Abstract
This paper demonstrates the application of mutual information based coregistration of radionuclide and magnetic resonance imaging (MRI) in an effort to use multimodality imaging for noninvasive localization of stem cells grafted in the infarcted myocardium in rats. Radionuclide imaging such as single photon emission computed tomography (SPECT) or positron emission tomography (PET) inherently has high sensitivity and is suitable for tracking of labeled stem cells, while high-resolution MRI is able to provide detailed anatomical and functional information of myocardium. Thus, coregistration of PET or SPECT images with MRI will map the location and distribution of stem cells on detailed myocardium structures. To validate this coregistration method, SPECT data were simulated by using a Monte Carlo-based projector that modeled the pinhole-imaging physics assuming nonzero diameter and photon penetration at the edge. Translational and rotational errors of the coregistration were examined with respect to various SPECT activities, and they are on average about 0.50 mm and 0.82°, respectively. Only the rotational error is dependent on activity of SPECT data. Stem cells were labeled with 111 Indium oxyquinoline and grafted in the ischemic myocardium of a rat model. Dual-tracer small-animal SPECT images were acquired, which allowed simultaneous detection of 111In-labeled stem cells and of [99mTc]sestamibi to assess myocardial perfusion deficit. The same animals were subjected to cardiac MRI. A mutual-information-based coregistration method was then applied to the SPECT and MRIs. By coregistration, the 111 In signal from labeled cells was mapped into the akinetic region identified on cine MRIs; the regional perfusion deficit on the SPECT images also coincided with the akinetic region on the MR image.
Keywords: SPECT, MRI, Coregistration, Myocardial ischemia, Stem cells, Mutual information
Introduction
The potential for stem cells to repair damaged heart tissue is being studied intensively. The rationale is to restore the contractile function of the heart by replacing the cardiomyocytes that are lost as a result of an ischemic insult. Noninvasive imaging would play a unique role to unfold the potential of stem cells for treatment of myocardial infarction. Furthermore, the combination of multiple imaging modalities would allow the collection of anatomical, functional, and metabolic information from the same subject. Thus, positron emission tomography (PET)/computed tomography (CT) and single photon emission computed tomography (SPECT)/CT hybrid scanners have been made commercially available for more than seven years. Compared to CT, magnetic resonance imaging (MRI) is more suitable for soft tissue (e.g., heart) delineation. It provides superior spatial resolution and is routinely used for the measurement of global cardiac function and regional contractile function. Because of its unique capability of applying noninvasive fiducial markers (or tags) on the myocardium, tagged MRI allows quantification of regional contractile function both in humans [1, 2] and in small animals [3, 4]. On the other hand, radionuclide imaging modalities including PET and SPECT are inherently more sensitive than MR detection, and as such only tracer quantity of radiolabeled material has to be used for various functional assays (i.e., receptor binding, and cardiac perfusion) although the spatial resolution achievable on PET/SPECT is 1 order-of-magnitude lower than MRI. Therefore, the combination of MRI and PET/SPECT would achieve optimized spatial resolution and detection sensitivity, respectively.
The multimodality approach requires coregistration of images acquired from different modalities in order to bring into the same anatomical reference all the available functional measurements [5]. In this paper, we presented our initial effort to coregister cardiac MRIs with SPECT images by using a mutual-information-based algorithm [6–15]. These images were acquired from the same rat that was subjected to myocardial infarction and grafting of stem cells that were labeled with [111In]Indium oxyquinoline [3]. Mutual information is defined as the amount of information that one image contains about the other. Mutual-information-based coregistration methods [6–10, 15] assume the statistical dependence between the intensity values of two images, and the mutual information is maximized after two images have been completely aligned. The majority of mutual-information-based methods are designed for rigid and affine registration. We will examine this approach in the context of localization of 111In signal that is associated with the labeled cells to the myocardium on the MRI image by coregistering the simultaneously acquired 99mTc(Sestamibi) SPECT image of the heart to the MR image.
Materials and Methods
Multimodality Image Coregistration
The intensity-based registration methods are the popular approaches for coregistration of multimodality images. They work by maximizing a similarity measure such as mutual information of the two images acquired from two different modalities, and assuming the statistical dependence between the intensity values of two images [15]. That is, the intensities of the myocardial region in the SPECT and MRIs are assumed to be statistically correlated. The coregistration is completed after one image has been optimally aligned with the other one, when the mutual information has been maximized.
The mutual information I(A,B) of the two images A and B can be defined as a combination of two separate marginal entropies, H(A) and H(B), and the joint entropy, H(A,B). In addition, to make the coregistration algorithm robust to changes in overlap, a normalized measure of mutual information [10] has been proposed:
This normalized mutual-information measure is used in our study for coregistration of SPECT and MR images [15].
