Abstract
The daily biological clock regulates the timing of sleep and physiological processes that are of fundamental importance to human health, performance, and well-being. Environmental parameters of relevance to biological clocks include (i) daily fluctuations in light intensity and temperature, and (ii) seasonal changes in photoperiod (daylength) and temperature; these parameters vary dramatically as a function of latitude and locale. In wide-ranging species other than humans, natural selection has genetically optimized adaptiveness along latitudinal clines. Is there evidence for selection of clock gene alleles along latitudinal/photoperiod clines in humans? A number of polymorphisms in the human clock genes Per2, Per3, Clock, and AANAT have been reported as alleles that could be subject to selection. In addition, this investigation discovered several novel polymorphisms in the human Arntl and Arntl2 genes that may have functional impact upon the expression of these clock transcriptional factors. The frequency distribution of these clock gene polymorphisms is reported for diverse populations of African Americans, European Americans, Ghanaians, Han Chinese and Papua New Guineans (including five subpopulations within Papua New Guinea). There are significant differences in the frequency distribution of clock gene alleles among these populations. Population genetic analyses indicate that these differences are likely to arise from genetic drift rather than from natural selection.
Keywords: Circadian, Biological Clock, Sleep, FASPS, Depression, Arntl, Bmal, Mop, Per, Clock, AANAT
Introduction
Eukaryotic and prokaryotic organisms manifest daily (circadian) rhythms controlled by endogenous biochemical oscillators [Dunlap et al. 2004]. In mammals, this biological clock regulates the timing of sleep and physiological processes–including feeding behavior, lipid and carbohydrate metabolism, sleep, and blood pressure control–that are of fundamental importance to human health, performance, and well-being [Dunlap et al. 2004]. These daily oscillations are controlled by a circadian clock composed of autoregulatory transcription/translation feedback loops of the expression of central clock genes, especially Clock, Bmal1/Arntl, Bmal2/Arntl2, Per1, Per2, Per3, Cry1, and Cry2 [Dunlap et al., 2004; Lowrey & Takahashi 2004]. The clock-controlled hormone melatonin also plays an important role in circadian and sleep systems [Lewy et al., 2006]. Perturbation of the central clock genes by mutation or knockout in mice elicits phenotypes ranging from arhythmicity [Bunger et al. 2000; van der Horst et al. 1999; Vitaterna et al. 1994] to metabolic syndrome/obesity [Staels 2006], altered insulin/glucose responsiveness [Rudic et al. 2004], and increased susceptibility to cancer [Fu et al. 2005].
In humans, disorders of the biological clock have been strongly implicated in sleep disorders, shiftwork, alertness/performance, cardiovascular disease, and mental health [Dunlap et al. 2004; Kryger et al. 2005]. For example, in depression, there is often a daily variation in mood (depression worse in morning) with altered phasing of the cortisol secretion rhythm (and other rhythms)[Dunlap et al. 2004; McClung 2007]. Recent studies have shown that clock gene polymorphisms are associated with several syndromes, including sleep and mood disorders, for example: (i) variants in Per2 and CKIδ with "Familial Advanced Sleep Phase Syndrome" (FASPS, [Toh et al. 2001; Xu et al. 2005]), (ii) variants of Per3 with delayed sleep phase syndrome [Ebisawa et al. 2001] and extreme diurnal preference [Archer et al. 2003], (iii) a variable number of tandem repeats in exon 18 of Per3 (VNTR) with sleep structure and dysfunction [Viola et al. 2007], (iv) a single nucleotide polymorphism (SNP) in the promoter of the human arylalkylamine N-acetyl-transferase gene (AANAT) with short sleep duration [Wang et al. 2004], and (v) a polymorphism in the 3’ UTR of Clock with diurnal preference [Katzenberg et al. 1998], and sleep disorders [Serretti et al. 2005].
A previous paper reported that a biallelic tandem repeat polymorphism (VNTR) in the human Per3 gene exhibited significantly different allelic frequencies between Papua New Guineans (0.19) versus East Asians (0.80–0.89), whereas European/American/African populations had intermediate frequencies (0.6~0.7) [Nadkarni et al. 2005]. We therefore tested whether these geographical/ethnic differences in allelic frequency were common in other biological clock genes or whether this phenomenon was specific to the VNTR polymorphism of Per3. This study reports the frequency distribution of previously reported polymorphisms of potential functional significance in PER2, PER3, CLOCK, and AANAT in African Americans, European Americans, Han Chinese, Papua New Guineans, and Africans from Ghana. Because Papua New Guinean samples may be genetically diverse, we also perform analyses on subpopulations within Papua New Guinea. In addition, we performed systematic screening and characterization of common exonic and promoter polymorphisms in the ARNTL (aka hBmal1 or hMop3 [Hogenesch et al. 1998]) and ARNTL2 (aka hBmal2 or hMop9 [Hogenesch et al. 2000]) genes by single-stranded conformation polymorphism (SSCP) analyses and sequencing, and report the discovery of several novel polymorphisms. We use population genetic analyses on these clock gene polymorphisms (i) to understand population stratification, (ii) to assess selection/drift, and (iii) as a basis for future association analyses of clock gene polymorphisms with phenotypes of behavioral and/or medical significance.
Materials and Methods
Study subjects
DNA samples were collected from unrelated human subjects from various global populations, including: African-American (n = 48 from the Coriell Cell Repository), European-American (n = 422 collected at Vanderbilt), Han Chinese (n = 48 from the Coriell Cell Repository), New Guinean (n = 66), and Ghanaian (n = 48, randomly selected from a cohort of unrelated individuals of the Ga ethnic group that were collected as part of an ongoing study of the genetic basis of hypertension and cardiovascular disease [Williams et al. 2000]). The PNG samples are further subdivided into separate sub-populations as follows: Sepik (n = 18), Madang (n = 4), Gimi (n = 10), Goroka (n = 11), and Nasioi/Bougainville (n = 23). The study was approved by all relevant institutional review boards and participating individuals.
