SUMMARY
CTCF plays a central role in vertebrate insulators and forms part of the Fab-8 insulator in Drosophila. dCTCF is present at hundreds of sites in the Drosophila genome, where it is located at the boundaries between bands and interbands in polytene chromosomes. dCTCF co-localizes with CP190, which is required for proper binding of dCTCF to chromatin, but not with the other gypsy insulator proteins Su(Hw) or Mod(mdg4)2.2. Mutations in the CP190 gene affect Fab-8 insulator activity, suggesting that CP190 is an essential component of both gypsy and dCTCF insulators. dCTCF is present at specific nuclear locations forming large insulator bodies that overlap with those formed by Su(Hw), Mod(mdg4)2.2 and CP190. The results suggest that Su(Hw) and dCTCF may be the DNA-binding components of two different subsets of insulators that share CP190 and cooperate in the formation of insulator bodies to regulate the organization of the chromatin fiber in the nucleus.
Keywords: CTCF, transcription, insulator, chromatin
INTRODUCTION
Insulators are DNA regulatory sequences that maintain functionally distinct chromatin domains from improperly interacting with each other. Based on their properties and mechanism of action, these sequences can be classified into barrier insulators, when their role is to stop the spreading of heterochromatin, or enhancer blocking insulators, when they interfere with enhancer-promoter communication (Gaszner and Felsenfeld, 2006). Enhancer blocking insulators have been found in a variety of organisms but have been studied in detail in vertebrates and Drosophila (Capelson and Corces, 2004).
The best characterized vertebrate insulator is cHS4, a sequence found in the 5′ region of the chicken β-globin locus (Felsenfeld et al., 2004). This insulator displays both barrier and enhancer-blocking activities that reside in different sequence elements. The enhancer-blocking activity is associated with binding sites for the Zn finger protein CTCF (Bell et al., 1999). In fact, most characterized vertebrate enhancer-blocking insulators are composed of sequences that interact with CTCF, and this protein is essential for their function. CTCF insulators play important roles in genomic imprinting at the Igf2/H19 locus (Bell and Felsenfeld, 2000; Hark et al., 2000) as well as in X-chromosome inactivation (Chao et al., 2002). In humans, CTCF binding sites at the DM1 locus form a functional insulator and loss of binding of CTCF may contribute to the phenotype of myotonic dystrophy (Filippova et al., 2001). These observations suggest an important role for enhancer-blocking insulators in the regulation of gene expression.
Contrary to vertebrates, which seem to rely on CTCF as the main source of insulator activity, Drosophila has been shown to possess a rich variety of enhancer-blocking insulators utilizing different proteins to accomplish their function (Capelson and Corces, 2004). The scs and scs’ sequences were first found to flank the hsp70 heat shock puffs; these sequences interact with the ZW5 and BEAF-32 proteins respectively, which are essential for their insulator activity (Gaszner et al., 1999; Zhao et al., 1995). Fab-8, a second insulator element first found in the Drosophila Abdominal-B (Abd-B) gene of the bithorax complex, has been shown to interact with the Drosophila homolog of CTCF, dCTCF (Moon et al., 2005). A third enhancer-blocking insulator named SF1 was first found in the Scr-ftz intergenic region and is made up of binding sites for the GAGA protein (Belozerov et al., 2003). The variety of sequences and associated proteins present in Drosophila enhancer-blocking insulators in contrast to the widely present CTCF insulator found in vertebrates raises the question of whether vertebrates have lost most insulators except for CTCF or whether other types of insulators remain to be identified in vertebrates.
The gypsy enhancer-blocking insulator of Drosophila, first found in the gypsy retrotransposon, is composed of several proteins that play distinct roles in regulating insulator function (Capelson and Corces, 2004). Su(Hw) is an 11 zinc finger DNA binding protein that interacts with insulator sequences; mutations in the su(Hw) gene cause female sterility but do not result in lethality (Harrison et al., 1993). Mod(mdg4)2.2 is a BTB domain protein that interacts with Su(Hw). The mod(mdg4) gene encodes approximately 30 different isoforms that arise by alternative cis- and trans-splicing (Buchner et al., 2000; Labrador and Corces, 2003); null mutations in the gene result in lethality, but mutations affecting the Mod(mdg4)2.2 isoform are viable and show defects in gypsy insulator function (Gerasimova et al., 1995). CP190 is a third protein component of the gypsy insulator. It contains a BTB domain as well as 3 zinc fingers and it interacts with Su(Hw) and Mod(mdg4)2.2. CP190 binds DNA but it does not interact directly with gypsy insulator sequences present in the gypsy retrotransposon, where it is recruited through interactions with Su(Hw) and Mod(mdg4)2.2 instead. Mutations in the CP190 gene are lethal (Pai et al., 2004).
Although enhancer-blocking insulators interfere with enhancer-promoter interactions, this may not be their primary function in nuclear biology. Instead, their primary role may be to establish independent domains of gene expression such that regulatory sequences in one domain cannot affect the expression of genes located in a different domain. Their effect on enhancer-promoter interactions may then be a byproduct of this primary role. This conclusion comes in part from the observation that enhancer-blocking insulators can form chromatin loops. For example, the scs and scs’ insulators have been shown to pair in vivo and, as a consequence, loop out the intervening chromatin fiber (Blanton et al., 2003). The chicken CTCF insulator can also can also mediate loop formation and at least some of these loops attach to the nucleolus through the nucleophosmin protein (Yusufzai et al., 2004). In the case of the gypsy insulator, interactions between multiple insulator sites form structures called insulator bodies that in turn attach to the nuclear lamina (Byrd and Corces, 2003). The formation of these bodies requires, in addition to the three core proteins Su(Hw), Mod(mdg4)2.2 and CP190, functional RNA silencing machinery. The Rm62 protein is a putative RNA helicase that may interact with small RNAs to regulate insulator function (Lei and Corces, 2006). The gypsy insulator can also be regulated by ubiquitination and sumoylation of some of the core proteins by the dTopors ubiquitin E3 ligase (Capelson and Corces, 2005; Capelson and Corces, 2006).
The three core protein components of the gypsy insulator colocalize in the insulator bodies but their distribution in polytene chromosomes, which is presumably a representation of their genomic distribution in the chromatin of interphase cells, does not show an exact overlap. Insulator proteins in Drosophila are present at the boundaries between bands (containing condensed chromatin and non-transcribed genes) and interbands (containing de-condensed chromatin and actively transcribed genes). Bands and interbands may thus represent chromatin domains defined and maintained my insulators (Labrador and Corces, 2002). CP190 is present at all or most band/interband boundaries in polytene chromosomes, whereas Su(Hw) and Mod(mdg4)2.2 co-localize with each other precisely but are present at only a subset of the CP190 sites (Pai et al., 2004). This distribution suggests that CP190 may be a general component of a widespread insulator present at every band/interband boundary. CP190 may use the Su(Hw) DNA binding protein as a tether to be recruited to a subset of these insulators but may use other DNA binding proteins or bind DNA itself to be recruited to the rest of the insulators where Su(Hw) is absent. This hypothesis is also supported by the observation that mutations affecting the Su(Hw) or Mod(mdg4)2.2 proteins are viable, but mutations affecting CP190 are lethal, suggesting a more general function for this protein (Pai et al., 2004).
Given the widespread role of CTCF in insulator function in vertebrates and its structural similarity to Su(Hw), we decided to investigate the possibility that dCTCF cooperates with the gypsy insulator in genome organization and regulation of gene expression. Here we show that this is indeed the case. dCTCF interacts with CP190 at a subset of genomic sites different from those occupied by Su(Hw), but it is present at the same insulator bodies formed by Su(Hw). The results suggest that Su(Hw) and dCTCF are components of two different subsets of insulators that cooperate in the formation of larger insulator bodies to regulate the organization of the chromatin fiber in the nucleus of Drosophila cells.