Entropy can be interpreted as a measure of uncertainty, variability, or complexity [7, 15]. The entropy of an image can be computed by first estimating its probability distribution of intensities {pi}, and then measuring the Shannon entropy, . Similarly, the joint entropy of the two images is estimated by computing a normalized joint histogram of paired intensities {pi,j} and further the Shannon measure of entropy, . Notably, only the overlapped regions of two images are used to compute the probability distribution of intensities in each image, as well as the joint histogram of paired intensities. Actually, we only need to compute the joint histogram of the paired intensities, and the marginal distribution of each image can be directly obtained by summing over the rows or the columns of the joint histogram, respectively.
Mathematically, the mutual-information-based coregistration of two images A and B can be completed by searching for an optimal transformation T (from image A to image B) that maximizes the mutual information between the original image A(x) and the transformed image B(T(x)):
where x is a voxel in image A, and its estimated corresponding voxel in image B is T(x). Suppose that the imaging spaces for images A and B are VA and VB, respectively, then the overlap region of images A and B is VA ∩ T−1 (VB), where T−1 is an inverse transformation of T. Therefore, the marginal entropies, H(A(x)) and H(B(T(x))), and the joint entropy, H(A(x), B(T(x))), should be computed from the overlap space VA ∩ T−1 (VB), in order to make the coregistration method robust to changes in overlap.
The performance of the coregistration method depends not only on the definition of objective cost function [15], but also on the optimization technique used. Most mutual-information-based coregistration methods use a gradient-based optimization method, which might lead to local minima if not regularized. To overcome this limitation, multiresolution implementation of the coregistration method is always used. However, the commonly used multiresolution local optimization methods can still get trapped in local minima as demonstrated by previous studies [12]. To reduce the chances of being captured by local minima, we adopted a novel hybrid global–local optimization method [12] for coregistration of cardiac SPECT and MRI.
To further improve the robustness of mutual-information-based coregistration method to noisy background particularly in SPECT images, we used a preprocessing step of extracting a region of interest, such as a cardiac region, from SPECT and MRI, respectively, before coregistration. Currently, we extract a region of interest by manually placing a boundary box.
Validation
To evaluate the performance of mutual-information-based coregistration method used in this study, we simulated a pinhole SPECT system and generated six sets of volumetric SPECT images from MR phantoms by using a variety of noise realizations and transformations. The simulated system was based on the Prism 3000XP system we used to acquire real data. Compared to the real system, the simulated system used the same pinhole (tungsten knife-edge, 3-mm diameter, 120° opening angle), same focal length (24 cm), but a slightly larger radius of rotation (6 cm) to allow various transformations of the phantom in space. MR images acquired from different time-points of the cardiac cycle are summed to create an average MRI (Fig. 1a). This procedure is necessary for coregistration with SPECT images because the SPECT images were not gated. The voxel size in average MR image was 0.27 mm in the xy plane and 1.0 mm along the z-axis. This average MRI (Fig. 1a) was manually segmented into five components: chest, liver, lung, myocardium, and ventricles (Fig. 1b), based on their uptake of the tracer (mibi) estimated from a realistic 99mTc MIBI study in mice [16]. Finally, the pinhole SPECT data were simulated from the phantom for various activity levels, using a Monte Carlo method that modeled the pinhole-imaging physics (nonzero diameter and photon penetration through the edge) by analytically computing the length of the path in the collimator of each photon [17]. Images were reconstructed from the data by using a model-based ordered-subsets expectation maximization (OSEM) algorithm with resolution recovery. Projections from 120 angles were divided to eight subsets. Twenty iterations were used without any postreconstruction filtering or prior regularization.
Fig. 1.