DNA isolation and primer design
For the European-American (EA) samples collected at Vanderbilt, 5 cc whole blood was collected from each subject and DNA was isolated from lymphocytes by the Vanderbilt DNA core facility. For the Nasioi, Gimi, and Goroka PNG samples, DNA was isolated from lymphoblast cell lines grown at Yale University. Previously reported primers were used for the Per2, Per3 and Clock variants (Table S1A). For Arntl and Arntl2, forward and reverse primers yielding ~200 bp amplicons were generated to cover the promoter regions, exons, and intronic regions immediately flanking exons in a series of overlapping fragments (Table S1A). These primers were designed with an annealing temperature of ~55°C.
SSCP screening and genotyping
Each amplicon was PCR amplified from the genomic DNA using one of two protocols: Platinum PCR Supermix (Invitrogen, Carlsbad, CA, USA) was used for Per2, Per3 and Clock. For Arntl and Arntl2, AmpliTaq Gold (Applied Biosystems, Foster City, CA, USA) was used with the provided PCR Buffer II and MgCl2. Loading dye (4X = 40 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol prepared in deionized formamide) was added to all PCR products. These products were then denatured at 94°C for 3 min, rapidly cooled to 4°C, and separated in MDE electrophoresis gels (Lonza, Basel, Switzerland) at 15 W in 0.6x TBE running buffer at 4°C for 8–14 h. Gels were developed by silver staining to visualize bands [Nataraj et al. 1999]. In the case of the Arntl and Arntl2 genes, we used SSCP to screen for common polymorphisms in the EA population (defined as those polymorphisms present in 5% or more of the population). As an initial screening cohort, we chose 20 random EA subjects (~ 5% of the total population) to look for novel variants and to verify the existence of known polymorphisms within each of the amplicons analyzed (80 amplicons for Arntl and 62 amplicons for Arntl2). The screening set was estimated to provide 98% power to detect a polymorphism with a minor variant frequency of 0.10, and 87% power with a frequency of 0.05. Each amplicon was amplified by PCR and then run on an acrylamide gel for SSCP analysis. Samples that represented the most common SSCP banding pattern and also any samples that exhibited an uncommon band shift pattern were sequenced as described below to confirm the common sequence and to identify polymorphisms (for the less common patterns).
In the cases of the Per2, Per3, Clock, and AANAT genes, we used previously reported primers or redesigned primers to produce ~200 bp amplicons that are roughly centered upon the site of SNPs that have already been described (Table S1A). For the Per2 and Clock genes, we discovered novel SNPs that were located within the amplicons in addition to the targeted SNP. Finally, SNP allele frequencies in all genes were determined on all the samples in the indicated five populations using the SSCP patterns as confirmed by sequencing. Results from the genotyping analyses using SSCP were concordant with the existing data in dbSNP for allele frequencies in the different populations, indicating the fidelity of this method.
Sequence analysis
Amplicons with band-shifts were PCR amplified as stated above, cleaned with the QIAQuick PCR Purification Kit (QIAGEN, Valencia, CA), and sequenced with an ABI 3730xl DNA Analyzer (Applied Biosystems). Control and shifted sequences were aligned using Clustal W, and variants were compared to dbSNP build 126 for Homo sapiens (NCBI, Bethesda, MD, USA).
Statistical analyses
Allelic and genotypic distributions were compared between ethnic groups using Fisher’s exact tests. Hardy-Weinberg Equilibria (HWE) were calculated for each marker using Fisher’s exact tests. Haplotype frequencies were estimated using an expectation maximization (EM) algorithm that reconstructs haplotypes and determines frequencies that maximize the likelihood of the genotypic data [Excoffier & Slatkin 1995; Hawley & Kidd, 1995]. Differences in haplotype distributions between populations were determined using a regression approach [Zaykin et al. 2002]. Wright’s FST estimates were calculated between each population and within the Papua New Guinea sub-populations using a method developed by Weir and Cockerham (1984). Wright’s FST is a measure of population differentiation that ranges from 0 to 1 with 0 indicating no differentiation and 1 indicating highly differentiated populations. The above analyses were performed with either Stata version 9, Powermarker [Liu & Muse 2005], or Tools for Population Genetic Analysis (TFPGA, version 1.3 available at http://www.marksgeneticsoftware.net/); all gave virtually the same results.
Results
SSCP screening of polymorphisms in Arntl and Arntl2
Based on the previous study of the geographical frequency distributions of the Per3 VNTR, we assessed the allele frequency of the clock gene SNPs in five populations: European Americans (EA), African Americans (AA), Ghanaians (GA), Han Chinese (HC), and Papua New Guineans (PNG)[Nadkarni et al. 2005]. Because PNG populations are known to be genetically heterogeneous [Friedlaender et al. 2007], we assayed several subpopulations within PNG to determine if the study of Nadkarni and coworkers [2005] provided data that reflected PNG as a whole or only certain subpopulations within PNG.
In addition to genotyping previously reported SNPs within the clock genes Clock, Per2, Per3, and AANAT, we identified several novel SNPs in the promoter and exonic regions of the key clock genes Arntl and Arntl2 using SSCP gel analyses [Nataraj et al. 1999]. Our criterion for a "common" polymorphism was frequency greater than five percent in the initial screening cohort. This screening strategy identified 11 SNPs in Arntl and Arntl2 that were chosen for further characterization (Table S1A). Of these 11 SNPs, three were novel variants in Arntl and three others were unreported variants in Arntl2 (Table 1). Not only did our SSCP method reveal these six novel polymorphisms in the Arntl and Arntl2 genes, it also revealed one new SNP in Clock and three new SNPs in Per2 in the amplicons designed to uncover the previously reported SNPs (Table 1). Two previously reported SNPs in Per2 and Per3 for which rs# have not been assigned are referred herein as c.1984A>G [Toh et al. 2001] and c.2460A>G [Ebisawa et al. 2001](see Table S1A for the location of all SNPs discussed in this paper along with the primer pairs we used).