RESULTS
Mutations in dCTCF affect the function of the Fab-8 insulator
The Fab-8 insulator contains two binding sites for the dCTCF protein. Mutations in the sites that disrupt binding of dCTCF also affect the ability of Fab-8 to interfere with enhancer-promoter interactions (Moon et al., 2005). To directly test the role of dCTCF in the function of the Fab-8 insulator, we made mutations in the dCTCF gene. A strain carrying an insertion of a P transposable element in the first non-coding exon of the dCTCF gene, P{EPgy2}CTCFEY15833 produces reduced levels of dCTCF protein (Figure 1A) but is viable and lacks any observable phenotype. In order to produce null mutations in the dCTCF gene, we mobilized the P element using the y1w1118; P{Δ2-3}Sb/TM6 strain as a source of transposase. The P element in P{EPgy2}CTCFEY15833 harbors the yellow gene reporter located adjacent to the 5′ region of dCTCF and the white gene reporter adjacent to the 3′ region. In order to obtain P element transpositions that result in deletion of the dCTCF gene, we selected for flies displaying a y+ w− phenotype. These deletions extend from the site of the P element insertion in the first exon towards the 3′ end of the dCTCF gene and complement mutations in the quemao gene; quemao is adjacent to the 3′ of dCTCF, suggesting that the new mutations only affect dCTCF. Figure 1A shows a Western analysis of protein extracts from pupae carrying three different dCTCF deletion mutations, dCTCFy+1, dCTCFy+2 and dCTCFy+6. All three strains show no detectable levels of dCTCF, which, together with the fact that they carry a deletion of the gene, suggest that the mutations are null. All three strains display late pupal lethality, although some individuals survive to the pharate adult stage and can be pulled from the pupal case for observation.
Figure 1. Mutations in the dCTCF gene affect Fab-8 insulator activity.
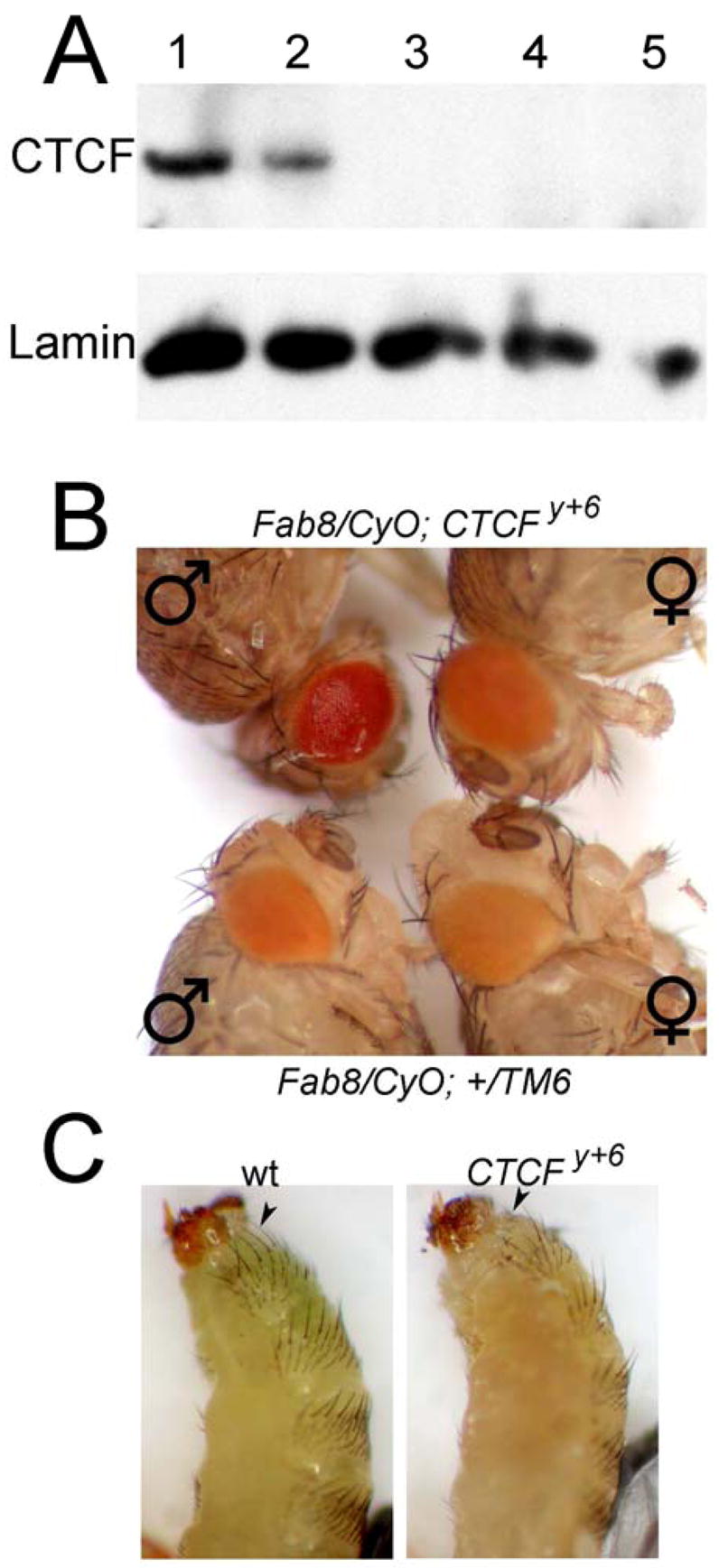
(A) Protein extracts from larvae of various fly strains were subjected to Western analysis. Lane 1, wild type OreR; lane 2, P{EPgy2}CTCFEY15833; lane 3, dCTCFy+1; lane 4, dCTCFy+2; lane 5, dCTCFy+6.
(B) Eyes of male and female flies of the genotypes Fab-860.39.2/CyO; dCTCFy+6 (top) and Fab-860.39.2/CyO; +/TM6 (bottom).
(C) Abdomens of pharate adult flies pulled from the pupal cases. The left panel shows the abdomen of a wild type fly whereas the right panel shows the abdomen of a fly homozygous for the dCTCFy+6 allele; the arrow points to an additional abdominal segment posterior to A6.
In order to test the role of dCTCF in the activity of the Fab-8 insulator, we examined the phenotype of fly strains Fab-860.39.2/CyO or Fab-860.71.1/CyO carrying a transgene that includes the Fab-8 insulator positioned between the eye enhancer and the coding region of the white gene. The presence of Fab-8 at this position results in a non-functional enhancer and flies with light yellow eyes (Figure 1B). When the strain is homozygous for a mutation in the dCTCF gene, the color of the eye becomes darker. This effect is more pronounced in males, in which the eye color becomes close to wild type (Figure 1B). This result suggests that dCTCF indeed plays a role in the function of the Fab-8 insulator. If this is the case, mutations in the dCTCF gene should display homeotic transformations affecting the abdominal segments. Results shown in Figure 1C indicate that this is the case. Male flies homozygous for the dCTCFy+6 mutation display an additional abdominal A7 segment posterior to A6 whereas the A7 and A8 segments are not visible in wild type males; this phenotype is similar to that observed in Abd-B class I mutants (Estrada et al., 2002).
dCTCF is present at a subset of gypsy insulator sites
The Fab-8 insulator is present in the bithorax complex in the 5′ regulatory region of the Abd-B gene (Barges et al., 2000). To determine whether dCTCF plays a general role in the Drosophila genome rather than a specific role in the regulation of Abd-B, we examined the distribution of this protein in polytene chromosomes from third instar larvae. Figure 2A shows that dCTCF is present at many sites and that it is localized at the boundaries between the darkly stained DAPI bands and the light interbands. This distribution correlates with a possible general role of dCTCF in the function of insulators present throughout the Drosophila genome.
Figure 2. Distribution of dCTCF on polytene chromosomes of wild type third instar larvae in relation to DNA and gypsy insulator proteins. DNA is stained with DAPI (blue) in all immunofluorescence images.