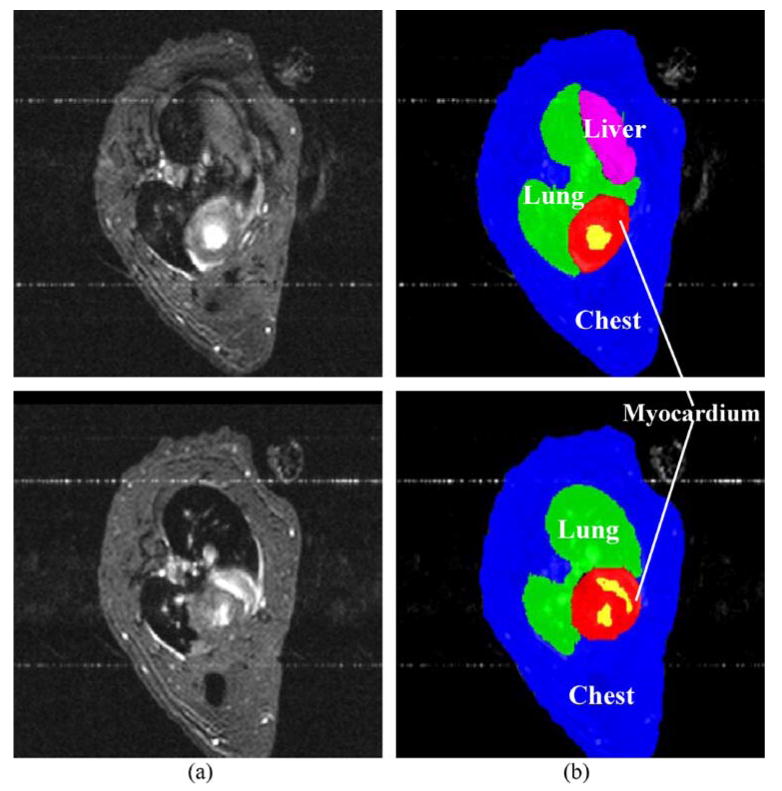
Representative time-averaged rat cardiac MR images from two levels, one close to apex (upper left panel) and the other close to base (lower left). The images were then manually segmented into five components and labeled with different colors (right).
Based on this phantom, six sets of volumetric SPECT images were generated, corresponding to various simulated injected doses (0.5, 1, 2, 3, 4, and 5 mCi). For the case of 185 MBq (5 mCi) total activity in the whole body, the uptake values in the five components are 5.32 MBq (chest), 1.55 MBq (liver), 0.31 MBq (lung), 4.67 MBq (myocardium), and 0.12 MBq (ventricles), respectively. Each dataset included 10 SPECT images, produced by shifting the phantom to various x and y locations along with a rotation around the z-axis. Random translations were independently chosen within the range 5–12mm, and random rotation was from −15° to 15°. A representative simulated SPECT image, corresponding to each activity level, is shown in Fig. 2. Note that the coregistration performance is evaluated only on x,y translations and z rotation. This is because our simulated data is obtained from the manually painted MRI, which has very low resolution in z dimension. Transforming images along low-resolution dimension such as z will significantly distort images under transformation, thereby affecting the fair evaluation of coregistration algorithm. On the other hand, because only rigid transformation is considered in our coregistration algorithm, the performance of our coregistration algorithm obtained on x,y translations and z rotation can be generally extended to other transformations.
Fig. 2.
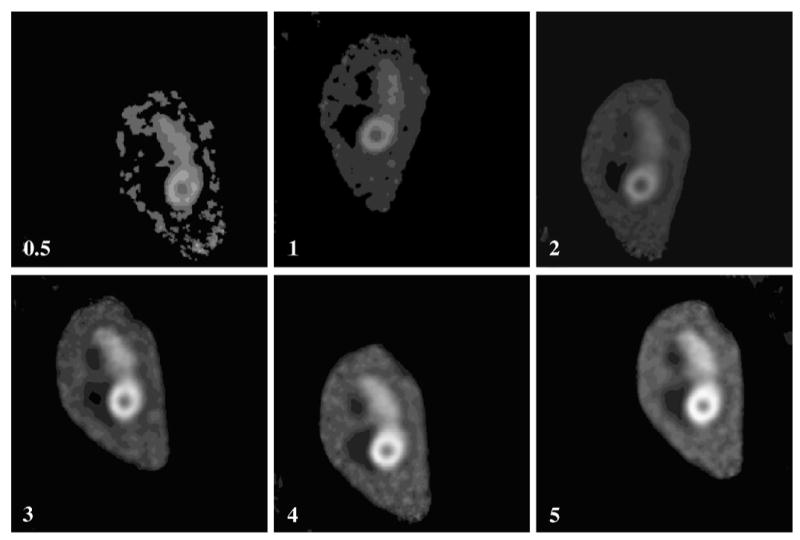
Representative SPECT images, corresponding to six simulated injected doses (0.5, 1, 2, 3, 4, and 5 mCi) and with a selection of random orientations.
Moreover, the performance of coregistration algorithm is relatively independent of the strength of signals, as long as the contrast between signals in heart and background is sufficiently high, because mutual information, and not simply the direct signal differences, are used for coregistration. However, the performance of coregistration algorithm is directly dominated by signals in the big regions, i.e., an almost entire region produced by 99mTc in our SPECT image. This is because the entire image is considered and only rigid transformation is applied in our coregistration algorithm. In contrast, the signals produced by 111In occupies a very small region, thereby the coregistration performance is less related to those signals. In this way, we directly measure the performance of our coregistration algorithm to the signals produced by 99mTc in this validation study.