Table 1.
Novel variants discovered in this study with their chromosomal locations
| Genes (OMIM) | Location | Chromosomal position | SNPs | Placement | Protein Change | Base Change |
|---|---|---|---|---|---|---|
| Arntl (602550) | 11p15.2 | 13286395 | ss95215854 | intronic | n/a | T>G |
| 13288144 | ss95215855 | 5'UTR | n/a | A>C | ||
| 13335041 | ss95215856 | intronic | n/a | G>A | ||
| Arntl2 | 12p11.23 | 27375128 | ss95215857 | promoter | n/a | A>G |
| 27420525 | ss95215858 | intronic | n/a | T>C | ||
| 27464992 | ss95215859 | 3' flanking | n/a | A>G | ||
| Clock (601851) | 4q12 | 55996140 | ss95215860 | 3'UTR | n/a | A>G |
| Per2 (603426) | 2q37.3 | 238830546 | ss95215861 | intronic | n/a | G>T |
| 238830376 | ss95215862 | exonic | A>V | C>T | ||
| 238830273 | ss95215863 | intronic | n/a | C>T | ||
Previously unreported SNPs are referred to by ss numbers; these have been submitted to dbSNP and will be publicly available as rs numbers in Build 130. OMIM references for each gene are shown below the gene names. Chromosomal locations are based on the NCBI Build 36.1 for the human genome (release date March 2006). See Table S1A for complete list of variants studied and further information about reference sequences, flanking sequences, primer pairs, etc.
Among the Arntl SNPs, ss95215855 was the only variant found exclusively in the EA population (Table 2 and Figure S1A). The other two SNPs in Arntl were both intronic (Table 1) and occurred in non-EA populations at low frequency (Table 2 and Table S2A, Figure S1A). We screened eight polymorphisms in the Arntl2 gene in the EA population (Table 1, Table 2, Table S2A, Figure S1A), of which five were polymorphic in all populations (Table 2). The first (rs5797225) was a TTG repeat in the 5’ region that is likely to act as promoter of Arntl2. This polymorphism consisted of either 7, 8, 9, or 10 TTG repeats, with the 8 or 9 repeat structure being most common in all populations (Table 2, Figure S1A). The next two polymorphisms (rs7137588 and rs11048972) were within 19 bp of each other in the Arntl2 promoter region and were variable in all populations (Table 2, Figure S1A). One other common SNP occurs within the putative promoter of Arntl2 (rs10548381) and another SNP (rs4964059) occurs in the intron between exons 2 and 3 (ss95215857, ss95215858, and ss95215859 do not appear in Table 2 because they are very rare). One of the SNPs in Arntl2 departed from HWE at the p < 0.05 level (rs4964059) and two SNPs departed from HWE at the p < 0.01 level (rs10548381 and ss95215857, Table 2 and Table S2A). Each of these departures was limited to a single population, and for two of these three the populations may represent either recent admixture among distinct populations (rs10548381 in African Americans) or stratification (rs4964059 in PNG).
Table 2.
Allele frequencies of the common polymorphisms in all populations with deviations from Hardy/Weinberg Equilibrium (HWE).
| Gene | Allele | African American | European American | Ghanaian African | Han Chinese | Papua New Guinea |
|---|---|---|---|---|---|---|
| Arntl | ||||||
| ss95215854 | T | 1.00 | 0.99 | 0.93 | 1.00 | 1.00 |
| ss95215855 | A | 1.00 | 0.97 | 1.00 | 1.00 | 1.00 |
| Arntl2 | ||||||
| rs5797225 | GTT (7) | 0.01 | - | - | - | - |
| GTT (8) | 0.49 | 0.71 | 0.51 | 0.48 | 0.71 | |
| GTT (9) | 0.50 | 0.29 | 0.46 | 0.51 | 0.28 | |
| GTT (10) | - | - | 0.03 | 0.01 | 0.01 | |
| rs7137588 | G | 0.86 | 0.73 | 0.87 | 0.93 | 0.75 |
| rs11048972 | A | 0.98 | 0.94 | 0.87 | 0.98 | 1.00 |
| rs10548381 | +/CTA | 0.93+ | 0.90 | 0.98 | 0.98 | 0.99 |
| rs4964059 | A | 0.91 | 0.67 | 0.88 | 0.43 | 0.43* |
| Clock | ||||||
| rs1801260 | T | 0.86 | 0.74* | 0.83 | 0.92 | 0.83 |
| Per2 | ||||||
| rs2304669 | A | 0.97 | 0.87 | 0.99 | 0.94 | 0.94 |
| rs2304670 | G | 0.90 | 0.93 | 0.87 | 0.94 | 1.00 |
| rs2304671 | G | 0.96 | 0.93 | 0.96 | 0.89 | 1.00 |
| Per3 | ||||||
| rs228696 | C | 0.95* | 0.97 | 0.98 | 1.00 | 1.00 |
| rs228697 | C | 0.98 | 0.99 | 1.00 | 0.95 | 0.83 |
| AB047536 | 4-repeat | 0.59 | 0.66 | 0.64 | 0.81+ | 0.41 |
| rs10462021 | A | 0.93 | 0.83 | 0.96 | 1.00 | 0.98 |
| AANAT | ||||||
| rs4238989 | G | 0.74 | 0.53 | 0.87 | 0.53 | 0.51 |
Population analyses of Arntl, Arntl2, Clock, Per2, Per3, and AANAT genes
We also analyzed previously reported SNPs in the central clock genes Clock, Per2, and Per3, and in the AANAT gene that encodes the rate-limiting enzyme of melatonin biosynthesis (Table 1 and Table S1A). Our rationale for including these SNPs in the study was three-fold. The first was to compare our systematic analyses with that of the only previous population genetic study of a clock gene SNP, namely of the VNTR SNP in Per3 [Nadkarni et al. 2005]. The second reason for studying these previously reported SNPs is that many of them have been reported to have functional significance. Finally, our third rationale was that many of them are sufficiently common in human populations that they might have some value in explaining common syndromes that might be associated with clock gene polymorphisms (in contrast, while the SNPs in Per2 and CKIδ have potent effects upon the timing of sleep and activity [Toh et al. 2001; Xu et al. 2005], they are so uncommon that they are unlikely to have any real explanatory power for disorders in the general population). We again used the SSCP method to assess the frequency of these SNPs by designing primers to amplify ~200 bp amplicons around the reported SNPs. We then sequenced representatives of every different SSCP pattern we observed for every amplicon. This method was successful in identifying the reported SNPs and in addition, we discovered novel SNPs in Clock and Per2 within these amplicons (Table 1).
