Abstract
Objective
Aldose reductase (AR) is the rate-limiting enzyme of the polyol pathway. In diabetes, it is related to microvascular complications. We discovered AR expression in foam cells by gene chip screening and hypothesized that it may be relevant in atherosclerosis.
Methods and Results
AR gene expression and activity were found to be increased in human blood monocyte-derived macrophages during foam cell formation induced by oxidized LDL (oxLDL, 100 μg/mL). AR activity as photometrically determined by NADPH consumption was effectively inhibited by the AR inhibitor epalrestat. oxLDL-dependent AR upregulation was further increased under hyperglycemic conditions (30 mM D-glucose) as compared to osmotic control, suggesting a synergistic effect of hyperlipidemia and hyperglycemia. AR was also upregulated by 4-hydroxynonenal, a constituent of oxLDL. Upregulation was blocked by an antibody to CD36. AR inhibition resulted in reduction of oxLDL-induced intracellular oxidative stress as determined by 2′7′-dichlorofluoresceine diacetate (H2DCFDA) fluorescence, indicating that pro-inflammatory effects of oxLDL are partly mediated by AR. Immunohistochemistry showed AR expression in CD68+ human atherosclerotic plaque macrophages.
Conclusions
These data show that oxLDL-induced upregulation of AR in human macrophages is pro-inflammatory in foam cells and may represent a potential link between hyperlipidemia, atherosclerosis and diabetes mellitus.
Keywords: Atherosclerosis, diabetes mellitus, lipoproteins, macrophage, plaque
Diabetes mellitus represents one of the major risk factors for atherosclerosis 1. Mechanisms by which diabetes perturbs cellular integrity include formation of advanced glycation end products (AGE), activation of protein kinase C (PKC), upregulation of 12/15-lipoxygenase and increased flux through the hexosamine or the polyol pathway 2, 3. AGE formation may result in cross-linking and subsequent conformational changes of many proteins, increased inflammatory signaling and oxidative stress in various cell types including macrophages and endothelial cells 2. Similarly, activation of PKC isoforms leads to endothelial dysfunction by activation of NADPH oxidase, enhanced vascular permeability through upregulation of vascular endothelial growth factor and thrombosis by increasing plasminogen activator inhibitor-1 levels 4. The contribution of the polyol pathway to diabetes-induced atherogenesis is currently not completely understood.
The polyol pathway includes aldose reductase (AR, AKR1B1), which is the rate-limiting enzyme, and sorbitol dehydrogenase (SDH, SORD) 3. Its physiological role is controversial. AR activity has been proposed to be protective against oxidative stress by detoxifying lipid peroxidation products, e.g. in vasculitis 5, 6, but inhibiting AR does not increase lipid peroxidation products 7. By contrast, AR activity has also been related to greater oxidative stress and increased damage during brain ischemia in mice 8. In keeping with a pro-inflammatory role of AR, AR null mice exhibit reduced oxidative stress and are protected from ischemic injury 8.
The main physiological substrates of the polyol pathway are aldehydes, but sugars like glucose or galactose have also been shown to be metabolized by AR 9. With rising intracellular glucose levels as seen in diabetics, glucose can be shunted into the polyol pathway resulting in sorbitol production, which is subsequently metabolized by sorbitol dehydrogenase to fructose 3. This leads to increased AGE formation due to increased fructose levels and increased oxidative stress due to consumption of NADPH, which is needed to regenerate glutathione 3. Thus, the polyol pathway has been implicated in microvascular diabetic complications like cataract 10, nephropathy 11, and neuropathy 12. Pharmacologic inhibition of AR has been shown to improve nerve conduction velocity and symptoms of polyneuropathy 13. Recently, the polyol pathway has been linked to atherosclerosis, as overexpression of human AR in LDL receptor deficient diabetic mice (which physiologically display only low AR expression) increased atherosclerotic lesions 14. However, the mechanisms by which AR promotes atherosclerosis remain still to be elucidated.
In a study of gene expression during foam cell formation by gene chip analysis, we found a significant increase of AR mRNA expression after treatment of primary human blood monocyte-derived macrophages with oxidized low density lipoprotein (oxLDL) 15. We therefore hypothesized that increased AR expression in human foam cells may represent a novel pro-inflammatory mechanism by which oxLDL increases oxidative stress and which may link two of the major risk factors for atherosclerosis, diabetes and hyperlipidemia.
Materials and methods
Macrophages and foam cells
Macrophages were derived from monocytes isolated from either peripheral blood or buffy coats provided by the Virginia Blood Services (Richmond, VA) as described previously 15. The study was approved by the institutional review committee. All subjects gave informed consent. Blood was drawn from fifteen healthy individuals (8 female, 7 male, age 20–40 years). Blood monocytes were differentiated to macrophages by macrophage colony-stimulating factor (M-CSF) or platelet factor-4 (CXCL4) for six days and exposed to vehicle, oxidized LDL (Biomedical Technologies, Stoughton, MA) or 4-hydroxynonenal (4-HNE, Sigma, St. Louis, MO) as indicated. For receptor blocking experiments, cells were pre-incubated with mAb to CD36 antibody (clone CB38) or IgM control (0.5 μg/1×106 cells, both BD Biosciences, San Diego, CA) for 30 minutes before addition of oxLDL 16 (see supplementary information).