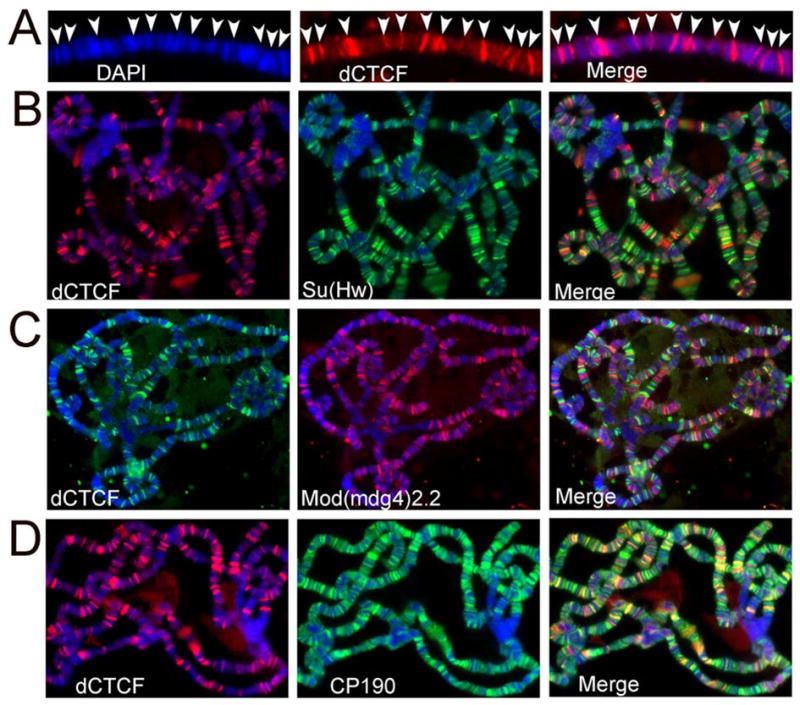
Immunolocalization of (A) dCTCF (red) on polytene chromosomes. Arrows point to the location of dCTCF sites at the boundary of DAPI band and interbands.
(B) dCTCF (red) and Su(Hw) (green) on polytene chromosomes.
(C) dCTCF (green) and Mod(mdg4)2.2 (red) on polytene chromosomes.
(D) dCTCF (red) and CP190 (green) on polytene chromosomes.
The gypsy insulator is also present at several hundred sites in polytene chromosomes at the boundaries between bands and interbands (Pai et al., 2004). To test the existence of a possible relationship between the dCTCF and gypsy insulators, we examined the distribution in polytene chromosomes of the Su(Hw) gypsy insulator protein and dCTCF. The two proteins localize to non-overlapping sets of sites on the chromosomes (Figure 2B). A similar result was obtained with Mod(mdg4)2.2, a second component of the gypsy insulator (Figure 2C). Su(Hw) and Mod(mdg4)2.2 overlap at every chromosome site; a third protein, CP190, overlaps with the other two but is also present at additional sites (Pai et al., 2004). We then tested whether CP190 and dCTCF co-localize in polytene chromosomes. Figure 2D shows that CP190 is present at every dCTCF site in the chromosomes, although not all CP190 sites contain dCTCF. This result suggests that CP190 may be a component of both the gypsy and dCTCF insulators.
dCTCF interacts with the gypsy insulator protein CP190
To further test the possibility that CP190 is a component of dCTCF-containing insulators, we determined the presence of CP190 at the site of the Fab-8 insulator in the bithorax complex. To this end, we carried out FISH with a DNA probe containing Fab-8 insulator sequences simultaneously with immunolocalization using α-dCTCF antibodies. Figures 3A and 3B show that, within the limits of resolution afforded by polytene chromosomes, both dCTCF and CP190 localize to the cytological region where Fab-8 is present. In order to further test whether dCTCF and CP190 are part of a complex, we carried out affinity chromatography using antibodies to the CP190 protein. We have previously used this strategy to isolate a protein complex containing CP190 (Lei and Corces, 2006). CP190, Su(Hw), Mod(mdg4)2.2 and dCTCF each associate with the α-CP190 but not preimmune column (Figure 3C). Since Su(Hw) and Mod(mdg4)2.2 do not overlap with dCTCF in polytene chromosomes, this result suggests that CP190 may form two different complexes, one containing Su(Hw)/Mod(mdg4)2.2 and one including dCTCF. To further test this hypothesis, we carried out chromatin immunoprecipitation (ChIP) analyses in Drosophila S2 cells using primers in the Fab-8 insulator as well as adjacent sequences as a control. Figure 3D shows that both dCTCF and CP190 are present at the Fab-8 insulator but absent from adjacent sequences, whereas Su(Hw) is not present in Fab-8. On the other hand, both CP190 and Su(Hw) are present at the gypsy insulator whereas dCTCF is absent. These results support the conclusion that dCTCF and CP190 can form an active insulator without Su(Hw) and, therefore, CP190 is part of two different insulators, gypsy and Fab-8.
Figure 3. Interaction of dCTCF with gypsy insulator proteins.
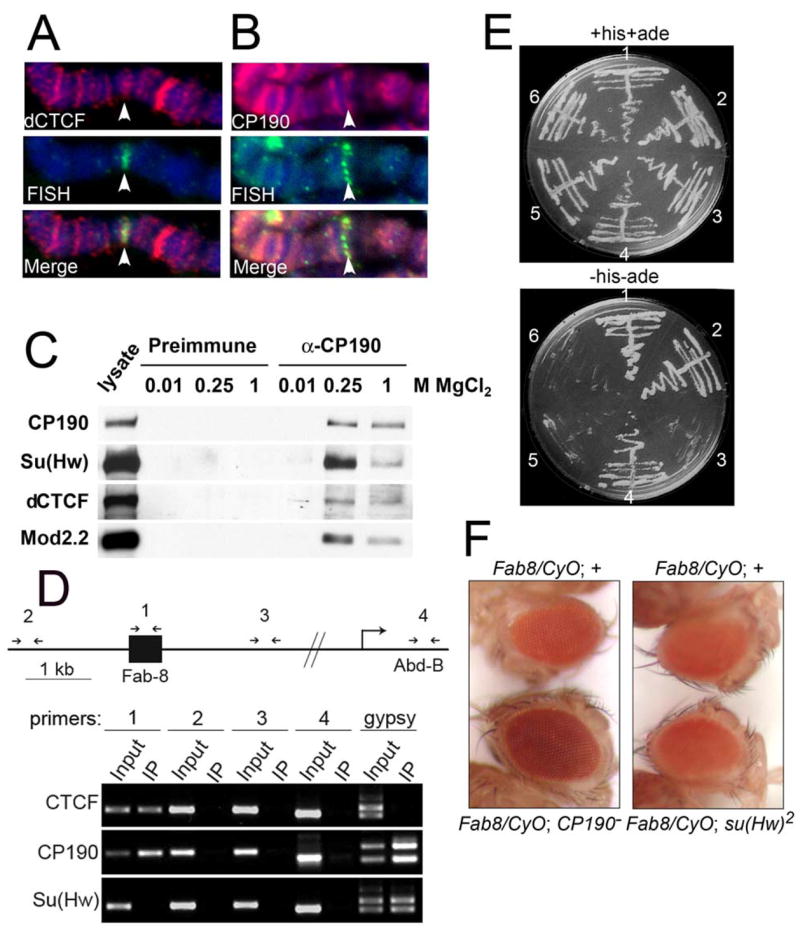
(A) dCTCF is present at the cytological location of the Fab-8 insulator. Red indicates the immunolocalization of dCTCF, green indicates the localization of Fab-8 sequences by FISH and DNA is stained with DAPI in blue. Arrow points to the location of Fab-8.
(B) CP190 is present at the cytological location of the Fab-8 insulator. Red indicates the immunolocalization of CP190, green indicates the localization of Fab-8 sequences by FISH and DNA is stained with DAPI in blue. Arrow points to the location of Fab-8.