Stem Cell Injection and Image Acquisition
Rat embryonic cardiomyoblasts H9c2 cells were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA) and were maintained in Dulbecco's modified Eagle medium (DMEM) supplied with 10% fetal bovine serum. Cells were labeled with [111In]oxyquinoline (Amersham, Arlington Heights, IL, USA) as previously described [3]. Briefly, three to four million cells were incubated with serum-free DMEM media containing 11.1–14.8 MBq (300–400 μCi) [111In]oxyquinoline for 30 minutes. Cells were then washed three times and resuspended in 200 μl DMEM media for injection.
Male Sprague–Dawley rats (six to eight weeks old, 220–250 g) were purchased from Charles River Laboratories (Wilmington, MA, USA). Myocardial infarction was generated by surgical procedures as described previously [3]. Briefly, after opening the thoracic cavity and the pericardium, the left anterior descending (LAD) coronary artery was ligated by passing a piece of 6–0 silk suture underneath the vessel and the surrounding myocardium. Ligation was maintained in place for 45 minutes and LAD perfusion was then restored. 111In-labeled cells were injected in four to five locations surrounding the infarcted (pale) region and the thoracotomy incision was closed.
Cardiac SPECT imaging was performed on a Prism 3000XP triple-headed gamma camera (Philips Medical Systems, Cleveland, OH, USA), equipped with custom-made tungsten double-knife-edge pinhole collimators (Nuclear Fields, Des Plaines, IL, USA). The details of acquisition parameters and corrections for center of rotation error and cross talk were described previously [3]. The focal length of the collimators was 24 cm, with a radius of rotation of 5 cm and a pinhole diameter of 3 mm. The acquisition parameters included a continuous mode with 120 projection angles over a 360° arc to obtain data in a 128 × 128 matrix with a slice thickness of 3.56 mm. The images were then reconstructed by using 10 iterations of a simultaneous algebraic reconstruction technique (SART) [18]. Images consisted of a matrix of 128 × 128 × 128 with an isotropic voxel size of 0.74 mm. Thirty minutes after the intravenous injection of 74 MBq (2 mCi) [99mTc]sestamibi (Cardiolite), 99mTc perfusion and 111In images were simultaneously acquired by using two separate energy windows to detect the 140- and 247-keV photons from 99mTc and 111In, respectively. No cross-talk between energy windows was observed [3].
The MR images were acquired for the same rat five to seven days after SPECT imaging (when radioactivity was decayed close to background) on a 4.7-T magnet interfaced to a Varian INOVA console [4, 19]. An ECG-gated fast gradient echo sequence [19] was used to acquire a cine series in the short axis orientation of the left ventricle (LV). Imaging parameters for this sequence were as follows: flip angle 20°, repetition time (TR) 6 ms (the effective TR is the heart rate), echo time (TE) 3 ms, FOV 5–6 cm2, matrix size of 128 × 128 zero-filled to 256 × 256, slice thickness 1.5 mm, 11–12 cardiac phases per heart beat, and 4 signal averages. Because of the rapid heart rate (350–450 beats/minute), one k-space line for each cine image was acquired per heartbeat. Acquisition time was approximately two minutes per slice. Typically, 10–11 slices were required to span the entire LV. In total, six rats are studied.
Results
To evaluate the performance of our coregistration algorithm in aligning SPECT and MR cardiac images, both quantitative and qualitative measures on the same rat model are provided. The quantitative measure is obtained by applying our method on simulated SPECT images, which were generated from an MR image of the rat, by using various doses of 99mTc as described above. The qualitative measure is obtained by applying our method to the same rat, which was subjected to stem cell grafting, SPECT dual tracer detection, and subsequent MRI. These two measurements are detailed in following two subsections, respectively.