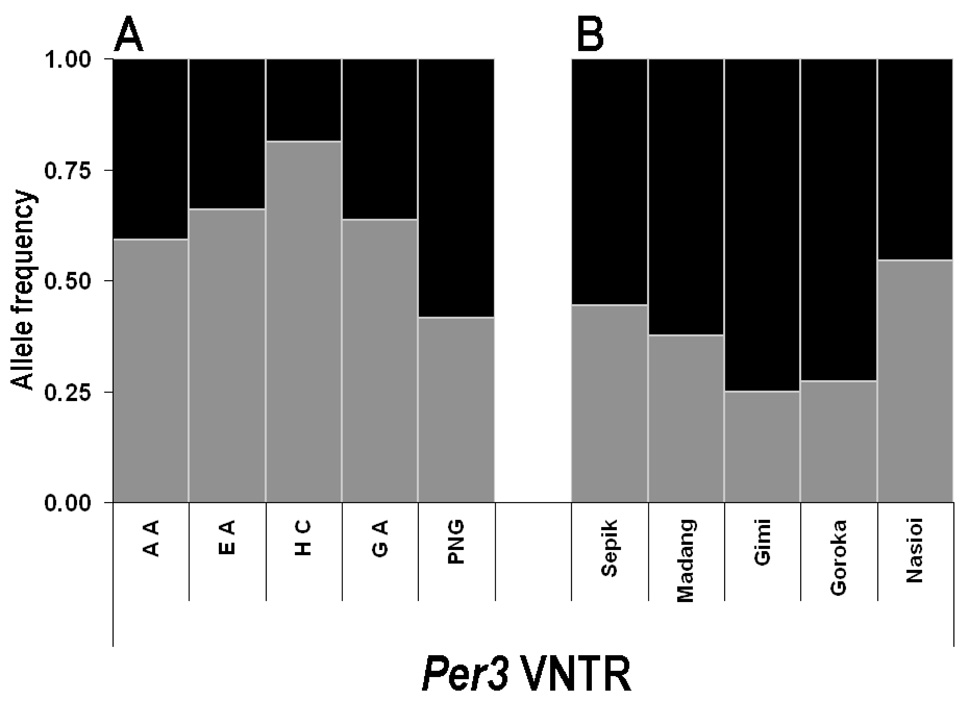
SNPs rs1801260 (in Clock) and rs4238989 (in AANAT) were polymorphic in all populations we studied (Table 2 and Figure S2A). SNP ss95215860 in Clock, a previously unreported SNP we identified in the PNG population was unique to PNG (Table S2A). The c.1984A>G SNP in Per2 that is associated with FASPS [Toh et al. 2001] was not detected in any of our populations (Table S2A). In the process of SSCP screening for c.1984A>G, however, we identified other Per2 SNPs, including three novel SNPs, ss95215861, ss95215862, and ss95215863 with extremely low frequencies (<0.01), and three SNPs that have been previously identified (rs2304669, rs2304670, and rs2304671)(Table 2). For most of the SNPs in Per3, there was a low but detectable frequency of polymorphisms in our populations (Table S2A). The most polymorphic of the Per3 variants was the VNTR repeat (AB047536) that has been associated with delayed sleep phase syndrome and sleep structure dysfunction [Ebisawa et al. 2001; Viola et al. 2007] and that has been reported to exhibit significantly different allelic frequencies among populations [Nadkarni et al. 2005]. This polymorphism is a four or five tandem repeat of a 54 bp sequence in exon 18 of Per3. We confirm that the 4-repeat allele is the most common in all populations except for the PNG population, where the 5-repeat is most common (Table 2, Figure 1A). The 5-repeat is least common in the Han Chinese population (19%), and is approximately the same (34–41%) in the EA, African-American, and Ghanaian populations.
Figure 1. Proportions of the VNTR SNP of hPer3 (AB047536).
A. Proportions of the VNTR SNP in all five populations (AA = African American, EA = European Americans, HC = Han Chinese, GA = Ghana Africans, and PNG = Papua New Guineans).
B. Proportions of the VNTR SNP in PNG sub-populations.
In both panels, gray is the 4-repeat and black is the five-repeat.
Deviation from Hardy/Weinberg Equilibrium (HWE) for all the SNPs of sufficient frequency are indicated in Table 2 and Table S2A (four SNPs were present in such low frequency that they were not studied further: ss95215861, ss95215863, rs35899625, and rs3760138). Including the Arntl2 SNPs that were out of HWE as noted above, there were two loci that deviated from HWE in the EA population, one of which had a nominal p-value < 0.01 (Table 2 and Table S2A). In the Han Chinese and Papua New Guinea populations there was one SNP in each that deviated from HWE; however, only the SNP in the Han population deviated with a nominal p < 0.01. In African-Americans there were three SNPs that deviated from HWE, two of which had a nominal p-value <0.01. None of these appears significant in the context of multiple tests. There were significant allele and genotype differences between many of the populations we compared (Table S3A and Table S4A). The populations that differed the most in both allelic and genotypic distributions were European Americans as compared with Ghanaians. In comparisons of these two populations, the common variant was more frequent in EA (except for the rs10462021 SNP in Per3). As expected, there were few allelic or genotypic differences between Ghanaians and African-Americans (Table S3A & Table S4A).