(C) Western analysis of control preimmune (lanes 2–4) and α-CP190 column (lanes 5–7) immunoaffinity purifications to verify the presence of CP190, Su(Hw), dCTCF and Mod(mdg4)2.2 in α-CP190 eluates at the indicated concentrations of MgCl2.
(D) Chromatin immunoprecipitation analysis of the Fab-8 insulator. The top part of the panel is a diagram of the 5′ region of the Abd-B gene, including the Fab-8 insulator. Numbered arrows above the diagram indicate the location of primers used in the PCR reactions. The lower portion of the panel shows the result of the electropheretic analysis of the PCR products after immunoprecipitation with antibodies to the proteins indicated on the left. For each primer pair, an input and IP lane are shown. Also shown is a control using primers to the insulator present in the gypsy retrotransposon.
(E) Growth of yeast strains expressing dCTCF and Su(Hw), CP190 or control on nonselective (+histidine +adenine) (top) or selective ( histidine -adenine) (bottom) media. Sector 1, Su(Hw)AD-CP190BD. Sector 2, dCTCFAD-CP190BD. Sector3, dCTCFAD-CP190-BTB domainBD. Sector 4, dCTCFAD-CP190-carboxytermBD. Sector 5, dCTCFAD-emptyBD. Sector 6, CP190BD-emptyAD.
(F) Left panel: Eyes of male flies of the genotype Fab-860.39.2/CyO;+/+ (top) and Fab-860.39.2/CyO; CP1904-1/CP190H31-2 (bottom). Right panel: Eyes of male flies of the genotype Fab-860.39.2/CyO;+/+ (top) and Fab-860.39.2/CyO; su(Hw)2 (bottom).
To more directly assess whether CP190 and dCTCF form a complex in the Fab-8 family of insulators, we tested whether the two proteins interact using the yeast two-hybrid assay. Figure 3E shows the result of this experiment. CP190 and dCTCF appear to interact and this interaction is similar to that observed between CP190 and Su(Hw). The interaction between dCTCF and CP190 does not take place through the BTB domain (residues 1–140) of this protein; rather, the Glu-rich carboxy-terminal region of CP190, between amino acid residues 605 and 1103, interacts with dCTCF (Figure 3E). If dCTCF and CP190 form part of a complex located at putative dCTCF insulator sites throughout the genome as well as the Fab-8 insulator, CP190 may be necessary for the proper function of this insulator. To test this possibility, we analyzed the effect of mutations in the CP190 gene on the activity of the Fab-8 insulator located between the white gene eye enhancer and promoter in the Fab-860.39.2 transgene. Figure 3F shows that this is the case. Mutations in CP190 (CP1904-1/CP190H31-2) partially reverse the phenotypic effect of Fab-8 on the expression of the white gene in the Fab-860.39.2/CyO strain (a similar result was obtained with Fab-860.71.1/CyO, data not shown), suggesting that CP190 is required for the proper function of Fab-8. On the other hand, mutations in the Su(Hw), protein, which is not present at Fab-8 or other dCTCF sites, have no effect on the expression of the same white transgene (Figure 3F). Since CP190 is present at all other genomic sites of dCTCF localization in addition to Fab-8, it is possible that these sites also represent dCTCF insulators and that CP190 is required for the function of dCTCF throughout the genome.
dCTCF and CP190 cooperate with each other to bind to chromatin
dCTCF and CP190 are zinc finger proteins and, in principle, have the ability to bind DNA. In vertebrates, CTCF has been shown to interact with a GC-rich loosely conserved sequence (Kim et al., 2007), and CP190 binds DNA with low affinity and specificity (Pai et al., 2004). In order to determine the role of dCTCF and CP190 in the dCTCF insulator, we analyzed the ability of each of these proteins to bind to polytene chromosomes in the absence of one another. Mutations in dCTCF result in a reduced number of CP190 sites in polytene chromosomes, whereas the binding of Rm62, used as a control, is not affected (Figure 4A). Although the total number of CP190 sites is lower than in wild type chromosomes, those sites present in the dCTCF mutant appear to be of a similar intensity to those in wild type, suggesting that lack of dCTCF affects the binding of CP190 at only a subset of the total number of CP190 sites. The sites where CP190 is still present in the chromosomes of dCTCF mutants are presumed to be those at which Su(Hw) overlaps with CP190. On the other hand, mutations in CP190 completely abolish binding of dCTCF at all sites in polytene chromosomes, suggesting that CP190 is essential for proper binding of dCTCF to DNA (Figure 4B). The same results were obtained whe the JIL-1 kinase or the PEP Zn finger protein were used as controls instead of Rm62 (data not shown).
Figure 4. Levels of dCTCF in wild type and mutant flies. DNA is stained with DAPI (blue) in all panels.
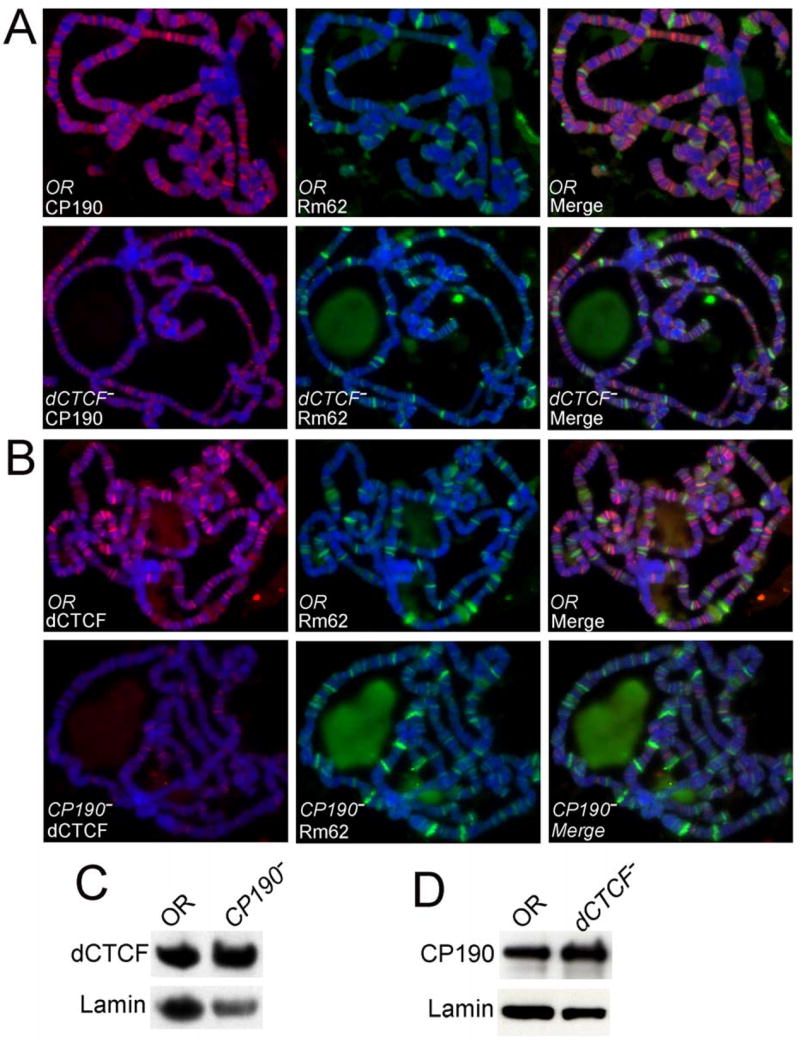
(A) Top row. Immunolocalization of CP190 (red) and of Lip (control, green) in polytene chromosomes from wild type OreR larvae. Bottom row. Immunolocalization of CP190 (red) and of Lip (control, green) in polytene chromosomes from dCTCFy+6/dCTCFy+6 mutant larvae.
(B) Top row. Immunolocalization of dCTCF (red) and of Lip (control, green) in polytene chromosomes from wild type larvae. Bottom row. Immunolocalization of dCTCF (red) and of Lip (control, green) in polytene chromosomes from CP190P1/CP190P11 mutant larvae.