Validation of the Coregistration Method Using Simulated Data
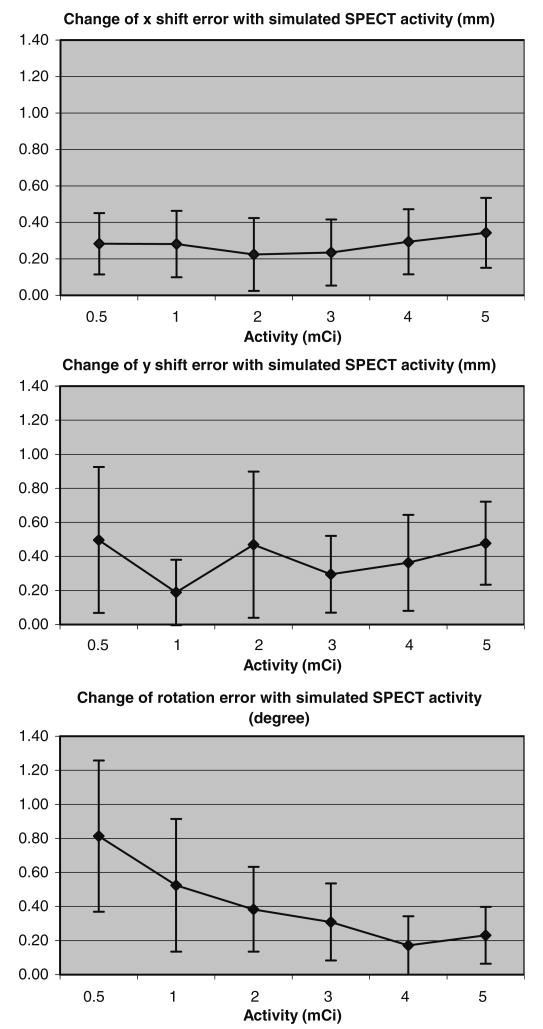
The performance of our coregistration method is evaluated with respect to various activities simulated in SPECT images. Six sets of volumetric SPECT images, corresponding to six activity levels (i.e., 0.5, 1, 2, 3, 4, and 5 mCi), were generated based on a phantom described above. Each dataset included 10 SPECT images, produced by shifting the phantom to various x and y locations along with a rotation around the z-axis. Thus, the performance of our coregistration method can be measured by computing the errors between our estimated shifts and rotations with the true values used in the simulations. The maximum of the average absolute shift error is 0.50 mm, and the maximum of the average absolute rotation error is 0.82°. Figure 3 shows the changes of average absolute estimation errors with respect to six different activity levels, for shifting along x- and y-axes and rotation around z-axis, respectively. The vertical bars in Fig. 3 denote the standard deviation of the absolute registration error for 10 different noise realizations. It shows that the shifting estimation is relatively independent of the simulated SPECT activity, whereas the rotation error decreases consistently with increasing SPECT activity. It is worth noting that the coregistration error within any specific region of the heart is the same as the provided global registration error, because rigid transformation is considered in our registration algorithm.
Fig. 3.
Variation of absolute registration error with respect to simulated SPECT activity. The absolute rotation error consistently decreased with increasing SPECT activity. On the other hand, absolute shifting errors along x- and y-axes are relatively independent of the simulated SPECT activity. The vertical bars denote the standard deviation of absolute registration error for 10 different noise realizations.
Coregistration of SPECT and MR Images
The coregistration between SPECT and MRI of the same heart was completed by the following steps. To speed up the coregistration algorithm and make it robust to noise, we extracted the cardiac regions from the SPECT and MRI. Figure 4a shows a region of interest, extracted from an MRI. SPECT images include 99mTc sestamibi image of the heart (mibi) and 111Indium image from the grafted stem cells (Indium). Because they were simultaneously acquired by using two different energy windows, these two images can be combined as a single SPECT image, to be coregistered with MRI. A mutual-information-based coregistration method and a rigid-body transformation was used to coregister the SPECT image with the MRI [12, 15]. Finally, the estimated rigid-body transformation was applied to the original mibi and Indium images, leading to the coregistration of these two SPECT images with the MRI. This coregistration algorithm is successfully applied to a group of rats in our study.
Fig. 4.
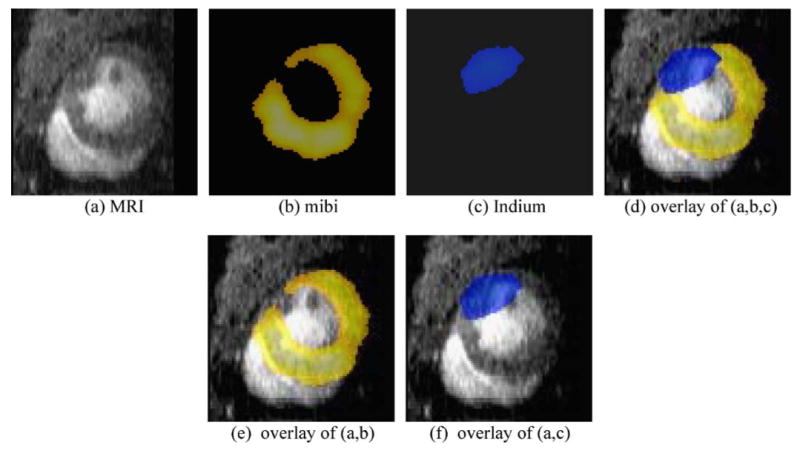
Coregistration of SPECT and MR images of the same rat heart. Mibi (b) and Indium (c) have been aligned to the space of the MR image (a), by using a mutual-information-based coregistration method, with the constraint of rigid-body transformation. The overlay of these three images is shown in (d), while the respective overlays between (a) and (b) and between (a) and (c) are given in panels (e) and (f). These results demonstrate that our coregistration algorithm can relatively accurately align SPECT images to the MR image, thus providing a way of noninvasive estimation of stem cells grafted in infarcted myocardium.