Gene chip experiments
RNA was extracted from cells using the RNEasy kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Gene expression was measured in duplicates at the University of Virginia Gene Expression Core Facility using Affymetrix equipment and H133A chips as described. Gene expression data are available at the NCBI gene expression and hybridization array data repository (http://www.ncbi.nlm.nih.gov/geo/, series GSE7138).
Real-time PCR
Total RNA was isolated from cultured macrophages and foam cells using the RNEasy Mini Kit with DNase treatment. Reverse transcription was performed with the Omni-script RT Kit (all Qiagen, Valencia, CA). Gene expression was measured using iQ SYBR green supermix in an iCycler iQ Real-Time Detection System (both BioRad, Hercules, CA). Product specificity was ascertained by melting curve analysis and initially by electrophoresis on agarose gels. Primers were purchased from Integrated DNA Technologies (Coralville, IA). Primer sequences were as follows: Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward 5′-GGCTCATGACCACAGTCCAT-3′, reverse 5′-GCCTGCTTCACCACCTTCT -3′ 17, aldose reductase (AKR1B1) forward 5′-AGTCGGGCAATGTGGTTCCC-3′, reverse 5′-GGATTAACTTCTCCTGAGTG-3′ 14. All samples were run in duplicates. Values were determined using iCycler iQ Real-Time Detection System software (version 3.1; Bio-Rad, Hercules, CA). Relative gene expression was calculated as described previously using GAPDH as housekeeping gene 18.
Aldose reductase activity assay and inhibition experiments
AR activity was measured photometrically as described previously 5, 19 (see supplementary information). For AR inhibition experiments, epalrestat (Sequioa Research Products, Pangbourne, UK) was used. The dosage was chosen based on the IC50 (ranging from 0.012 20 to 0.021 μM 21) and effective inhibition of AR activity in monocyte-derived macrophages. For AR inhibition experiments, epalrestat was added to the culture medium at a concentration of 0.2 μM 24 hours before addition of oxLDL, which completely inhibited AR activity.
Oxidative stress assay
Generation of reactive oxygen species was measured as described previously using 2′7′-dichlorofluoresceine diacetate (H2DCFDA), a dye which converts to fluorescent 2′7′-dichlorfluorescein (DCF) in the presence of reactive oxygen species 22 (see supplementary information).
Immunohistochemistry
Human coronary arteries were obtained from the University of Vir-ginia Department of Pathology/Tissue bank and were stained for AR, CD68 or smooth muscle α-actin. Immunofluorescence was performed to determine colocalization of AR with CD68 or smooth muscle α-actin (for details of staining procedure see supplemental materials). Coronary arteries from eight donors were studied, two of whom were diabetic.
Statistics
Differences between the groups were evaluated by two-tailed t test or 1-way ANOVA followed by a post-hoc Tukey test. The exact details of gene array data analysis have been described previously 15 (see supplementary information). Generally, P<0.05 was considered statistically significant. Data are presented as mean ± SEM. Statistical analysis was performed using PRISM (GraphPad, San Diego, CA).
The authors had full access to the data and take responsibility for its integrity. All authors have read and agreed to the manuscript as written.
Results
AR expression in human macrophages is increased during oxLDL-dependent foam cell formation
Cell culture of human blood-derived monocytes with M-CSF (100 ng/mL) or CXCL4 (100 ng/mL) for six days resulted in macrophage formation. Large scale analysis of gene expression using Affymetrix gene HU133A arrays revealed a significant increase of AR mRNA in macrophages generated with either M-CSF or CXCL4 as compared to peripheral blood mononu-clear cells (PBMC) or monocytes (P<0.0001). Incubation of monocyte-derived macrophages with 100μg/mL mmLDL or oxLDL for 48 hours reproducibly resulted in foam cell formation as assessed by oil-O-red staining (Figure S1B). AR gene expression was further increased during foam cell formation caused by oxLDL but not by mmLDL (P=0.10 for M-CSF, P<0.0001 for CXCL4) (Figure S2A). A full set of other genes significantly regulated during foam cell formation by Affymetrix chip analysis is given in supplementary table S1.
The time course of oxLDL-dependent upregulation of AR mRNA was investigated by real-time PCR revealing an early peak four to six hours after addition of oxLDL. The peak reached about 60 and 80 % above baseline in macrophages generated with M-CSF or CXCL4, respectively (P<0.01 in both cases, Figure 1A and S2B). A second increase was seen between 24 and 48 hours.