(C) Western analysis of protein extracts from wild type (OR) and CP190P1/CP190P11 larvae. Lamin was used as a control.
(D) Western analysis of protein extracts from wild type (OR) and dCTCFy+6/dCTCFy+6 larvae. Lamin was used as a control.
The absence of dCTCF protein in polytene chromosomes of larvae carrying a mutation in the CP190 gene could be due to an effect on the expression of the dCTCF gene. An effect on CP190 transcription could also be an explanation for the reduced number of CP190 sites in chromosomes of dCTCF mutant larvae. To test whether this is the case, we carried out western analyses of protein extracts obtained from larvae bearing each of the two mutations, using lamin as a loading control. Levels of dCTCF appear to be unchanged in larvae carrying a mutation in CP190 compared to wild type (Figure 4C). Likewise, larvae mutant in the dCTCF gene accumulate similar levels of CP190 than wild type larvae (Figure 4D). These results suggest that the observed differences in the presence of the dCTCF and CP190 proteins in polytene chromosomes of mutant flies are due to decreased binding to DNA rather than lower levels of the proteins, suggesting that the two proteins cooperate in some fashion in their interaction with DNA.
dCTCF may be the DNA binding component in a subset of insulators that share the CP190 protein
The Su(Hw) protein component of the gypsy insulator and the dCTCF component of the Fab-8 insulator are both present at the boundaries between bands and interbands in polytene chromosomes but the two proteins do not overlap, suggesting that their sites of localization are different (Figure 2). Nevertheless, both sets of sites share the CP190 protein (Figures 5A and 5B). To further analyze the pattern of overlap between these proteins we carried out immunolocalization experiments in which the CP190 protein was detected using FITC-conjugated secondary antibodies whereas Su(Hw) and dCTCF were simultaneously detected using secondary antibodies conjugated to Texas red (Figure 5C). When Su(Hw) and dCTCF are visualized together, the number of sites where CP190 is present alone decreases considerably when compared to the results obtained with each protein separately (Figure 5). An approximate quantification of the sites of overlap suggests that Su(Hw) and dCTCF are each present at 1/3 of all sites of CP190 localization in the genome and, therefore, approximately 1/3 of CP190 sites are not occupied by either the Su(Hw) or dCTCF proteins. These results support a model in which each of these two proteins may be part of a distinct insulator and that the dCTCF and gypsy insulators may be subclasses of a more general insulator defined by the CP190 protein. A prediction of this model is that, in dCTCF mutants, the extent of overlap between Su(Hw) and CP190 should be more extensive than in wild type flies, since the sites were CP190 and dCTCF are normally present should be absent in these mutants. Figure 5D shows that this is indeed the case, and that Su(Hw) is able to bind normally to the chromatin in the absence of dCTCF, supporting the conclusion that gypsy and dCTCF insulators share the CP190 protein but are independent. Figure 5D also shows the existence of sites in which CP190 does not overlap with Su(Hw), suggesting the existence of at least a third protein required for proper binding of CP190 to a third subclass of insulators that lack Su(Hw) and dCTCF. A second prediction of the model is that mutation of the su(Hw) and dCTCF genes simultaneously will have an additive effect in the phenotype with respect to single mutations in each of the two genes separately. This is indeed the case. Null mutations in su(Hw), such as su(Hw)2, are viable but display female sterility; null mutants in dCTCF, such as dCTCFy+6, show late pupal lethality. Flies carrying both mutations die during late larvae/early pupae stages, an earlier stage of development (data not shown).
Figure 5. Co-localization of dCTCF and gypsy insulator proteins on polytene chromosomes from wild type larvae. DNA is stained with DAPI (blue) in all panels.
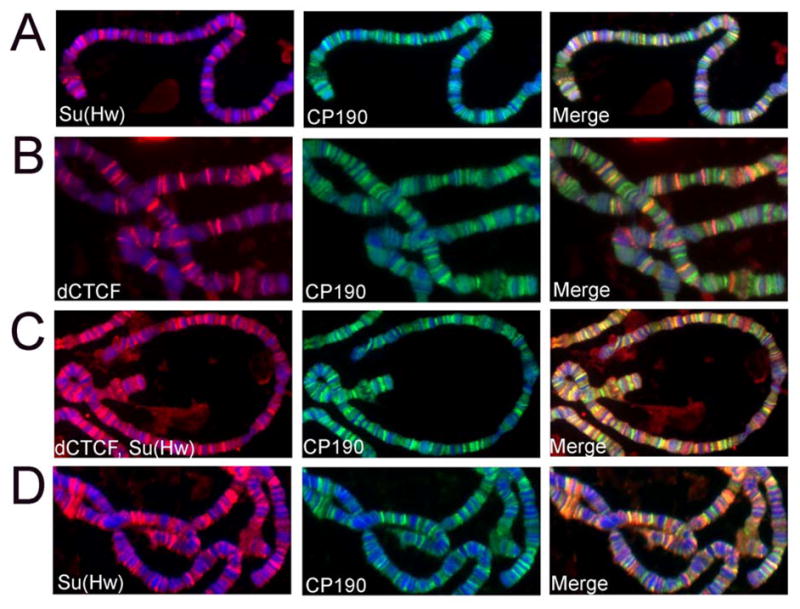
(A) Su(Hw) (red) and CP190 (green) on polytene chromosomes.
(B) dCTCF (red) and CP190 (green) on polytene chromosomes.
(C) dCTCF plus Su(Hw) (red) and CP190 (green) on polytene chromosomes.
(D) Su(Hw) (red) and CP190 (green) on polytene chromosomes of dCTCFy+6 mutants.
dCTCF is present in the same insulator bodies as gypsy insulator proteins
The observed genetic interaction between mutations in su(Hw) and dCTCF is suggestive of parallel or related functions for the two subclasses of insulators. In the case of the gypsy insulator, it has been shown that insulator activity correlates closely with the formation of insulator bodies (Capelson and Corces, 2005; Capelson and Corces, 2006), which are sites in the nucleus where several insulator sites come together while causing the chromatin fiber to form loops that are presumed to coincide with independent domains of gene expression (Byrd and Corces, 2003; Gerasimova et al., 2000). To test whether the dCTCF protein is present in insulator bodies with a nuclear distribution similar to those of gypsy insulator proteins, we examined the pattern of dCTCF localization in nuclei from diploid imaginal disc cells. Figure 6A shows that dCTCF is present in the nucleus in a pattern similar to that of gypsy insulator proteins, suggesting that dCTCF may also form insulator bodies. Interestingly, the pattern of dCTCF localization overlaps perfectly with that of CP190 (Figure 6A). Since CP190 forms insulator bodies where other gypsy insulator proteins are also located, this result suggests that dCTCF and gypsy insulator bodies may overlap. To further test this possibility, we examined the distribution of a second protein component of the gypsy insulator, Mod(mdg4)2.2. Although this protein does not overlap with dCTCF in polytene chromosomes, the two proteins coincide perfectly in nuclei of diploid cells (Figure 6B), supporting the idea that dCTCF and gypsy insulator bodies are the same or are located closely in the nucleus.
Figure 6. Localization of dCTCF and gypsy insulator proteins in diploid nuclei of wild type and mutant larvae. DNA is stained with DAPI (blue) in all images.
