole brain was used for analysis.
Fig. 2.
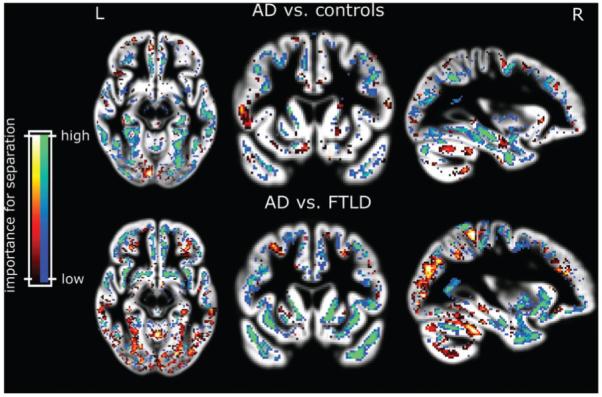
Voxels most relevant for classification of patients from group I after SVM training with the data from group I (upper panel). The blue and green areas indicate higher grey matter volume increasing the likelihood of classification into normal. Red and yellow show regions where higher grey matter volume indicates the opposite. The lower panel depicts relevant areas for the separation from AD from FTLD. Blue and green indicate areas where lower grey matter volume indicates FTLD. Results are overlaid on the mean grey matter compartment image from all subjects.
Subjects from group II were correctly assigned in 92.9% of trials with the leave-one out method using whole brain images (sensitivity 100%, specificity 85.7%). The two oldest controls were misclassified.
When group I and group II were combined in a single data-set, patients were correctly assigned to the appropriate group in 95.6% of trials with the leave-one out method using whole brain images (sensitivity 97.1%, specificity 94.1%).
Finally, when group I was used to train the data and group II was used to test, 96.4% of patients were correctly assigned to the appropriate group (sensitivity 100%, specificity 92.9%). Conversely, when group II was used to train and group I to test, 87.5% of patients were correctly assigned to the appropriate group (sensitivity 95.0%, specificity 80.0%).
Mild AD versus controls
Subjects from group III were correctly assigned to the appropriate group in 81.1% of trials using whole brain images (sensitivity 60.6%, specificity 93.0%). A further improvement to 85.6% (sensitivity 75.8%, specificity 91.2%) was obtained when analysis was restricted to the medial temporal lobe region defined in the methods section. Combining the matrix kernels from the whole brain and medial temporal lobe region improved the overall classification to 88.9%.
AD versus FTLD
AD and FTLD subjects were correctly assigned to the appropriate group in 89.2% of trials using whole brain images (sensitivity 94.7% specificity 83.3%; three AD and one FTLD subject misclassified). There were no identifiable associations with age or MMSE in misclassified subjects. The misclassified FTLD had the behavioural variant of the disease. Tables 2 and 3 summarise all classification results with and without the antero-medial region of interest.
Table 2. Results of SVM classification using grey matter from the whole brain for image analysis.
| Group | Correctly classified (%) | Sensitivity (%)* | Specificity (%)* |
|---|---|---|---|
| AD and controls Group I | 95.0 | 95.0 | 95.0 |
| AD and controls Group II | 92.9 | 100 | 85.7 |
| AD and controls Group III | 81.1 | 60.6 | 93.0 |
| Dataset I for training, set II for testing | 96.4 | 100 | 92.9 |
| Dataset II for training, set I for testing | 87.5 | 95.0 | 80.0 |
| Group I + II | 95.6 | 97.1 | 94.1 |
| AD from Dataset II and FTLD Group IV | 89.2 | 83.3 | 94.7 |
Considering a correctly identified AD case as a true positive.