Figure 1. AR gene expression and activity during foam cell formation.
(A) Gene expression in macrophages exposed to oxLDL (100 μg/mL) for indicated periods.
(B) AR activity in macrophages after exposure to oxLDL (48 hours) with or without epalrestat treatment (means+SEM, *** P<0.001, ** P<0.01, * P<0.05).
To investigate whether increased AR gene expression was accompanied by functional changes, AR activity was determined in macrophages generated with M-CSF or CXCL4 before and 48 hours after addition of oxLDL. AR activity was significantly increased in oxLDL-treated macrophages irrespective of the growth factor used for differentiation (P<0.01). Treatment of macrophages with epalrestat (0.2 μM) resulted in complete inhibition of AR activity in both cases (Figure 1B and S2C).
Molecular mechanism of oxLDL-induced AR upregulation
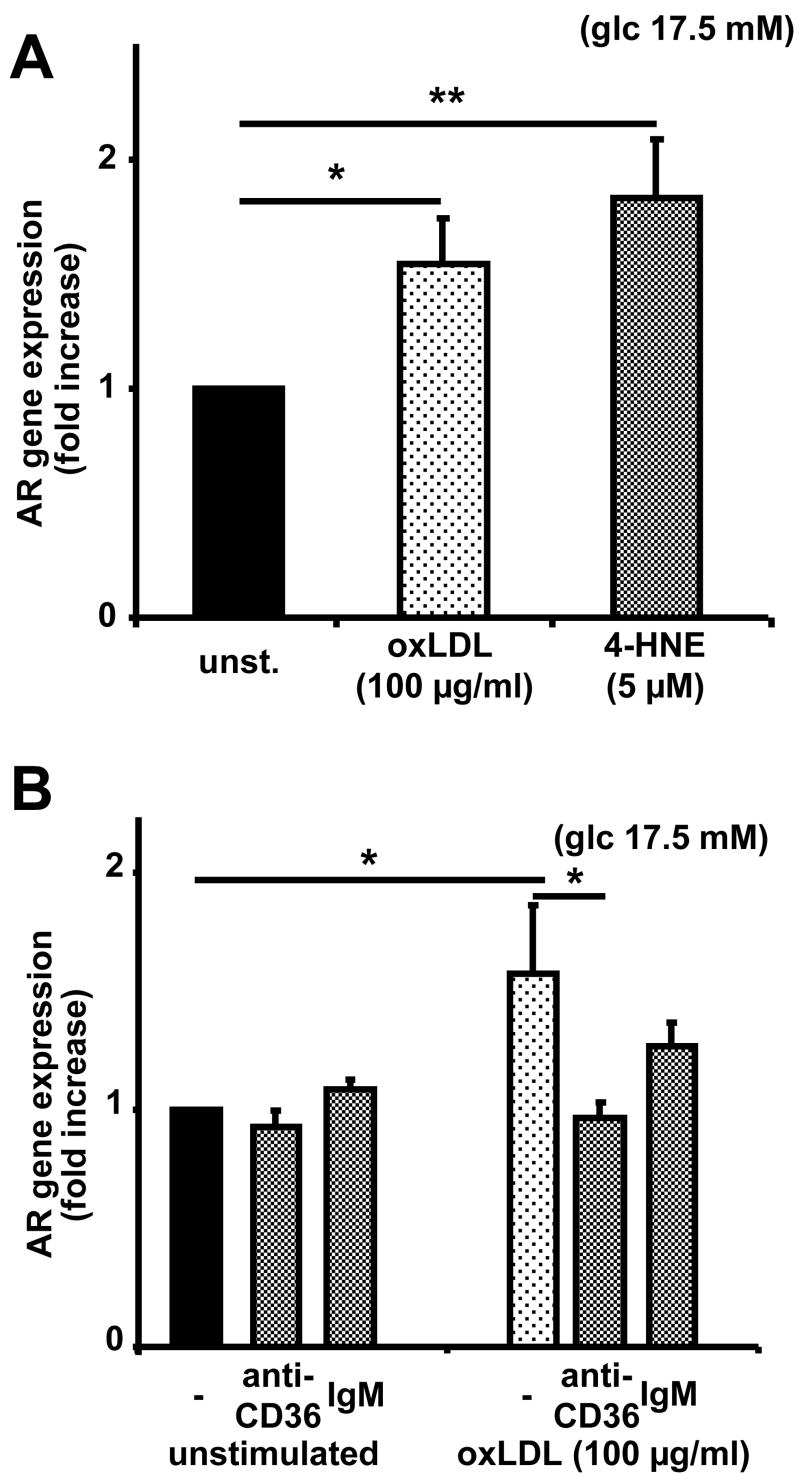
To investigate the mechanisms by which oxLDL upregulates AR gene expression, different constituents of oxLDL were tested for their ability to upregulate AR in macrophages. Previous studies have reported increased AR expression in rat vascular smooth muscle cells or human lymphocytes after stimulation with 4-hydroxynonenal (4-HNE), a constituent of oxLDL, displaying a peak at a 4-HNE concentration of 5 μM 5, 6. We confirm this finding in macrophages (Figure 2A). To identify the receptor involved, we used a blocking mAb against the scavenger receptor CD36, which accounts for up to two thirds of oxLDL uptake by macrophages 23. Anti-CD36 antibody significantly reduced oxLDL-induced AR upregulation (Figure 2B).