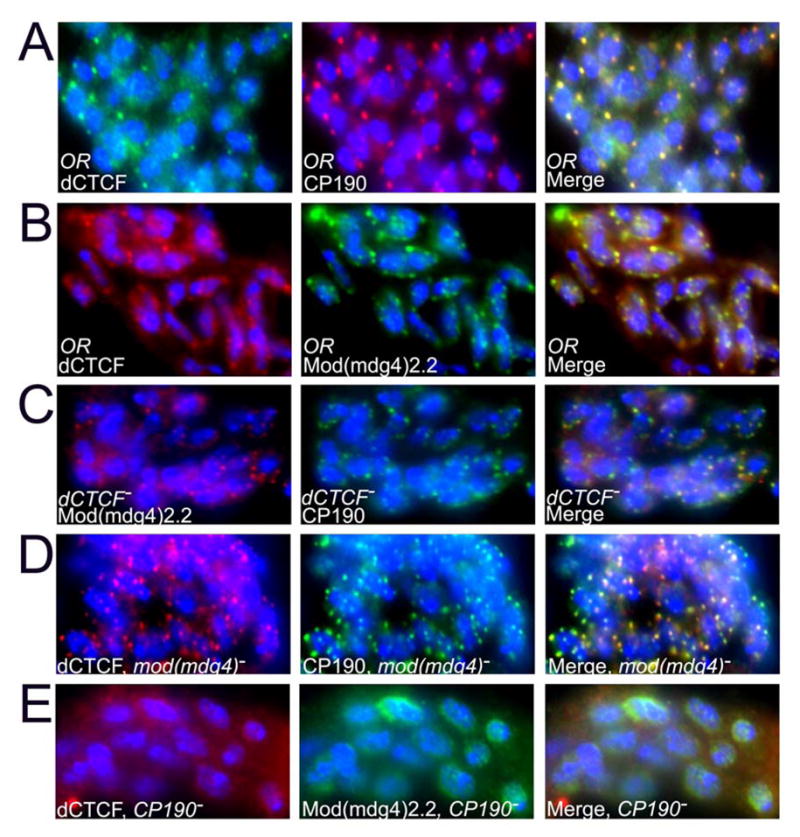
(A) Immunolocalization of dCTCF (green) and CP190 (red) in diploid nuclei from imaginal disc cells of wild type larvae.
(B) Immunolocalization of dCTCF (red) and Mod(mdg4)2.2 (green) in diploid nuclei from imaginal disc cells of wild type larvae.
(C) Immunolocalization of Mod(mdg4)2.2 (red) and CP190 (green) in diploid nuclei from imaginal disc cells of dCTCFy+6/dCTCFy+6 mutant larvae.
(D) Immunolocalization of dCTCF (red) and CP190 (green) in diploid nuclei from imaginal disc cells of mod(mdg4)T6 mutant larvae.
(E) Immunolocalization of dCTCF (red) and Mod(mdg4)2.2 (green) in diploid nuclei from imaginal disc cells of CP190P1/CP190P11 mutant larvae.
To further determine the relationship between dCTCF and Mod(mdg4)2.2 at insulator bodies, we analyzed the effect of mutations in the genes encoding the two proteins on insulator body integrity. Mutations in dCTCF do not affect the presence of either Mod(mdg4)2.2 or CP190 at insulator bodies (Figure 6C). Similarly, the mod(mdg4)T6 mutation, which affects the Mod(mdg4)2.2 isoform, does not affect the distribution of dCTCF or its co-localization with CP190 at insulator bodies. These results suggest that dCTCF and gypsy insulators co-localize in the nucleus but the integrity of dCTCF insulators is not necessary for gypsy insulator body formation and vice-versa. To the contrary, mutations in CP190 affect the integrity of the insulator bodies formed by dCTCF and Mod(mdg4)2.2; in the absence of CP190 protein, both dCTCF and Mod(mdg4)2.2 fail to form insulator bodies, have an aberrant cellular localization and dCTCF appears distributed throughout the cytoplasm (Figure 6E). These results suggest that, although dCTCF and gypsy insulator bodies are present at the same nuclear location and share the CP190 protein, they can form independent of each other. This observation on the integrity of insulator bodies formed by dCTCF and Mod(mdg4)2.2 in the presence of mutations in either gene correlates well with analyses of these mutations on the function of each insulator. Mutations in Mod(mdg4)2.2 or Su(Hw) affect the function of the gypsy insulator, as determined by the phenotypic reversion of gypsy-induced mutations such as y2, but they do not affect the phenotypic effect of the Fab-8 insulator on the expression of the white gene (data not shown). Mutations in dCTCF do not affect the function of gypsy insulator in gypsy-induced mutations (data not shown) but they affect the Fab-8 insulator. On the other hand, mutations in CP190 impair the function of both the gypsy and dCTCF insulators. These results suggest that Su(Hw)/Mod(mdg4)2.2 and dCTCF form two subfamilies of insulators that share the CP190 protein. Both subfamilies form insulator bodies that colocalize in the nucleus but can form and function independently of each other.
DISCUSSION
Insulators are thought to compartmentalize the genome into independent domains of gene expression by creating chromatin loops between individual insulator sites and physically tethering the chromatin fiber to a fixed nuclear substrate such as the nuclear matrix (Capelson and Corces, 2004). In the case of the gypsy insulator, the formation of loops is manifested in the formation of insulator bodies, structures in which multiple individual insulators coalesce in specific nuclear locations (Gerasimova and Corces, 1996). Formation of large insulator bodies has not been shown for other Drosophila or vertebrate insulators. However, the chicken CTCF insulator interacts with the nucleolus through the nucleophosmin protein and CTCF is present in the nuclear matrix fraction, suggesting the need for a nuclear scaffold for its function (Yusufzai and Felsenfeld, 2004; Yusufzai et al., 2004).
In order to address this issue, we analyzed the properties and functional characteristics of the Drosophila dCTCF protein. dCTCF in Drosophila has been shown to interact with the Fab-8 insulator in the Abd-B gene of the bithorax complex; mutations in the binding site that abolish dCTCF binding also affect Fab-8 insulator function (Moon et al., 2005). Using antibodies to Drosophila dCTCF, we find that this protein is located at several hundred sites in polytene chromosomes from third instar larvae, suggesting a general role for dCTCF in nuclear biology rather than a specific function in the regulation of Abd-B expression. In addition, we find that dCTCF is located at the borders of bands and interbands in polytene chromosomes. This pattern of localization appears to be a hallmark of insulator proteins in Drosophila, suggesting that dCTCF may be part of an insulator with broad function similar to that of CTCF in vertebrates, where CTCF is the main insulator found thus far.
Gypsy insulator proteins such as Su(Hw) or Mod(mdg4)2.2 are also present at the boundaries between bands and interbands, but only in a subset of all possible boundaries. This observation prompts the question of whether boundaries that lack Su(Hw)/Mod(mdg4)2.2 contain a different insulator. If bands and interbands correspond to domains of gene expression established or maintained by insulators, and the creation of these domains requires the formation of chromatin loops between adjacent insulators, the proteins present at each insulator site must be able to interact in order to form these loops. Therefore, insulator proteins located at non-gypsy insulators should then interact with Su(Hw), Mod(mdg4)2.2 or CP190. A solution to this issue is the finding that, although dCTCF does not co-localize with Su(Hw) or Mod(mdg4)2.2 in polytene chromosomes, sites of dCTCF also contain CP190. Therefore, CP190 may be a component of both gypsy and dCTCF insulators that can mediate interactions between the two. Support for the conclusion that CP190 is a component of the dCTCF insulator comes from the observation that dCTCF co-purifies with CP190 and that the two proteins are present at the Fab-8 insulator and interact in the yeast two-hybrid assay. These interactions have in vivo significance, as mutations in CP190 affect the function of the Fab-8 insulator. These results suggest that, although dCTCF and Su(Hw)/Mod(mdg4)2.2 are not present at the same chromosomal locations, they share the CP190 insulator protein. Su(Hw) and dCTCF are very similar proteins, containing 11 Zn fingers that can mediate DNA binding, and they both interact with CP190. It is possible that the Zn fingers in CP190 mediate protein interactions rather than binding to DNA, and that this protein is tethered to DNA through interactions with Su(Hw) or dCTCF. This is supported by the fact that CP190 is absent in two different subsets of chromosomal locations in larvae carrying mutations in either the su(Hw) or dCTCF genes, and suggests that the Su(Hw) (gypsy) and dCTCF insulators are two subfamilies of a much larger class of insulators whose defining characteristic is the presence of CP190. In agreement with this model, mutations in su(Hw) affect the function of the gypsy insulator but not Fab-8, whereas mutations in dCTCF affect Fab-8 but not gypsy and mutations in CP190 affect both. Nevertheless, the role of CP190 in the dCTCF insulator appears more complex than to passively be recruited to DNA by this protein. In a CP190 mutant, Su(Hw) binds normally to polytene chromosomes (Pai et al., 2004) but dCTCF is absent, suggesting that CP190 is necessary for proper binding of dCTCF. It would be interesting to examine whether CTCF in vertebrates also requires a CP190-like protein for proper DNA binding.