The coregistration of SPECT and MRI was completed in three-dimensional space, while cross-sectional images (2D) were displayed for convenience. Shown in Fig. 4b and c are the color-coded mibi and Indium signal maps, after alignment with the MRI in Fig. 4a. Figure 4d shows the overlay of these three images, whereas Fig. 4e and f respectively show the overlay of Fig. 4b or c with Fig. 4a. The overlay of the three images in Fig. 4d clearly shows the region of perfusion deficit on 99mTc sestamibi image, which is coincident with the akinetic region identified on the cine MRI and thus confirmed that 111Indium oxine-labeled stem cells were injected into the infarcted region.
Discussion
We have shown that a mutual-information-based coregistration method is effective for coregistration of SPECT and cardiac MRI of the rat heart, for our designed experiments. Registration on both experimental and simulated data shows the relatively high accuracy of our registration method. The simulation results (Fig. 3) indicate that the maximal translational error (0.50 mm) is comparable to an MR pixel size (0.27 mm) and only about 1/6 of the SPECT image resolution (3–4 mm), and it is not dependent on the SPECT activity. The errors in x and y dimensions are not correlated (correlation coefficient 0.09). This suggests that locations of stem cells (detected by SPECT) can be mapped onto the high-resolution MRI with relatively high accuracy. In both data simulation and coregistration, rotation of images usually introduces more intensity and geometric errors to the images, compared to shifting images. These errors become significant when the SPECT activity is low. Therefore, the estimated rotation error of our coregistration algorithm is highly related to the SPECT activity level, whereas translational error is not (Fig. 3). It is worth noting that the performance of our coregistration algorithm was evaluated only on x, y translations and z rotation, because of the limitation of image resolution along the z dimension in our simulators. However, we believe that the accuracy of our coregistration algorithm would not be affected when the simulators shifted and rotated in the other planes, as long as the simulators also have high resolution in z dimension.
The coregistration of the experimental data (Fig. 4) has shown a good alignment of MIBI signal to the LV myocardium on the MRI, whereas a spillover of Indium signal to the ventricular lumen is apparent. This could result from the high-energy gamma rays from 111Indium giving more penetration of the pinhole edges, leading to worse resolution and partial volume effects. It is also noted that the acquisition of MRI and SPECT images occurred several days apart during which the myocardium was undergoing remodeling, resulting in errors in the coregistered images. We found that by summing the MRI acquired at different cardiac cycle into one time-averaged image reduces the registration error significantly. We suspect that once we apply cardiac gating during SPECT image acquisition, the error of coregistration with MRI, which are cardiac gated, may be further reduced.
The simulation study was performed by using a tracer (99mTc sestamibi) that distributes uniformly throughout the heart except for the regions (e.g., infarcted region) having perfusion deficit. The performance of our coregistration algorithm is mainly determined by 99mTc signals; this is consistent with our application because 111In signal was only emitted by stem cells grafted in the infarcted region.
The simulation study conducted to validate the new coregistration algorithm was based on a simple, but effective small-animal pinhole SPECT model that assumes that the scattering and attenuation are negligible. In contrast to clinical situations, the gamma-ray scatter and attenuation are usually found to be secondary and relatively negligible in small animal SPECT imaging. In a previous study by Deloar et al. [27], for example, photons scattered by the pinhole collimator were only 3.0% of total counts, relative to the edge penetration of 23% and primary counts of 61%. A refined simulation model that accounts for the scattering and attenuation would represent the future direction of this study.
In the past, two approaches have been developed to coregister radionuclide (e.g., PET) and cardiac MRI. The first approach is based on surface registration: Mäkelä et al. [20] presented a deformable model based on the segmentation of the main thorax structures for coregistration of cardiac PET and MRI; Sinha et al. [21] proposed to coregister gated cardiac PET images to the coordinates of gated MRI, by matching the contours of the left ventricular wall that are correspondingly segmented from both imaging modalities. The second approach is based on intensity similarity measures, which can be divided into methods based on cross-correlation measure [22, 23] and methods based on mutual information. The majority of mutual-information-based algorithms are designed for rigid and affine registration, whereas a few are for nonrigid registration [9, 24]. Mutual-information-based criteria can be used with other criteria to achieve better registration [15], for example, mutual information has been proposed to associate gradient information with the smoothness of registration transformation for coregistration of PET and MRI [24].