Figure 2. 4-hydroxynonenal upregulates AR mRNA expression to the same extent as oxLDL.
(A) Gene expression in macrophages treated with vehicle (unst.), oxLDL or 4-hydroxynonenal for 6 hours.
(B) Gene expression in macrophages treated with vehicle or oxLDL in the presence of IgM to CD36 (anti-CD36) or irrelevant IgM (means+SEM, * P<0.05, ** P<0.01)
oxLDL-dependent upregulation of AR depends on hyperglycemic conditions
As mentioned previously, AR expression has been shown to be of clinical importance in diabetic individuals. To assess the effects of oxLDL on AR expression and activity not only under standard cell culture conditions with a glucose concentration of 17.5 mM 15, but also under normoglycemic and hyperglycemic conditions, AR expression and activity were measured with 5 mM glucose or 30 mM glucose in the culture media.
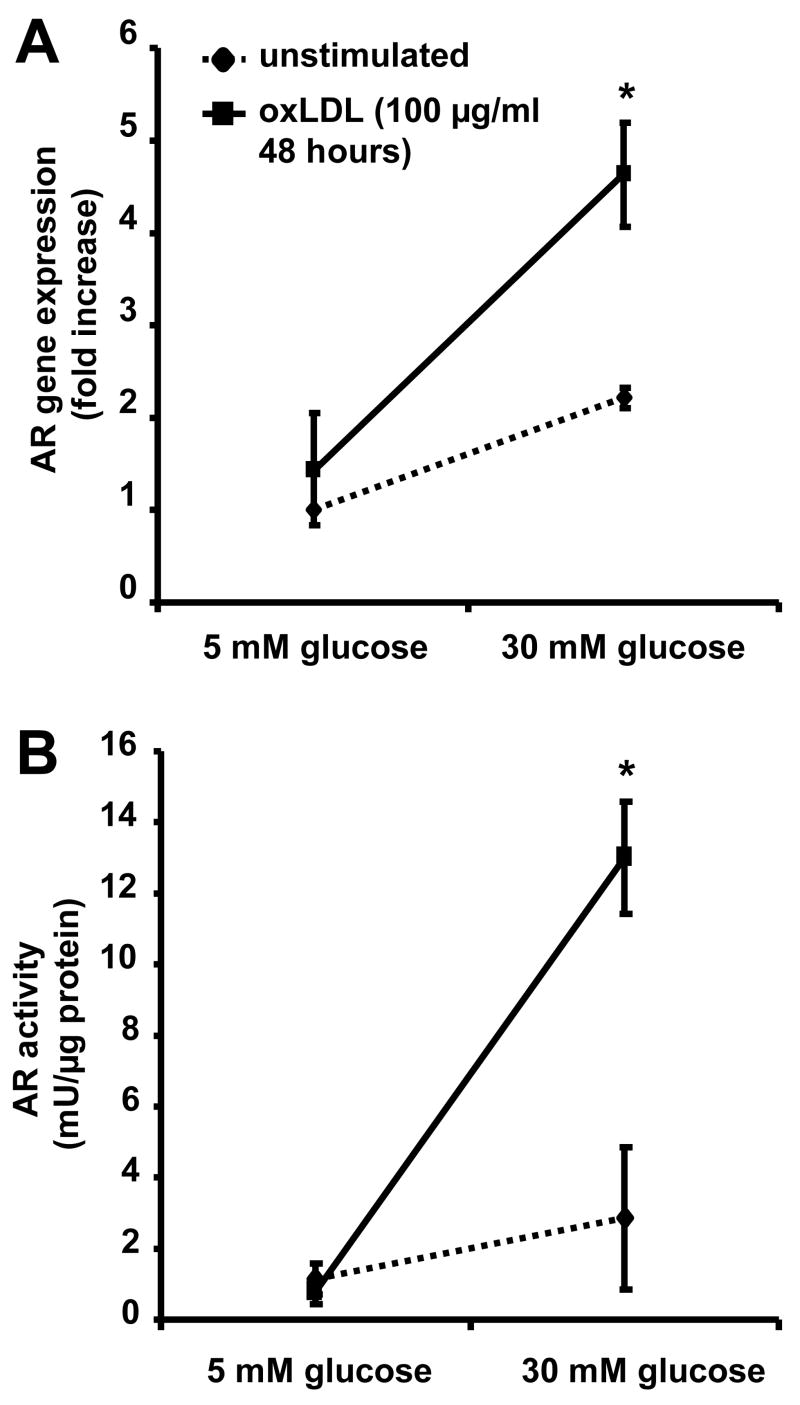
Real-time PCR revealed a significant increase of AR gene expression under hyperglycemic as compared to normoglycemic conditions (P<0.05). Under hyperglycemic conditions, stimulation of macrophages with oxLDL resulted in a significant increase of AR gene expression (P<0.05; Figure 3A). Exposure of macrophages to oxLDL resulted in a tenfold increase of AR activity under hyperglycemic, but not normoglycemic conditions (P<0.01). The activity increase exceeded the increase in mRNA expression levels (Figure 3B).