The formation of chromatin loops representing independent domains of gene expression can be visualized in the case of the gypsy insulator by the formation of insulator bodies that attach to the nuclear matrix. These structures represent nuclear sites where multiple individual insulators coalesce. Insulator bodies have not been detected for the CTCF protein in vertebrate cells, although CTCF is present in the nuclear matrix fraction and may attach the chromatin fiber to this structure (Yusufzai and Felsenfeld, 2004). We find that in Drosophila the dCTCF protein forms insulator bodies that appear similar to those formed by gypsy insulator proteins in number and distribution, suggesting that the dCTCF insulator may function by mechanisms similar to that of the gypsy insulator. The reason for the apparent discrepancy between the formation of insulator bodies by the Drosophila and vertebrate CTCF proteins is not clear. It is possible that CTCF insulator bodies in vertebrates are made of a lower number of individual insulators, making the insulator bodies difficult to resolve by light microscopy. Alternative, or in addition, the increased complexity in DNA content and number of chromosomes in chicken or mouse cells may result in a large number of insulator bodies that make their detection challenging. Finally, it is possible that the different appearance of insulator bodies in Drosophila versus vertebrate cells is due to the quality of available antibodies to the CTCF prtein, as we have observed a punctate pattern of very small dots with poor quality antibodies to gypsy insulator proteins.
dCTCF insulator bodies in Drosophila coincide with those formed by CP190, as one would expect from their pattern of co-localization in polytene chromosomes. Surprisingly, dCTCF insulator bodies also co-localize with those formed by Su(Hw) and Mod(mdg)2.2 in spite of their presence at different genomic sites, and each can be formed in the absence of the other. CP190 is nevertheless essential for the formation of both types of insulator bodies; in its absence, dCTCF and Mod(mdg4)2.2 are miss-localized. These observations can be explained by two different models that have different implication for the role of nuclear architecture in insulator function. One possibility is that Su(Hw) and dCTCF insulators form independent insulator bodies but both co-localize in the same regions of the nucleus. A corollary of this model is the possible existence of special nuclear compartments where insulator bodies are located. A second model would suggest that Su(Hw) and dCTCF insulators form the same insulator bodies through interactions between the CP190 protein present in both insulators. This model would not require the existence of a specific nuclear compartment where insulator bodies form, since the co-localization is due to the presence of CP190 in both types of insulators. In the second model, although the Su(Hw) and dCTCF insulators interact at insulator bodies through CP190, each type of insulator can still form nuclear bodies in the absence of the other.
Su(Hw) and dCTCF co-localize with CP190 at approximately two thirds of all the sites where this protein is present in polytene chromosomes, suggesting that at least one additional DNA binding protein may tether CP190 to chromosomes. This would suggest the existence of a third subclass of CP190 insulators similar to Su(Hw) and dCTCF. If CP190 is the critical protein in the establishment of insulator bodies, why have three different proteins to recruit CP190 to the DNA? One explanation could be that the use of three different DNA binding proteins affords additional possibilities in the regulation of insulator activity during cell differentiation and development. For example, lack of expression of one of the DNA binding proteins, or a covalent modifications that would interfere with its ability to bind DNA or interact with CP190, would render one subclass of insulators inactive while allowing normal function of the other two subclasses. The flexibility in the regulation of insulator activity provided by the use of three different DNA binding proteins raises the question of why vertebrates appear to use mostly CTCF. Although a vertebrate homologue of Su(Hw) does not appear to exist, it is possible that vertebrates do have a homologue of the third DNA binding protein. Identification of this protein in Drosophila may provide additional insights into possible mechanisms of regulating insulator function and the role of insulators in the control of nuclear organization during vertebrate development.
EXPERIMENTAL PROCEDURES
Fly strains and yeast two hybrid assays
Fly stocks were maintained in standard medium at 25°C. A P element insertion into the 5′ UTR of the dCTCF gene, P{EPgy2}CTCFEY15833, was obtained from the Indiana Stock Center. Flies carrying this insertion are viable and produce reduced but detectable levels of dCTCF (see Figure 1). In order to produce null mutations in the dCTCF gene, we mobilized the P element using the y1w1118; P{Δ2–3}Sb/TM6 strain as a source of transposase. Three different alleles named dCTCFy+1, dCTCFy+2 and dCTCFy+6 were obtained and analyzed further. The Fab-860.39.2/CyO and Fab-860.71.1/CyO strains carrying a white transgene with the Fab-8 insulator inserted between the eye enhancer and the promoter were obtained from Dr. Paul Schedl.
Two hybrid assays were performed using yeast strain pJ694A and plasmids and protocols obtained from Clontech. Bait and target fusion proteins were produced constitutively under the control of the ADH1 promoter. Co-transformants were plated on media lacking tryptophan and leucine (non-selective plates for the reporter gene) or on plates lacking tryptophan, leucine, histidine and adenine (selective plates for the reporter gene). Single colonies were streaked out on selective plates to obtain the plates showed in Figure 3.
Protein analyses
In order to obtain dCTCF protein for antibody production, the carboxy terminus of the dCTCF gene was amplified from cDNA GH14774 (Research Genetics) by PCR using primers 5′ GARGATTCGGCGAATGCTCGCGAA 3′ and 5′ CTAAGAGTCCTGCTCAATCATATCCAT 3′ and cloned into the pET100/D-TOPO vector (Invitrogen) in-frame with the N-terminal His6 tag. Purified His-dCTCF was used to immunize rats and rabbits and antibodies were affinity purified by standard procedures.
Immunoaffinity purification of CP190 complexes was carried out as previously described (Lei and Corces, 2006). For western analysis, larvae were homogenized in either HEMGN buffer (25 mM Hepes-KOH pH 7.6, 0.1 mM EDTA, 12.5 mM MgCl2, 10% glycerol, 0.1% NP40, 1 mM DTT, 0.1 mM PMSF, 0.3 M KCl) or in cold sodium dodecyl sulfate (SDS) sample buffer (20 mM sodium phosphate, 2% SDS, 0.001% bromophenol blue, 0.2 M DTT, 2% glycerol). In the former case an equal volume of 2X SDS reducing buffer was added and then boiled for 5 minutes. The extract was spun, the supernatant run on a SDS-10% polyacrylamide gel and transferred onto a polyvinylidene difluoride (PVDF) membrane. The presence of dCTCF and CP190 was detected using the Super signal West Pico chemiluminescent substrate from Pierce.
Immunocytochemistry and in situ hybridization of polytene chromosomes
Salivary glands were fixed in 3.7% formaldehyde, 45% acetic acid mix for 2 min and squashed in 45% acetic acid on subbed slides. The slides were incubated overnight in dilution buffer containing primary antibodies at 1:1000 dilution for α-CP190, 1:100 for α-Mod(mdg4)2.2 and 1:50 for α-CTCF and α-Su(Hw). Slides were then washed in antibody dilution buffer three times and incubated with FITC- or Texas red-conjugated goat anti-rabbit IgG (Jackson Laboratories) at 1:250 dilution for 1 h at room temperature or 30′ at 37°C. Slides were stained for 2 min with 4′,6-diamidino-2-phenylindole (DAPI), 0.5 μg/ml and mounted in Vectashield anti fade mounting medium (Vector Laboratories).