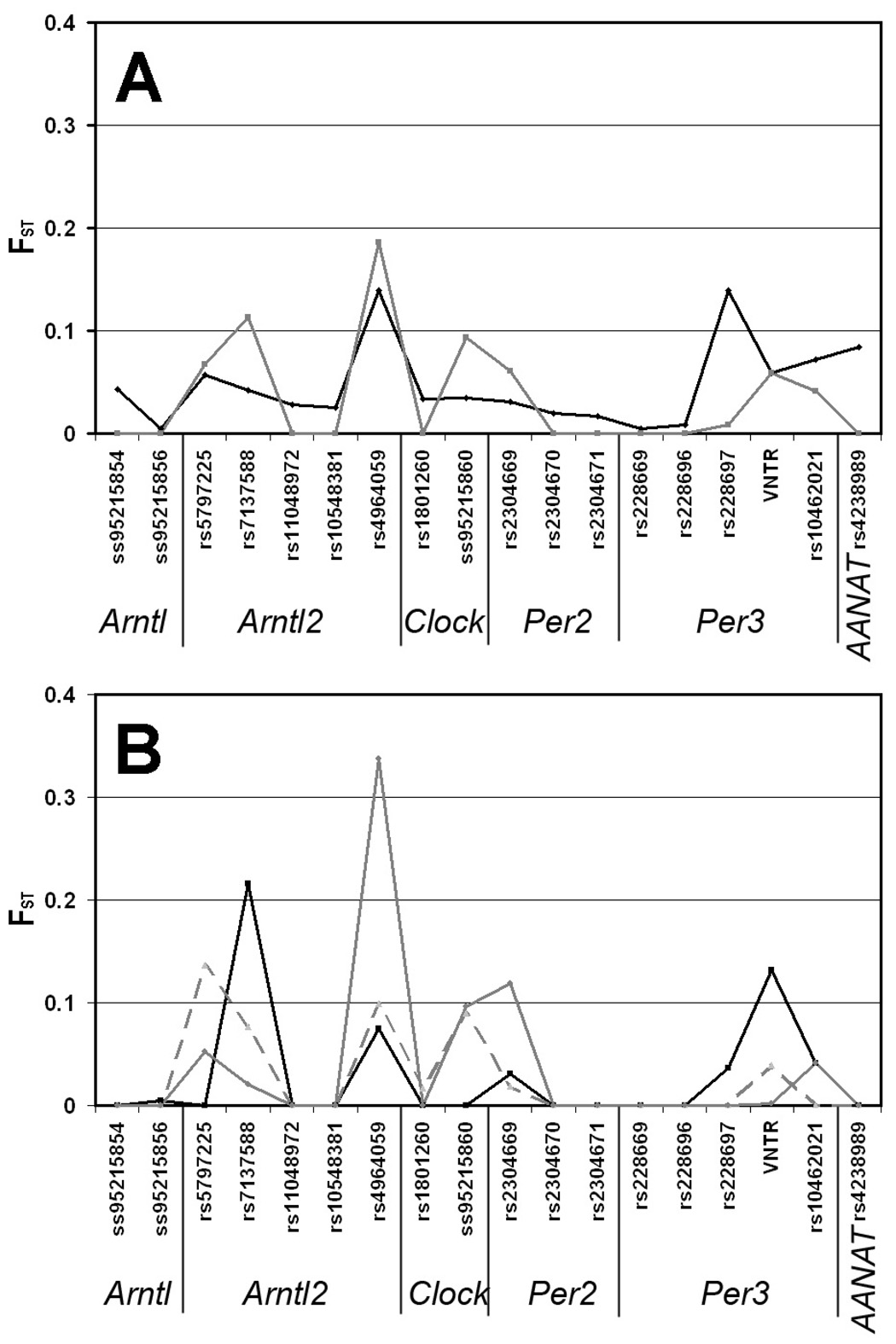
Loci were excluded from haplotype and FST analyses if four or more populations had only one of the two alleles. Haplotypes for Arntl2, Per2, and Per3 differ in frequency among the populations (Table S5A and Table S6A). In particular, for Arntl2 the comparisons revealed significant differences (p < 0.001) for all of the population comparisons except when comparing Ghanaians to African-Americans. There were also significant differences in haplotype comparisons when comparing PNG to the other populations (Table S5A & Table S6A). FST averaged across all five populations was below 0.1 for all polymorphisms (Table S7A & Figure 2A, black trace), with the exception of Arntl2 (rs4964059) and Per3 (rs228697) (FST = 0.139 for both SNPs). Comparing the PNG subpopulations among each other shows two SNPs (rs7137588 and rs4964059 in Arntl2) for which FST exceeds 0.1 and one that nearly exceeds 0.1 (ss95215860 in Clock), indicating greater genetic differentiation among PNG populations than across the global samples (Figure 2A, gray trace).
Figure 2. FST values for SNPs (theta P).
Two comparisons are shown—the five populations together (panel A) and the PNG subpopulations (panel B). In panel A, the black line is an average FST across all populations with all PNG sub-populations combined into one; the gray line is the FST across all PNG sub populations compared simultaneously. Panel B is the pairwise comparison of PNG sub-populations. Black solid traces are EH vs. NA, dashed traces are EH vs. Coastal, gray solid traces are NA vs. Coastal. EH = Eastern Highlands population (Gimi & Goroka pooled), NA = Nasioi (Bougainville), and Coastal = Sepik & Madang pooled.