Table 3. Results of SVM classification using only grey matter of antero-medial lobe volume of interest for analysis.
| Group | Correctly classified (%) | Sensitivity (%)* | Specificity (%)* |
|---|---|---|---|
| AD and controls Group I | 90.0 | 85.0 | 95.0 |
| AD and controls Group II | 92.9 | 92.9 | 92.9 |
| AD and controls Group III | 85.6 | 75.8 | 91.2 |
| Dataset I for training, set II for testing | 71.4 | 50 | 92.9 |
| Dataset II for training, set I for testing | 70.0 | 95.0 | 45.0 |
| Group I + II | 94.1 | 97.1 | 91.2 |
Considering a correctly identified AD case as a true positive.
Discussion
Our results indicate that supervised machine learning techniques can aid the clinical diagnosis of AD. The analytical technique presented here promises to distinguish disease-specific atrophy from that of normal aging in a standard T1 weighted structural MRI scan. Furthermore, the study provides evidence that the method can be developed to correctly differentiate between different forms of dementia.
Before comparing the method to other approaches and a discussion of translation into clinical practice with prospects for future studies, we discuss some methodological aspects.
We used linear SVMs. They allow a localization of voxels relevant to separation of scans into two groups. The voxels which a whole brain SVM classification of AD from controls depended on most were clustered around the parahippocampal gyrus and parietal cortex (Fig. 2, upper panel). A similar distribution was found for all the classifications of AD from normal scans we report here. Classification of FTLD from AD depended on voxels in frontal as well as parietal areas (Fig. 2, lower panel) for group assignation. A recent study using cortical thickness also found parietal areas important in differentiating these two dementia types (Du et al., 2007). Figure 2 also shows cortical voxels scattered throughout the brain without any regionally specific pattern. They are however specific in the sense that they also contributed to a differentiation between two groups. It can therefore be argued that they too reflect differences in overall brain shape resulting from degeneration of specific structures. We tested the performance of non-linear kernels (such as radial-basis functions) but these failed to improve performance suggesting a linear approach is both valid and adequate. The excellent results obtained using scans for training and testing from different centres that used different scanners shows that linear SVMs generalize well.
The results we obtained are comparable or better than other classification methods described in the literature based on MR images (Gosche et al., 2002; Jack et al., 2002; Barnes et al., 2004; Csernansky et al., 2004; Wahlund et al., 2005), most of which restrict analysis to temporal lobe structures. Our method also performs as well as or better than the average reported diagnostic accuracy of clinicians using clinical exam, history, neuropsychological testing and classical image reporting as outlined in NINCDS-ADRDA or DSM-III-R (Knopman et al., 2001). However, to make this conclusion, a formal comparison with modern conventional clinical assessment is required. The construction of a ‘library’ of SVM’s for all dementia types can be envisaged to help differentiate other conditions that can be confused with AD e.g. vascular dementia, FTLD, Lewy body disease, etc. The preliminary result from our attempt to separate FTLD and AD is very promising because FTLD is a group of degenerative diseases that also affect frontal and temporal lobes but differ in extent and neuropathological characteristics. AD and FTLD can be difficult to separate clinically and patients with confirmed AD pathology have been shown to present with a focal clinical syndrome. A recent postmortem study found that up to 30% of patients diagnosed with the language subtype of FTLD (progressive non-fluent aphasia or semantic dementia) had AD pathology (Knibb et al., 2006). One limitation of our FTLD versus AD classification is that we did not test pure pathological subtypes of FTLD separately. It is possible that the performance of a suitably trained SVM classifier will be better for distinguishing certain subtypes of FTLD from AD than others. Our sample size is too small to explore this question further.
Unlike methods that include an expert-dependent hippocampal tracing step (Jack et al., 1992), the SVM technique is fully automated and can use all the information in a brain scan. Automation eliminates observer/experimenter bias completely, generates totally reproducible results with the same image set and makes the method much less labour-intensive. These are important characteristics for a method proposed for clinical use.