Figure 3. High glucose levels and oxLDL have synergistic effects on AR expression and activity.
(A) Gene expression in macrophages treated with oxLDL for 48 hours under normoglycemic (glucose 5 mM) or hyperglycemic (glucose 30 mM) conditions (means+SEM, * P<0.05)
(B) AR activity in macrophages treated with oxLDL under normoglycemic or hyperglycemic conditions (means+SEM, * P<0.05)
Ox-LDL dependent AR upregulation is not related to increased gene expression of pro-inflammatory mediators but to oxidative stress in macrophages
AR expression has been related to expression of pro-inflammatory cytokines induced by LPS in mouse peritoneal macrophages 24. However, exposure of macrophages to oxLDL under the conditions applied in this study did not result in increased gene expression of inflammatory mediators like tumor necrosis factor-α (TNαF), interleukin-6 (IL-6) or IL-1β (Figure S3).
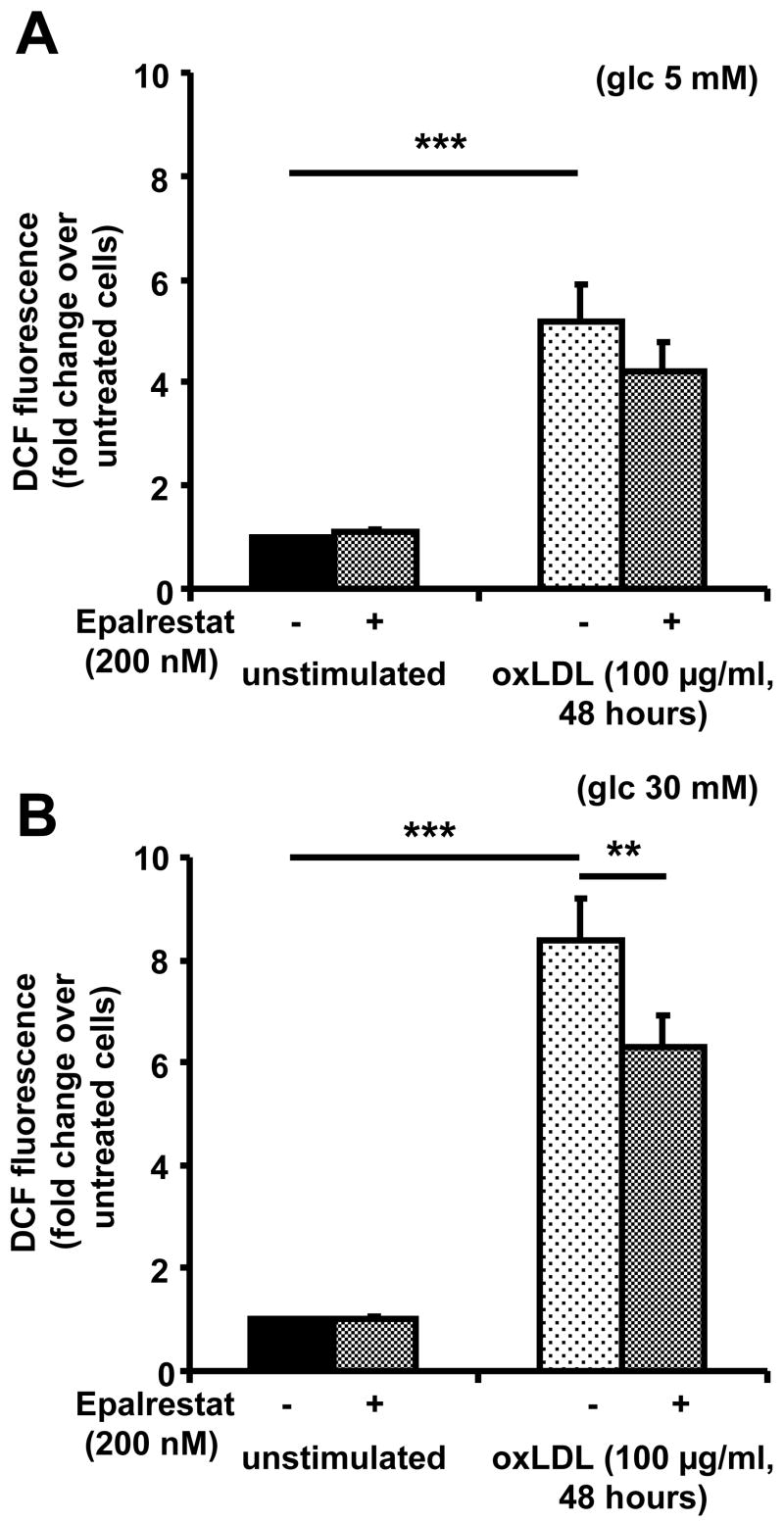
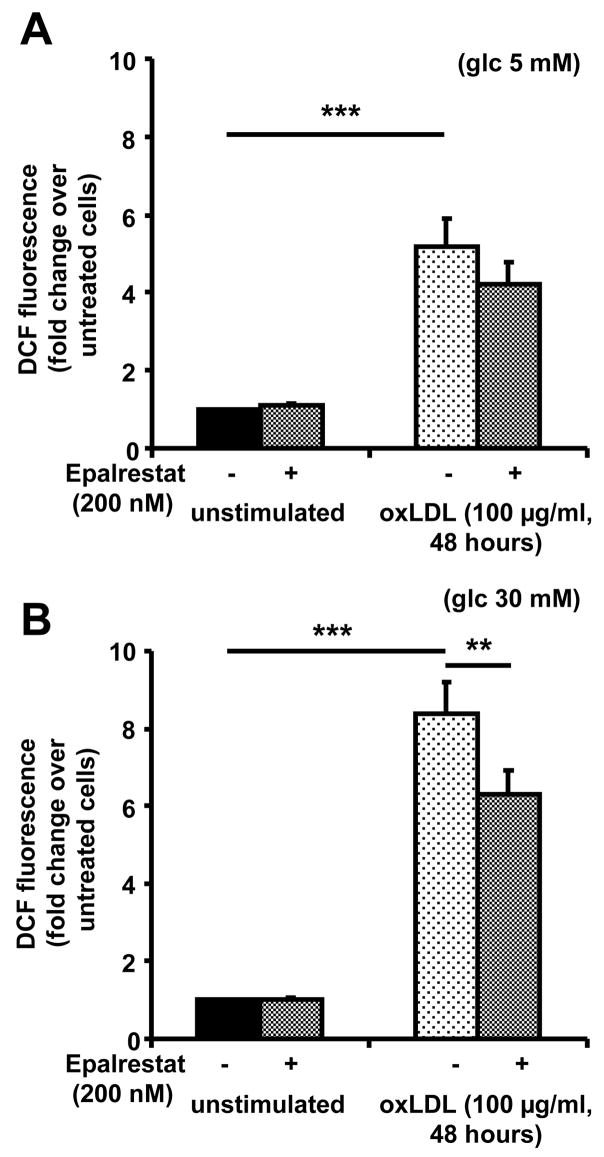
OxLDL has been described to increase production of reactive oxygen species in macrophages 25. AR activity in foam cells may represent a pro-inflammatory mechanism, by which oxLDL increases oxidative stress in the macrophage. To test this hypothesis, intracellular oxidative stress was measured in macrophages under different conditions using 2,7-dichlorfluorescein, a dye