Stratification within the PNG sub-populations
Nadkarni and coworkers [2005] reported that the VNTR polymorphism of Per3 (AB047536) exhibited significantly different allelic frequencies in PNG (0.19), as compared to East Asians (0.80–0.89) and European/American/African populations (intermediate frequencies of 0.6~0.7). However, PNG populations can be very diverse genetically [Friedlaender et al. 2007], and the previous study did not give the origin(s) of its PNG study population. We therefore performed our analyses using 3 populations within Papua New Guinea (Sepik/Madang, Gimi, and Goroka) and one population from the nearby island of Bougainville (Nasioi). We found that there is very significant variation of clock gene polymorphisms among the PNG sub-populations. For example, the 4-repeat variant of VNTR ranged in allele frequency from 25% (Gimi) to 54% (Nasioi)(Figure 1B). This result implies that Nadkarni and coworkers [2005] might have studied a distinct sub-population within PNG and that their results might not be reflective of PNG populations in general. The VNTR was not the only polymorphism that showed significant variation among PNG subpopulations, as implied by the allele frequencies and FST values for PNG (Figure 2A and Table 3, Table S7A, and Table S8A) and as is more explicity shown by the FST comparisons among the PNG sub-populations (Figure 2B and Table S9A, including pairwise comparisons between the PNG population and the other populations, Table S10A). There are numerous FST values greater than 0.10 when the PNG subpopulations are compared, e.g., in the rs5797225, rs7137588, rs4964059, rs2304669, and VNTR polymorphisms. In fact, the FST values for most of those SNPs exceed that of the VNTR polymorphism when compared among the PNG sub-populations (FST is only greater than 0.1 for the VNTR polymorphism when the Eastern Highlands sub-population is compared with the Nasioi sub-population, Table S9A). These data support the idea that genetic differentiation among the PNG populations is greater than that across much of the rest of the world.
Table 3.
Allele frequencies for selected polymorphisms in the Papua New Guinea (PNG) subpopulations with deviations from Hardy/Weinberg Equilibrium (HWE).
| Gene | Allele | Eastern Highlands | Nasioi | Coastal |
|---|---|---|---|---|
| Arntl | ||||
| ss95215856 | G | 0.93 | 0.98 | 0.98 |
| Arntl2 | ||||
| rs5797225 | GTT (7) | - | - | - |
| GTT (8) | 0.83 | 0.74 | 0.57+ | |
| GTT (9) | 0.17 | 0.24 | 0.43 | |
| GTT (10) | - | 0.02 | - | |
| rs7137588 | G | 0.57 | 0.89 | 0.78 |
| rs4964059 | C | 0.57 | 0.35 | 0.80 |
| Clock | ||||
| rs1801260 | T | 0.88 | 0.85 | 0.77 |
| ss95215860 | A | 1.00 | 1.00 | 0.89 |
| Per2 | ||||
| rs2304669 | A | 0.95 | 1.00 | 0.86 |
| Per3 | ||||
| rs228697 | C | 0.76 | 0.89 | 0.84+ |
| AB047536 | 5-repeat | 0.74 | 0.46 | 0.57 |
| rs10462021 | A | 1.00 | 0.93 | 1.00 |
| AANAT | ||||
| rs4238989 | G | 0.55 | 0.48 | 0.50 |
indicates deviation from HWE at p < 0.05
indicates deviation from HWE at p < 0.01
Eastern Highlands = Gimi and Goroka
Nasioi = Bouganville
Coastal = Sepik and Madang
Discussion
The function of biological clocks is to provide temporal information to the organism so that physiological and/or behavioral responses can be coordinated during the daily cycle to maximize adaptiveness. Environmental parameters of relevance to biological clocks include daily fluctuations in light intensity and temperature and seasonal changes in daylength and temperature [Dunlap et al. 2004]. These parameters vary dramatically as a function of latitude and locale. Therefore, biological clocks within wide-ranging species may be genetically “tuned” by natural selection to optimize adaptiveness along latitudinal clines or other location-dependent factors. Indeed, Drosophila melanogaster exhibits such genetic polymorphisms in circadian clock genes along latitudinal clines [Sawyer et al. 2006; Tauber et al. 2007]. Homo sapiens is a species that is distributed to practically every terrestrial niche on earth. Therefore it is of interest to study levels and patterns of variation in human circadian clock genes to assess the evolutionary mechanism that may have shaped these patterns.
Our study is the first systematic report of the frequency distribution of polymorphisms in multiple clock genes. We chose two groups of polymorphisms to study: (i) previously reported polymorphisms of potential functional significance in Per2, Per3, Clock, and AANAT, and (ii) polymorphisms in Arntl and Arntl2 identified by a systematic SSCP screen of promoter and exonic regions. This screen revealed several novel polymorphisms in Arntl and Arntl2. We studied five diverse populations (including subpopulations within Papua New Guinea) from very different latitudes and environments. Significant differences between the populations were found for allelic and genotypic frequencies. For the reported clock gene polymorphisms that also appear in HapMap, our allele frequencies are nearly identical to the HapMap data for most of the assays (Table 2 and Table S2A), confirming the accuracy of the SSCP method. There were 7 out of 145 SNP/population combinations that were out of Hardy Weinberg Equilibrium (HWE)–well within the range expected for a Type 1 error at the p < 0.05 level (7/145 = 4.8%). Moreover, none of the SNPs show more than one population deviating from HWE (Table 2 and Table S2A). Of note, one of these is in the combined PNG population that is probably not a single panmictic population (Friedlaender et al., 2008), as shown by the large FST values among the discrete populations.
On the basis of the SNPs for which allelic and genotypic frequency differences could be calculated for all five populations, EA and Ghanaians differed the most. In contrast, there were very few allelic or genotypic differences between the Ghanaian and African-American samples. As expected, haplotype comparisons revealed similar patterns (Table S5A and Table S6A). It is interesting that the most common haplotype for all populations was the same for Per2 and Arntl, but that this was not the case for Arntl2 and Per3. For Arntl2, the most common haplotype in the Han population was exceedingly rare in all other populations except PNG, where it was the second most common haplotype. In Per3, the most common haplotype in PNG was second most common in all of the other populations (Table S5A).