Our findings warrant application of the proposed methods to larger image sets such as those being collected for the Alzheimer’s Disease Neuroimaging Initiative (ADNI—Mueller et al., 2005) for several reasons. The cases from group I are more typical of community based samples, with a later age of onset, whereas cases from group II are more typical of referral centres with greater numbers of early onset cases. That we could get comparable results and even use one scanner’s images to train and another to test suggests the technique will generalize for use in clinical settings. However, it is clear that when the relatively younger subjects from group II are used for training, specificity goes down; a result attributable to the fact that group II included more early onset AD who may show a somewhat different patterns of degeneration, i.e. relatively more parietal involvement (Schott et al., 2006; Frisoni et al., 2007). Because of their younger age, subjects from group II are also less likely to have co-morbidity (e.g. subtle vascular changes) but possibly more AD related atrophy for the same MMSE. A limitation of the population used in Group III is that the clinical diagnoses were likely not 100% accurate as no pathological verification was available for this group. Previous studies have shown that a clinical diagnosis is inaccurate, compared to pathological diagnosis, in about 11% of mild cases in which similar diagnostic criteria to those in our sample were used (Salmon et al., 2002). We therefore speculate that some of the misclassification is in fact due to misdiagnosis in mildly affected AD-patients. The ability to generalize across image-sets from different centres is very important in this respect as it could facilitate the generation of SVMs for rarer forms of dementia based on reliable diagnoses.
It will be a matter of judgement and empirical verification whether to use whole brain or partial data or a combination of the two for diseases other than AD. We recommend an exploratory whole brain approach as a necessary initial step for the time being. We expect that the earlier the stage or more localized a disease, the more a well-placed VOI will improve categorization. In group III, classification by SVM improved substantially by restricting analysis to medial temporal lobes because non-contributory, noisy brain areas were excluded from analysis. However, reduction of the brain volume analysed risks excluding potentially important differential image features. Therefore, combined kernels from whole brain and VOI serve to retain information obtained from the whole brain while weighting the classification to the area of brain most relevant at early stages of disease. The implication for more generalized diseases is that the opposite will be true. In this perspective we tested groups I and II using a medial temporal area analysis and found that classification was slightly worse for leave-one-out and much worse when using one data-set to train and another to test (Table 3). As many forms of dementia involve hippocampal atrophy, accurate differential classification of the dementias may well need whole brain analysis.
A goal of machine learning based automated MR image analysis that we believe achievable, is better sensitivity and specificity of ante-mortem diagnosis than is currently possible. The method we have described clearly has potential in achieving more accurate dementia diagnosis in clinical practice. Although the processing and preparation of a training dataset is relatively time consuming (around a week for all data-sets in this study on a standard PC at 2.4 MHz), this is unlikely to be a limiting factor. Firstly, this represents computer processing time without user interaction. Secondly, once a training dataset is prepared, spatial normalization and classification of any new scan can be done in a matter of minutes. The time required is likely to shorten further with the advent of faster computers. The current implementation still requires a user to check intermediate results for misregistration. This step can be further automated by the introduction of thresholds to alert an operator to check image quality. Although it has been suggested that MRI is likely to help diagnosis in specialty clinics only (Wahlund et al., 2005), we see no reason why the method cannot be translated to a more general setting, since a training set of pathologically confirmed cases can come from a specialist centre and because the method is computer-based, automated and does not require expert anatomical knowledge.
Future studies will focus on the application of SVMs to aid differential diagnosis in situations where more than two diagnoses are possible. Stratification of patients by anatomical severity can be envisaged. The limits of sensitivity, for example in predicting which MCI patients will transform into AD also need definition. Encouraging results with other multivariate classification methods have recently been reported (Teipel et al., 2007).