which displays increased fluorescence intensity in the presence of reactive oxygen species. Exposure of macrophages to oxLDL (100 μg/mL) for 48 hours resulted in a significant increase of DCF fluorescence under normoglycemic (5 mM) as well as under hyperglycemic (30 mM) conditions (P<0.001). Under hyperglycemia, oxLDL induced an 8-fold increase of DCF fluorescence, compared to fivefold under normoglycemic conditions (Figure 4). Pre-treatment of macrophages with epalrestat (0.2 μM) for 24 hours before and throughout exposure to oxLDL resulted in a statistically significant (P<0.01) reduction of ROS formation under hyperglycemic conditions. Treatment of macrophages with epalrestat alone did not have any effect. These findings suggest that under hyperglycemic conditions about 20–30 % of the oxidative stress in macrophage foam cells is due to AR activity.
Figure 4. oxLDL-induced AR upregulation causes increased formation of reactive oxygen species.
Macrophages generated under (A) normoglycemic or (B) hyperglycemic conditions were treated with vehicle (unstimulated) or oxLDL with or without 24 hour pre-treatment with epalrestat. Cells were stained with 2′7′-dichlorofluoresceine diacetate (10 μM) for 30 minutes, and fluorescence was determined by flow cytometry (means+SEM of fold change over untreated cells, ** P<0.01, *** P<0.001).