The PNG population was less different from the EA, African-American, and Ghanaian populations than the EA-Ghanaian difference when the PNG samples were pooled, in contrast to expectations based on the previous report [Nadkarni et al. 2005]. As might be expected in terms of geographic distance, the PNG population as a whole was most similar to the Han Chinese in terms of individual SNP allele frequencies (Table S3A), but this difference was not as clear from the haplotype analyses. This might indicate a very different history of the PNG founding chromosomes. Upon examining sub-populations within PNG, it became clear that even the VNTR SNP–studied by Nadkarni & coworkers [2005] and used to argue that PNG is a genetic outlier–has very different allele frequencies among sub-populations. For example, in the Nasioi sub-population, the VNTR allele frequency is close to that in the non-PNG populations (EA/GA/AA). Therefore, the very low VNTR frequency (19% for the 4-repeat allele) reported by Nadkarni & coworkers [2005] does not reflect PNG as a whole. Moreover, the FST analyses that compare the three PNG sub-groups indicate much larger FST values among the PNG subgroups (Figure 2B and Table S9A) than among any of the five global populations (Figure 2A and Table S7A). Because it is difficult to imagine that the environment within the PNG region is more heterogeneous than the environments inhabited by EA vs. Han Chinese vs. African-Americans vs. Ghana Africans vs. Papua New Guineans, it is unlikely that the differences in allele frequency we observe are due to natural selection acting in response to environmental pressures. Genetic similarities based on large numbers of autosomal markers place the Nasioi subpopulation sample close to the East Asian populations and far from the European and African populations [Tishkoff & Kidd, 2004]. Some, but not all, of the markers we examined follow this pattern.
Therefore, it seems likely that the differences in the clock gene allele distributions that we have observed are due to genetic drift (e.g., possibly a founders’ effect). Most of the genetic heterogeneity among Papua New Guineans has likewise been attributed to genetic drift [Friedlaender et al. 2007]. In particular, our data do not support selection of clock gene alleles on the basis of latitude/photoperiod, which might be predicted to affect the evolution of circadian clock genes (as in Drosophila [Sawyer et al. 2006; Tauber et al. 2007]). Our Ghana and PNG samples are the best defined in terms of latitude and both are close to the equator where the annual photoperiod will be nearly constant at 12 h light/12 h dark (LD 12:12). Nevertheless, the allele frequencies are very different between the Ghana and PNG populations. Our other populations (EA, AA, and HC) are less well defined geographically and have been subject to admixture. Nevertheless, these populations come from latitudes ranging between 22–70°N. At 22°N, the annual photoperiod ranges between LD 13.2:10.8 (summer) and LD 10.4:13.6 (winter), whereas around 60°N photoperiods range between LD 18.4:5.6 (summer) and LD 5.4:18.6 (winter). (At 70°N there is no night in mid-summer and no day in mid-winter.) Therefore, EA and HC populations will experience a broad range of annual photoperiodic changes and there is no evidence from an EA/HC comparison to GA/PNG to suggest a selective pressure for annual photoperiod. Our analysis is of central clock genes (and of a melatonin synthesis gene); perhaps the central clock mechanism has been mechanistically constrained in humans and has therefore not been subject to natural selection along latitudinal clines (in contrast to the situation in Drosophila [Sawyer et al. 2006; Tauber et al. 2007]). However, because the most important input signal for the entrainment of circadian pacemakers is light, it is very possible that the light input pathway has been optimized by natural selection for specific latitudes/photoperiods. Therefore, while we have not found evidence for natural selection acting on central clock genes, our data do not exclude the possibility that genes encoding input pathway components (e.g., melanopsin) will show genetic evidence for natural selection.
Finally, we discovered several new SNPs, including some relatively common polymorphisms in Arntl and Arntl2. These polymorphisms may affect the expression of these two critical clock genes. For example, because 5’UTR regions are often involved in translational control, the novel ss95215855 SNP of Arntl could affect the translational efficiency of Arntl mRNA. In addition, common SNPs in the Arntl2 gene affect regulatory regions such as the promoter (rs5797225, rs7137588, rs11048972, ss95215857, and rs10548381) and intronic regions (rs4964059 and ss95215858). For example, the rs7137588 SNP is in the recognition motif for the EBP-80 transcriptional factor, and therefore this SNP could affect transcriptional regulation of Arntl2. In addition to being key components of the central circadian clockwork, the ARNTL/BMAL1 and ARNTL2/BMAL2 transcriptional factors also mediate a large number of output pathways controlled by the clock via E-box containing promoters. Therefore, polymorphisms in the regulatory regions of the Arntl and Arntl2 genes have the potential to affect many clock-regulated output processes.
Our study unveiled significant differentiation among populations for clock gene allele, genotype and haplotype frequencies that can serve as a basis for future association analyses of clock gene polymorphisms with phenotypes of behavioral and/or medical significance. Although it is impossible based on the present data to determine the motivating factors in the patterns of differentiation, it appears unlikely that these differences are due to consistent patterns of natural selection but rather are more likely to be caused by random processes. This conclusion is based on the large scale differences among the Ghana and PNG populations that should have similar photoperiod and temperature regimes as compared to the other populations studied.
Supplementary Material
Acknowledgements
We thank Matthew Pullen and Kontip C. Mahautmr for technical assistance, the Vanderbilt University DNA Core for DNA storage and sequencing, and Vanderbilt’s General Clinical Research Center (supported in part by grant M01 RR-00095 from the National Center for Research Resources, NIH) for DNA isolation and storage. This research was supported by a Discovery Grant from Vanderbilt University, the Vanderbilt Joint Center for Nursing Research, U01 HL065962 (to Dr. Dan Roden), and grants from the National Institute of Mental Health (R01 MH043836 to CHJ and R01 MH063341 to DGM). CMC was supported by T32 and P50 awards from the National Institute of Mental Health (T32 MH064913; P50 MH078028).