In this study, mutual-information measure was used for coregistering SPECT and cardiac MRI of the rat heart. Because both SPECT images and high-resolution MRI were acquired from the same rat, rigid body transformation was assumed between these two modality images. In SPECT images, signals are usually strong in the cardiac regions, but low and very noisy in other regions. To facilitate image coregistration, a manual bounding box was placed around cardiac region in MRI, thus only cardiac region was considered during the coregistration procedure. This is a limitation of current coregistration algorithm.
For robust and automated coregistration, the current coregistration method will be extended in the future in a number of ways. The region of interest, such as the cardiac region, will be extracted automatically. The current coregistration procedure was guided by the mutual information computed from the intensities of the two images. It can also be guided by the mutual information between other features of the two images, i.e., geometric features [25, 26], because intensities in the myocardial region are relatively uniform and thus the shape of the myocardial region becomes more important and can be exactly captured by geometric features [25, 26]. By incorporating this geometric-feature-based similarity measure into the objective function, our coregistration method will have more confidence to judge how well the two images are actually coregistered, based not only on the matching of intensities but also on the matching of geometric features. After coregistering a number of SPECT and MRI pairs acquired from a group of rats, a joint intensity distribution can be statistically collected from those aligned image pairs, and further used as a priori knowledge for future registration tasks [8]. For example, we can align any two images of different modalities by requiring that the observed joint intensity distribution be matched with the expected one seen in the training data. We believe that this strategy will make the multimodality image coregistration more efficient and robust.
In summary, we have applied a mutual-information-based coregistration method to the SPECT and cardiac MRI of the rat heart, thus providing a way of mapping the location and distribution of 111In-labeled stem cells into the akinetic region identified on cine MRI. We expect to utilize this technique to establish the relationship between the stem cell survival in infarcted myocardium estimated by radionuclide imaging and the therapeutic outcome resulting from stem cell grafting, i.e., improvement of contractile function, estimated by MRI. Ultimately, this technique will become important to assess the fate of stem cells and their contributions to the recovery of cardiac function in the same animals.
Acknowledgments
The authors would like to thank the National Institutes of Health for funding this work [EB-002473 (R.Z.) and R01-EB001809 (P.D.A) ], Dr. Girish Bal for providing the uptake values of the MIBI study to build the SPECT phantom from segmented MR image, and Drs. Daniel Thomas and Hui Qiao for their help with generation of the experimental data.
References
- 1.Osman NF, McVeigh ER, Prince JL. Imaging heart motion using harmonic phase (MRI) IEEE Trans Med Imag. 2000 March;19:186–202. doi: 10.1109/42.845177. [DOI] [PubMed] [Google Scholar]
- 2.Denney TS, Jr, Prince JL. Reconstruction of 3-D left ventricular motion from planar tagged cardiac MR images: an estimation theoretic approach. IEEE Trans Med Imag. 1995;14:625–635. doi: 10.1109/42.476104. [DOI] [PubMed] [Google Scholar]
- 3.Zhou R, Thomas DH, Qiao H, Bal H, Choi SR, Alavi A, Ferrari VA, Kung HF, Acton PD. In vivo detection of stem cells grafted in the infarcted rat myocardium. J Nucl Med. 2005;46:816–822. [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas DH, Ferrari VA, Janik M, Kim DH, Pickup S, Glickson JD, Zhou R. Quantitative assessment of regional myocardial function in a rat model of myocardial infarction using tagged MRI. Magma. 2004;17:179–187. doi: 10.1007/s10334-004-0051-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mäkelä TJ, Clarysse P, Sipilä O, Pauna N, Pham QC, Katila T, Magnin IE. A review of cardiac image registration methods. IEEE Trans Med Imag. 2002;21:1011–1021. doi: 10.1109/TMI.2002.804441. [DOI] [PubMed] [Google Scholar]