AR is expressed in macrophages in human atherosclerotic plaque
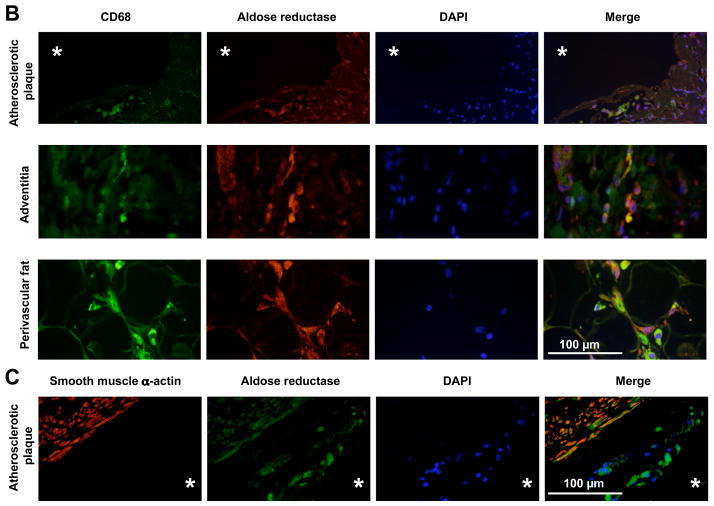
To assess the relevance of our findings for the pathogenesis of atherosclerosis, AR expression in human atherosclerotic plaque was investigated. Immunohistochemistry revealed that AR was abundantly expressed in a wide range of cells in the vascular wall, mostly vascular smooth muscle cells, including cells in atherosclerotic plaques (Figure 5A). AR expression in macrophage foam cells was verified by immunofluorescence demonstrating colocalization of CD68 and AR in atherosclerotic lesions (Figure 5B). AR was also colocalized with CD68 in macrophages in the vascular adventitia and perivascular adipose tissue (Figure 5B). Colocalization with smooth muscle α-actin confirmed AR expression in vascular smooth muscle cells, which had been described previously 26 (Figure 5C). Interestingly, not all CD68 positive cells in lesions expressed AR ranging from 75 to 100 %. The proportion of AR-expressing macrophages was similar in the adventitia, perivascular adipose tissue and within the atherosclerotic lesions (Figure S4).
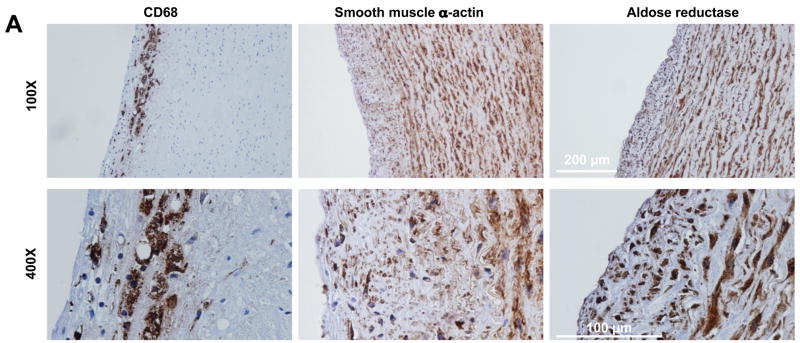
Figure 5. AR expression in atherosclerotic lesions.
(A) Immunohistochemistry demonstrating expression of CD68 (macrophages), smooth muscle α-actin (smooth muscle cells) and AR in atherosclerotic lesions of human coronary artery. 100X and 400X magnification.
(B) Immunofluorescence demonstrating colocalization of CD68 (green) and AR (red) in cells in atherosclerotic lesions, adventitia and perivascular adipose tissue of human coronary artery. DAPI (blue) is shown as nuclear stain. * indicates the lumen of the vessel, 400X magnification.
(C) Immunofluorescence demonstrating colocalization of smooth muscle α-actin (red) and AR (green) in cells in atherosclerotic lesions in human coronary artery. * indicates the lumen of the vessel, 400X magnification.
Discussion
This paper describes the discovery of increased aldose reductase (AR) gene expression and activity in macrophages during foam cell formation induced by oxLDL in the presence of high glucose. AR was similarly upregulated by 4-hydroxynonenal, a constituent of oxLDL. Most of the oxLDL-induced upregulation of AR expression was blocked by mAb to CD36. Inhibition of AR using the pharmacological inhibitor epalrestat indicates that AR upregulation accounts for about 20% of oxLDL-induced oxidative stress. AR expression in macrophages in atherosclerotic lesions and the vascular adventitia suggests that macrophage AR may represent a link between diabetes and atherosclerosis.
In diabetics, glucose is shunted into the polyol pathway and thus converted by AR into sorbitol and subsequently to fructose by sorbitol dehydrogenase 9. For several reasons, this is considered harmful: Firstly, activation of AR results in decreased NADPH levels leading to reduced regeneration of glutathione which is a key protective factor against oxidative stress 3. Reduced NADPH levels may also lead to increased levels of 12-hydroperoxyeicosatetraonic acid (12-HPETE), which cannot be reduced to less toxic 12-hydroxyeicosatetraenonic (12-HETE) by glu-tathione peroxidase 27. Increased fructose levels cause increased advanced glycation end products, which interfere with many cellular processes 3.