Footnotes
Additional Data (as Tables S1–S10 and Figures S1–S2) are available from the following websites:
Vanderbilt's Center for Human Genetics Research website: http://chgr.mc.vanderbilt.edu/
Johnson Laboratory website: http://www.cas.vanderbilt.edu/johnsonlab/
McMahon Laboratory website: http://sitemason.vanderbilt.edu/mcmahonlab
References
- Archer SN, Robilliard DL, Skene DJ, Smits M, Williams A, Arendt J, von Schantz M. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26:413–415. doi: 10.1093/sleep/26.4.413. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ, DeCoursey PJ, editors. Chronobiology: Biological Timekeeping. Sunderland, MA: Sinauer; 2004. p. 406. [Google Scholar]
- Ebisawa T, Uchiyama M, Kajimura N, Mishima K, Kamei Y, Katoh M, Watanabe T, Sekimoto M, Shibui K, Kim K, Kudo Y, Ozeki Y, Sugishita M, Toyoshima R, Inoue Y, Yamada N, Nagase T, Ozaki N, Ohara O, Ishida N, Okawa M, Takahashi K, Yamauchi T. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO Rep. 2001;2:342–346. doi: 10.1093/embo-reports/kve070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- Friedlaender J, Friedlaender F, Reed F, Kidd K, Kidd J, Chambers G, Lea R, Koki G, Merriwether DA, Weber J. The genetic structure of Pacific Islanders. PLoS Genetics. 2008;4:e19. doi: 10.1371/journal.pgen.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee CC. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Hawley ME, Kidd KK. HAPLO: a program using the EM algorithm to estimate the frequencies of multi-site haplotypes. J Hered. 1995;86:409–411. doi: 10.1093/oxfordjournals.jhered.a111613. [DOI] [PubMed] [Google Scholar]
- Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch JB, Gu YZ, Moran SM, Shimomura K, Radcliffe LA, Takahashi JS, Bradfield CA. The basic helix-loop-helix-PAS protein MOP9 is a brain-specific heterodimeric partner of circadian and hypoxia factors. J Neurosci. 2000;20:RC83. doi: 10.1523/JNEUROSCI.20-13-j0002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzenberg D, Young T, Finn L, Lin L, King DP, Takahashi JS, Mignot E. A CLOCK polymorphism associated with human diurnal preference. Sleep. 1998;21:569–576. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- Kryger MH, Roth T, Dement WC, editors. Principles and Practice of Sleep Medicine. Philadelphia: Elsevier/Saunders; 2005. p. 1517. [Google Scholar]
- Lewy AJ, Emens J, Jackman A, Yuhas K. Circadian uses of melatonin in humans. Chronobiol Int. 2006;23:403–412. doi: 10.1080/07420520500545862. [DOI] [PubMed] [Google Scholar]
- Liu K, Muse S. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian Circadian Biology: Elucidating Genome-Wide Levels of Temporal Organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA. Circadian genes, rhythms and the biology of mood disorders. Pharmacol Ther. 2007;114:222–232. doi: 10.1016/j.pharmthera.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadkarni NA, Weale ME, von Schantz M, Thomas MG. Evolution of a length polymorphism in the human PER3 gene, a component of the circadian system. J Biol Rhythms. 2005;20:490–499. doi: 10.1177/0748730405281332. [DOI] [PubMed] [Google Scholar]
- Nataraj AJ, Olivos-Glander I, Kusukawa N, Highsmith WE., Jr Single-strand conformation polymorphism and heteroduplex analysis for gel-based mutation detection. Electrophoresis. 1999;20:1177–1185. doi: 10.1002/(SICI)1522-2683(19990101)20:6<1177::AID-ELPS1177>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:1893–1899. doi: 10.1371/journal.pbio.0020377. e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer LA, Sandrelli F, Pasetto C, Peixoto AA, Rosato E, Costa R, Kyriacou CP. The period gene Thr-Gly polymorphism in Australian and African Drosophila melanogaster populations: implications for selection. Genetics. 2006;174:465–480. doi: 10.1534/genetics.106.058792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serretti A, Cusin C, Benedetti F, Mandelli L, Pirovano A, Zanardi R, Colombo C, Smeraldi E. Insomnia improvement during antidepressant treatment and CLOCK gene polymorphism. Am J Med Genet B Neuropsychiatr Genet. 2005;137:36–39. doi: 10.1002/ajmg.b.30130. [DOI] [PubMed] [Google Scholar]
- Staels B. When the clock stops ticking, metabolic syndrome explodes. Nat Med. 2006;12:54–55. doi: 10.1038/nm0106-54. [DOI] [PubMed] [Google Scholar]
- Tauber E, Zordan M, Sandrelli F, Pegoraro M, Osterwalder N, Breda C, Daga A, Selmin A, Monger K, Benna C, Rosato E, Kyriacou CP, Costa R. Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science. 2007;316:1895–1898. doi: 10.1126/science.1138412. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Kidd KK. Implications of biogeography of human populations for 'race' and medicine. Nat Genet. 2004;36:S21–S27. doi: 10.1038/ng1438. [DOI] [PubMed] [Google Scholar]
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Viola AU, Archer SN, James LM, Groeger JA, Lo JC, Skene DJ, von Schantz M, Dijk DJ. PER3 Polymorphism Predicts Sleep Structure and Waking Performance. Curr Biol. 2007;17:613–618. doi: 10.1016/j.cub.2007.01.073. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GY, Lee CGL, Lee EJD. Genetic Variability of Arylalkylamine-NAcetyl-Transferase (AA-NAT) Gene and Human Sleep/Wake Pattern. Chronobiol Int. 2004;21:229–237. doi: 10.1081/cbi-120037822. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Williams SM, Addy JH, Phillips JA, 3rd, Dai M, Kpodonu J, Afful J, Jackson H, Joseph K, Eason F, Murray MM, Epperson P, Aduonum A, Wong LJ, Jose PA, Felder RA. Combinations of variations in multiple genes are associated with hypertension. Hypertension. 2000;36:2–6. doi: 10.1161/01.hyp.36.1.2. [DOI] [PubMed] [Google Scholar]
- Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH. Functional consequences of a CKIδ mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.