To detect the location of Fab-8 sequences on polytene chromosomes we used a 1180 bp DNA fragment obtained by PCR of the bithorax locus using primers 5′AGTTTTGCCTCCCGACTTCGACGGC 3′ and 5′AGGTGTAGCGGGCTTTGGGCTACCA 3′. Digoxigenin-labeled DNA was prepared using the Prime-A-Gene random priming kit (Promega). The labeled probe was ethanol precipitated and stored in hybridization buffer [4x SSC (1X SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 50% formamide, 1X Denhardt’s containing 0.4 mg/ml of salmon sperm DNA] until ready to use. Polytene chromosomes from third instar larvae were prepared as described above, except that the glands were fixed in acetic acid-lactic acid mixture (1 acetic acid: 2 water: 3 lactic acid). Slides were heated at 65°C in 2X SSC for30 min, dehydrated using an ethanol series, and denatured in 0.07 M NaOH. For hybridization of DNA, boiled probes were added to the slide and covered immediately with a cover slip. The slide and cover slip were sealed with rubber cement and incubated at 37°C overnight in a humidified chamber. Following hybridization, cover slips were removed and the slides were washed in 2X SSC at 37°C, then at room temperature in 1XPBS and finally in antibody dilution buffer. Antibody detection and visualization were carried out as outlined above.
Chromatin Immunoprecipitation
Approximately 1×107 Drosophila Kc cells were crosslinked with 1% formaldehyde for 10min at room temperature. Nuclear lysates were sonicated to generate 200–1000bp DNA fragments. Chromatin immunoprecipitation was then performed with rabbit α-CTCF, α-CP190, or α-Su(Hw) antibodies. PCR amplification of the resulting DNA was performed using the following primers: Fab-8, 5′GGCACAATCAAGTTAATGTTGG 3′ and 5′-GCAAGCGAAGAGTTCCATTC 3′ (Moon et al., 2005); 1500bp upstream of Fab-8, 5′GAGAGTCCAGGCCCAAGGAC 3′ and 5′CCCACCAGGGGCGATATTTG 3′; 1500bp downstream of Fab-8, 5′GCCTGTTTCGCGGAACTGAT 3′ and 5′CGTGTGGAATTCTGTTCTGTCAC 3′; Abd-B, 5′CACTGGTGCATCTTGGCGGC 3′ and 5′GCAGCTGACCGAGCGACAGG 3′; gypsy, 5′CGCAAAAACATTGCATATTTTCGGC 3′ and 5′CGGCTAAATGGTATGGCAAGAAAAGG 3′.
Figure 7. Possible models to explain the formation of insulator bodies by dCTCF. In both panels insulator proteins are represented by differently colored ovals: Su(Hw) is shown in blue, Mod(mdg4)2.2 in green, dCTCF in purple and CP190 in pink.
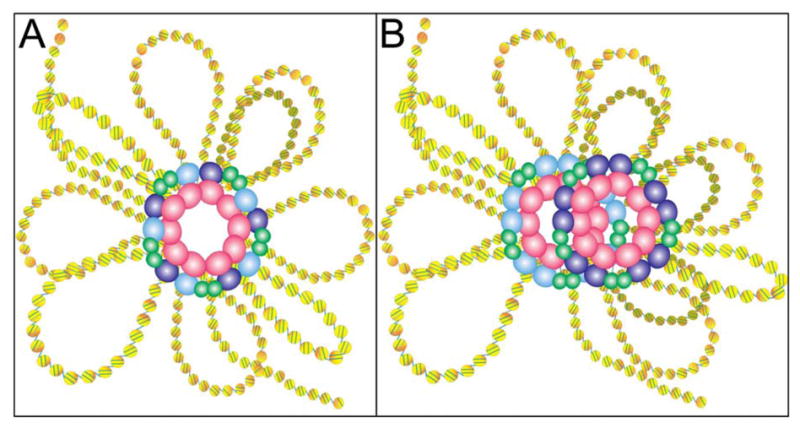
(A) Su(Hw) and dCTCF bind different insulator sequences but coalesce in the same insulator bodies through interactions between the CP190 protein present in both insulators
(B) Su(Hw) and dCTCF form independent insulator bodies but both co-localize in the same region of the nucleus. This may imply the existence of special nuclear compartments where insulator bodies are located.
Acknowledgments
We would like to thank Drs. P. Shedl and G. Shanower for transgenic flies with the Fab-8 insulator, Helen Baxter for technical assistance, Dr. Maya Capelson for helpful discussions and Dr. Elena Makeyeva for help in the purification of the dCTCF fusion protein. Work reported here was supported by U.S. Public Health Service Award GM35463 from the National Institutes of Health. E.L. was supported by a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research.
References
- Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab- 7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127:779–790. doi: 10.1242/dev.127.4.779. [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Belozerov VE, Majumder P, Shen P, Cai HN. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. Embo J. 2003;22:3113–3121. doi: 10.1093/emboj/cdg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 2003;17:664–675. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner K, Roth P, Schotta G, Krauss V, Saumweber H, Reuter G, Dorn R. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics. 2000;155:141–157. doi: 10.1093/genetics/155.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd K, Corces VG. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J Cell Biol. 2003;162:565–574. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Corces VG. Boundary elements and nuclear organization. Biol Cell. 2004;96:617–629. doi: 10.1016/j.biolcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell. 2005;20:105–116. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Capelson M, Corces VG. SUMO conjugation attenuates the activity of the gypsy chromatin insulator. Embo J. 2006;25:1906–1914. doi: 10.1038/sj.emboj.7601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT. CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002;295:345–347. doi: 10.1126/science.1065982. [DOI] [PubMed] [Google Scholar]
- Estrada B, Casares F, Busturia A, Sanchez-Herrero E. Genetic and molecular characterization of a novel iab-8 regulatory domain in the Abdominal-B gene of Drosophila melanogaster. Development. 2002;129:5195–5204. doi: 10.1242/dev.129.22.5195. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Burgess-Beusse B, Farrell C, Gaszner M, Ghirlando R, Huang S, Jin C, Litt M, Magdinier F, Mutskov V, et al. Chromatin boundaries and chromatin domains. Cold Spring Harb Symp Quant Biol. 2004;69:245–250. doi: 10.1101/sqb.2004.69.245. [DOI] [PubMed] [Google Scholar]
- Filippova GN, Thienes CP, Penn BH, Cho DH, Hu YJ, Moore JM, Klesert TR, Lobanenkov VV, Tapscott SJ. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet. 2001;28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–2107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Harrison DA, Gdula DA, Coyne RS, Corces VG. A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev. 1993;7:1966–1978. doi: 10.1101/gad.7.10.1966. [DOI] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador M, Corces VG. Setting the boundaries of chromatin domains and nuclear organization. Cell. 2002;111:151–154. doi: 10.1016/s0092-8674(02)01004-8. [DOI] [PubMed] [Google Scholar]
- Labrador M, Corces VG. Extensive exon reshuffling over evolutionary time coupled to trans-splicing in Drosophila. Genome Res. 2003;13:2220–2228. doi: 10.1101/gr.1440703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet. 2006;38:936–941. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Lei EP, Ghosh D, Corces VG. The Centrosomal Protein CP190 Is a Component of the gypsy Chromatin Insulator. Mol Cell. 2004;16:737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Shaffer CD, Wuller JM, Elgin SC. Preparation of Drosophila nuclei. Methods Cell Biol. 1994;44:185–189. doi: 10.1016/s0091-679x(08)60913-9. [DOI] [PubMed] [Google Scholar]
- Yusufzai TM, Felsenfeld G. The 5′-HS4 chicken {beta}-globin insulator is a CTCF-dependent nuclear matrix-associated element. PNAS. 2004;101:8620–8624. doi: 10.1073/pnas.0402938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF Tethers an Insulator to Subnuclear Sites, Suggesting Shared Insulator Mechanisms across Species. Molecular Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]