Acknowledgement
The authors would like to thank Geoffrey Tan and Eric Reiman for helpful suggestions in the preparation of the manuscript. This work was supported by the Wellcome Trust (grant 075696 2/04/2 to R.S.J.F. and J.A.), Mayo Clinic (grant to C.M.S.), the National Institute on Aging (grants P50 AG16574, U01 AG06786, and AG11378 to Mayo Clinic Rochester, MN), the Robert H. and Clarice Smith and Abigail Van Buren Alzheimer’s Disease Research Program of the Mayo Foundation (to Mayo Clinic Rochester, MN). N.C.F. and R.I.S. are supported by UK Medical Research Council grants G9626876 and G90/86, respectively. J.D.R. is supported by a Wellcome Trust Research Training Fellowship. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Abbreviations
- AD
Alzheimer’s disease
- DSM-IIIR
Diagnostic and Statistical Manual, 3rd edition, revised
- FTLD
frontotemporal lobar degeneration
- MCI
mild cognitive impairment
- MMSE
Mini Mental State Examination
- NINCDS-ADRDA
National Institute of Neurological and Communicative Disorders and Stroke/Alzheimer’s disease and related Disorders Association
- OSH
optimal separating hyperplane
- SVM
support vector machine
References
- American Psychiatric Association . Diagnostic, and statistical manual of mental disorders. American Psychiatric Press; Washington, DC: 1987. [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry—the methods. Neuroimage. 2000;11:805–21. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Barnes J, Scahill RI, Boyes RG, Frost C, Lewis EB, Rossor CL, et al. Differentiating AD from aging using semiautomated measurement of hippocampal atrophy rates. Neuroimage. 2004;23:574–81. doi: 10.1016/j.neuroimage.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Bishop C. Pattern recognition and machine learning. Springer; New York: 2006. [Google Scholar]
- Blennow K, de Leon MJ, Zetterberg H. Alzheimer’s disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–59. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Cairns NJ, Bigio EH, Mackenzie IR, Neumann M, Lee VM, Hatanpaa KJ, et al. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol (Berl) 2007;114:5–22. doi: 10.1007/s00401-007-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D, Fox NC, Scahill RI, Crum WR, Whitwell JL, Leschziner G, et al. Patterns of temporal lobe atrophy in semantic dementia and Alzheimer’s disease. Ann Neurol. 2001;49:433–42. [PubMed] [Google Scholar]
- Csernansky JG, Hamstra J, Wang L, McKeel D, Price JL, Gado M, et al. Correlations between antemortem hippocampal volume and postmortem neuropathology in AD subjects. Alzheimer Dis Assoc Disord. 2004;18:190–5. [PubMed] [Google Scholar]
- Davatzikos C, Fan Y, Wu X, Shen D, Resnick SM. Detection of prodromal Alzheimer’s disease via pattern classification of MRI. Neurobiol Aging. 2006 doi: 10.1016/j.neurobiolaging.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deFigueiredo RJ, Shankle WR, Maccato A, Dick MB, Mundkur P, Mena I, et al. Neural-network-based classification of cognitively normal, demented, Alzheimer disease and vascular dementia from single photon emission with computed tomography image data from brain. Proc Natl Acad Sci USA. 1995;92:5530–4. doi: 10.1073/pnas.92.12.5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du AT, Schuff N, Kramer JH, Rosen HJ, Gorno-Tempini ML, Rankin K, et al. Different regional patterns of cortical thinning in Alzheimer’s disease and frontotemporal dementia. Brain. 2007;130:1159–66. doi: 10.1093/brain/awm016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Shen D, Davatzikos C. Classification of structural images via high-dimensional image warping, robust feature extraction, and SVM. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2005;8:1–8. doi: 10.1007/11566465_1. [DOI] [PubMed] [Google Scholar]