- 6.Pluim JPW, Maintz JBA, Viergever MA. Mutual-information-based registration of medical images: a survey. IEEE Trans Med Imag. 2003;22:986–1004. doi: 10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- 7.Wells I, William M, Viola P, Atsumi H, Nakajima S, Kikinis R. Multi-modal volume registration by maximization of mutual information. Med Image Anal. 1996;1:35–51. doi: 10.1016/s1361-8415(01)80004-9. [DOI] [PubMed] [Google Scholar]
- 8.Gan R, Wu J, Chung ACS, Yu SCH, Wells MW., III Multiresolution image registration based on Kullback – Leibler distance. The Seventh International Conference on Medical Image Computing and Computer-Assisted Intervention 2004 (MICCAI04); Saint-Malo, France: Rennes. 2004. [Google Scholar]
- 9.Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ. Non-rigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imag. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 10.Studholme C, Hill DLG, Hawkes DJ. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recogn. 1998;32:71–86. [Google Scholar]
- 11.Viola P, Wells WM., III Alignment by maximization of mutual information. Presented at Proc. Int. Conf. Computer Vision; Los Alamitos, CA. 1995. [Google Scholar]
- 12.Jenkinson M, Bannister PR, Brady JM, Smith SM. Improved optimisation for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 13.Studholme C, Hill DLG, Hawkes DJ. Proc IPMI. Ile de Berder, France: 1995. Multiresolution voxel similarity measures for MR-PET registration; pp. 287–298. [Google Scholar]
- 14.Grova C, Biraben A, Scarabin JM, Jannin P, Buvat I, Benali H, Gibaud B. A methodology to validate MRI/SPECT registration methods using realistic simulated SPECT data. In: Niessen WJ, Viergever MA, editors. MICCAI 2001. Lecture Notes in Computer Science. Vol. 2208. Utrecht: Springer; 2001. pp. 275–282. [Google Scholar]
- 15.Luan H, Qi F, Shen D. Multi-modal image registration by Quantitative–Qualitative Measure of Mutual Information (Q-MI). International Conference on Computer Vision Workshop; Beijing, China. 2005. [Google Scholar]
- 16.Madar I, Weiss L, Izbicki G. Preferential accumulation of H-3-tetraphenylphosphonium in non-small cell lung carcinoma in mice: comparison with Tc-99m-MIBI. J Nucl Med. 2002;43:234–238. [PubMed] [Google Scholar]
- 17.Cao Z, Bal G, Accorsi R, Acton PD. Optimal number of pinholes in multi-pinhole SPECT for mouse brain imaging—a simulation study. Phys Med Biol. 2005;50:4609–4624. doi: 10.1088/0031-9155/50/19/013. [DOI] [PubMed] [Google Scholar]
- 18.Andersen A, Kak A. Simultaneous algebraic reconstruction technique (SART): a superior implementation of the art algorithm. Ultrason Imag. 1984 Jan;6:81–94. doi: 10.1177/016173468400600107. [DOI] [PubMed] [Google Scholar]
- 19.Zhou R, Pickup S, Glickson JD, Scott CH, Ferrari VA. Assessment of global and regional myocardial function in the mouse using cine and tagged MRI. Magn Reson Med. 2003;49:760–764. doi: 10.1002/mrm.10423. [DOI] [PubMed] [Google Scholar]
- 20.Mäkelä TJ, Clarysse P, Lötjönen J, Sipilä O, Lauerma K, Hänninen H, Pyökkimies EP, Nenonen J, Knuuti J, Katila T, Magnin IE. A new method for the registration of cardiac PET and MR images using deformable model based segmentation of the main thorax structures. MICCAI'01. 2001:557–564. [Google Scholar]
- 21.Sinha S, Sinha U, Czernin J, Porenta G, Schelbert HR. Noninvasive assessment of myocardial perfusion and metabolism: feasibility of registering gated MR and PET images. Am J Roentgenol. 1995;36:301–307. doi: 10.2214/ajr.164.2.7839959. [DOI] [PubMed] [Google Scholar]
- 22.Habboosh AW. A review of MRI and PET correlation. Proc IEEE Conf Bioengineering. 1992:16–17. [Google Scholar]
- 23.Kim R, Aw T, Bacharach S, Bonow R. Correlation of cardiac MRI and PET images using lung cavities as landmarks. Proc IEEE Conf Computers in Cardiology. 1991:49–52. [Google Scholar]
- 24.Lötjönen J, Mäkelä T. Elastic matching using a deformation sphere. MICCAI'01. 2001:541–548. [Google Scholar]
- 25.Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imag. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 26.Shen D. MICCAI'04. France: 2004. Image Registration by Hierarchical Matching of Local Spatial Intensity Histograms. [Google Scholar]
- 27.Deloar HM, Watabe H, Aoi T, Iida H. Evaluation of penetration and scattering components in conventional pinhole SPECT: phantom studies using Monte Carlo simulation. Phys Med Biol. 2003;48(8):995–1008. doi: 10.1088/0031-9155/48/8/303. [DOI] [PubMed] [Google Scholar]