- Ferri CP, Prince M, Brayne C, Brodaty H, Fratiglioni L, Ganguli M, et al. Global prevalence of dementia: a Delphi consensus study. Lancet. 2005;366:2112–7. doi: 10.1016/S0140-6736(05)67889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox NC, Schott JM. Imaging cerebral atrophy: normal ageing to Alzheimer’s disease. Lancet. 2004;363:392–4. doi: 10.1016/S0140-6736(04)15441-X. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Pievani M, Testa C, Sabattoli F, Bresciani L, Bonetti M, et al. The topography of grey matter involvement in early and late onset Alzheimer’s disease. Brain. 2007;130:720–30. doi: 10.1093/brain/awl377. [DOI] [PubMed] [Google Scholar]
- Gosche KM, Mortimer JA, Smith CD, Markesbery WR, Snowdon DA. Hippocampal volume as an index of Alzheimer neuropathology: findings from the Nun Study. Neurology. 2002;58:1476–82. doi: 10.1212/wnl.58.10.1476. [DOI] [PubMed] [Google Scholar]
- Herholz K, Salmon E, Perani D, Baron JC, Holthoff V, Frolich L, et al. Discrimination between Alzheimer dementia and controls by automated analysis of multicenter FDG PET. Neuroimage. 2002;17:302–16. doi: 10.1006/nimg.2002.1208. [DOI] [PubMed] [Google Scholar]
- Hirata Y, Matsuda H, Nemoto K, Ohnishi T, Hirao K, Yamashita F, et al. Voxel-based morphometry to discriminate early Alzheimer’s disease from controls. Neurosci Lett. 2005;382:269–74. doi: 10.1016/j.neulet.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Dickson DW, Parisi JE, Xu YC, Cha RH, O’Brien PC, et al. Antemortem MRI findings correlate with hippocampal neuropathology in typical aging and dementia. Neurology. 2002;58:750–7. doi: 10.1212/wnl.58.5.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, O’Brien PC, Tangalos EG. MR-based hippocampal volumetry in the diagnosis of Alzheimer’s disease. Neurology. 1992;42:183–8. doi: 10.1212/wnl.42.1.183. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Shiung MM, Gunter JL, O’Brien PC, Weigand SD, Knopman DS, et al. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62:591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki Y, Suzuki M, Kherif F, Takahashi T, Zhou SY, Nakamura K, et al. Multivariate voxel-based morphometry successfully differentiates schizophrenia patients from healthy controls. Neuroimage. 2007;34:235–42. doi: 10.1016/j.neuroimage.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Knibb JA, Xuereb JH, Patterson K, Hodges JR. Clinical and pathological characterization of progressive aphasia. Ann Neurol. 2006;59:156–65. doi: 10.1002/ana.20700. [DOI] [PubMed] [Google Scholar]
- Knopman DS, DeKosky ST, Cummings JL, Chui H, Corey-Bloom J, Relkin N, et al. Practice parameter: diagnosis of dementia (an evidence-based review). Report of the Quality Standards Subcommittee of the American Academy of Neurology. Neurology. 2001;56:1143–53. doi: 10.1212/wnl.56.9.1143. [DOI] [PubMed] [Google Scholar]
- Lao Z, Shen D, Xue Z, Karacali B, Resnick SM, Davatzikos C. Morphological classification of brains via high-dimensional shape transformations and machine learning methods. Neuroimage. 2004;21:46–57. doi: 10.1016/j.neuroimage.2003.09.027. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Pruessner J, Zijdenbos AP, Collins DL, Teipel SJ, Hampel H, et al. Automated cortical thickness measurements from MRI can accurately separate Alzheimer’s patients from normal elderly controls. Neurobiol Aging. 2006 doi: 10.1016/j.neurobiolaging.2006.09.013. [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ. Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick’s Disease. Arch Neurol. 2001;58:1803–9. doi: 10.1001/archneur.58.11.1803. [DOI] [PubMed] [Google Scholar]
- Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, et al. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology. 1991;41:479–86. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- Morris JC, Heyman A, Mohs RC, Hughes JP, van Belle G, Fillenbaum G, et al. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part I. Clinical and neuropsychological assessment of Alzheimer’s disease. Neurology. 1989;39:1159–65. doi: 10.1212/wnl.39.9.1159. [DOI] [PubMed] [Google Scholar]
- Mourao-Miranda J, Bokde AL, Born C, Hampel H, Stetter M. Classifying brain states and determining the discriminating activation patterns: Support Vector Machine on functional MRI data. Neuroimage. 2005;28:980–95. doi: 10.1016/j.neuroimage.2005.06.070. [DOI] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, et al. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am. 2005;15:869–77. xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–54. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- NIA-RIA Consensus recommendations for the postmortem diagnosis of Alzheimer’s disease. The National Institute on Aging, and Reagan Institute Working Group on Diagnostic Criteria for the Neuropathological Assessment of Alzheimer’s Disease. Neurobiol Aging. 1997;18:S1–2. [PubMed] [Google Scholar]
- Perneczky R, Wagenpfeil S, Komossa K, Grimmer T, Diehl J, Kurz A. Mapping scores onto stages: mini-mental state examination and clinical dementia rating. Am J Geriatr Psychiatry. 2006;14:139–44. doi: 10.1097/01.JGP.0000192478.82189.a8. [DOI] [PubMed] [Google Scholar]
- Salmon DP, Thomas RG, Pay MM, Booth A, Hofstetter CR, Thal LJ, et al. Alzheimer’s disease can be accurately diagnosed in very mildly impaired individuals. Neurology. 2002;59:1022–8. doi: 10.1212/wnl.59.7.1022. [DOI] [PubMed] [Google Scholar]
- Schott JM, Ridha BH, Crutch SJ, Healy DG, Uphill JB, Warrington EK, et al. Apolipoprotein e genotype modifies the phenotype of Alzheimer disease. Arch Neurol. 2006;63:155–6. doi: 10.1001/archneur.63.1.155. [DOI] [PubMed] [Google Scholar]
- Solomon PR, Murphy CA. Should we screen for Alzheimer’s disease? A review of the evidence for and against screening Alzheimer’s disease in primary care practice. Geriatrics. 2005;60:26–31. [PubMed] [Google Scholar]
- Stonnington CM, Tan G, Kloppel S, Chu C, Draganski B, Jack CR, Jr, et al. Interpreting scan data acquired from multiple scanners: A study with Alzheimer’s disease. Neuroimage. 2007 doi: 10.1016/j.neuroimage.2007.09.066. (in press) DOI: 10.1016/j.neuroimage.2007.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teipel SJ, Born C, Ewers M, Bokde AL, Reiser MF, Moller HJ, et al. Multivariate deformation-based analysis of brain atrophy to predict Alzheimer’s disease in mild cognitive impairment. Neuroimage. 2007;38:13–24. doi: 10.1016/j.neuroimage.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Valcour VG, Masaki KH, Curb JD, Blanchette PL. The detection of dementia in the primary care setting. Arch Intern Med. 2000;160:2964–8. doi: 10.1001/archinte.160.19.2964. [DOI] [PubMed] [Google Scholar]
- Vapnik V. Statistical Learning Theory. Wiley Interscience; New York: 1998. [Google Scholar]
- Wahlund LO, Almkvist O, Blennow K, Engedahl K, Johansson A, Waldemar G, et al. Evidence-based evaluation of magnetic resonance imaging as a diagnostic tool in dementia workup. Top Magn Reson Imaging. 2005;16:427–37. doi: 10.1097/01.rmr.0000245463.36148.12. [DOI] [PubMed] [Google Scholar]
- Wechsel D. Wechsler Memory Scale-Revised. Harcourt Brace Jovanovich; San Antonio, USA: 1987. [Google Scholar]
- Wolfson C, Oremus M, Shukla V, Momoli F, Demers L, Perrault A, et al. Donepezil and rivastigmine in the treatment of Alzheimer’s disease: a best-evidence synthesis of the published data on their efficacy and cost-effectiveness. Clin Ther. 2002;24:862–86. doi: 10.1016/s0149-2918(02)80004-2. discussion 837. [DOI] [PubMed] [Google Scholar]