AR expression and activity have been described in various tissues including the lens of the eye, kidney, and nerve in several animal models including rat, rabbit, and pig 28. A causal relation between metabolization of glucose by AR and diabetic complications was shown in several animal models of cataract 29 and renal dysfunction 30 or neuropathy 31. Similarly, AR activity has been associated with cataract 10, nephropathy 11, and polyneuropathy 12 in diabetic patients. Clinical studies have demonstrated that AR inhibition reduces symptoms of diabetic neuropathy 13. In the vasculature, AR is expressed in rat smooth muscle cells as demonstrated in rats 14 and human umbilical vein endothelial cells 32. Furthermore, AR expression was found in smooth muscle cells and mononuclear cells in vivo in human temporal artery specimens from patients with giant cell arteriitis 6.
A potential role for AR in atherosclerosis was demonstrated in LDL receptor knock-out mice overexpressing human AR 14. Normoglycemic AR overexpressing Ldlr−/− mice fed a Western diet did not display increased atherosclerotic lesions as compared to Ldlr−/− mice not expressing human AR. However, a significant increase in lesion size was demonstrated in the AR overex-pressing mice after streptozotocin (STZ) injection, which induces diabetes by selectively killing pancreatic β cells, supporting a pathogenetic role of the polyol pathway for atherogenesis in diabetes 14. The mechanism described here may at least partially account for increased atherogenesis in this mouse model. Macrophages harvested from human atherosclerotic plaques by laser capture microdissection showed 75% increased AR mRNA expression as measured by gene chip analysis (Volger OL et al., http://www.ncbi.nlm.nih.gov/geo/, series GSE7074), supporting a role for the polyol pathway in human atherosclerosis.
Our findings that oxLDL-induced AR upregulation increases oxidative stress suggest that the harmful effects clearly outweigh any potentially beneficial role of the polyol pathway. The protective effects of AR inhibition on ROS formation seem to be more pronounced under hyperglycemic conditions, suggesting that harmful effects of AR require oxLDL as well as elevated glucose levels. AR accounts for 20–30 % of oxidative stress as demonstrated by the significant reduction of oxidative stress after inhibition of AR with epalrestat. Other pro-oxidant pathways like lipoxygenases33, cyclooxygenases 33 and NADPH oxidase34 are likely to contribute the remaining oxidative stress.
AR gene regulation involves three osmotic response elements and one androgen response element; there may also be one glucose response element 35. Accordingly, increased AR expression under hyperglycemic conditions has been shown in different cell types 36, 37. Here, we extend this finding to human monocyte-derived macrophages. According to our results, 4-HNE is a key constituent of oxLDL responsible for AR upregulation. This is consistent with previous data demonstrating 4-HNE-dependent AR upregulation in PBL 6. The fact that blocking experiments using antibody against CD36 – a scavenger receptor responsible for about two thirds of oxLDL uptake into macrophages 23 – resulted in a significant inhibition of oxLDL-induced AR upregulation suggest that uptake of oxLDL mediated by CD36 is necessary for AR upregulation.
In conclusion, we demonstrate hyperglycemia-related upregulation of AR gene expression and activity as a new pro-inflammatory mechanism of oxLDL in human macrophages with potential relevance for atherogenesis, thereby identifying a potential link between two of the most important atherosclerotic risk factors, namely diabetes and hyperlipidemia.
Supplementary Material
Acknowledgments
We thank Dr. Nancy Harthun, University of Virginia, Charlottesville/VA for providing carotid endatherectomy specimens, Dr. Maria-Beatriz Lopes, University of Virginia, Charlottesville/VA for providing post mortem coronary artery specimens, and Dr. Aruni Bhatnagar, University of Kentucky, Louisville/KY for providing an antibody against human aldose reductase.
Sources of Funding
This work was supported by the Deutsche Forschungsgemeinschaft (grant GL599/1-1 to C.A.G.), by NIH grant HL58108 (to K.L.), and by NIH grant HL55798 (to K.L. and J.L.N.).
Footnotes
Disclosures
None.
References
- 1.Haffner SM. Coronary heart disease in patients with diabetes. N Engl J Med. 2000;342:1040–1042. doi: 10.1056/NEJM200004063421408. [DOI] [PubMed] [Google Scholar]
- 2.Natarajan R, Nadler JL. Lipid inflammatory mediators in diabetic vascular disease. Arterioscler Thromb Vasc Biol. 2004;24:1542–1548. doi: 10.1161/01.ATV.0000133606.69732.4c. [DOI] [PubMed] [Google Scholar]
- 3.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 4.Williams MD, Nadler JL. Inflammatory mechanisms of diabetic complications. Curr Diab Rep. 2007;7:242–248. doi: 10.1007/s11892-007-0038-y. [DOI] [PubMed] [Google Scholar]
- 5.Spycher SE, Tabataba-Vakili S, O’Donnell VB, Palomba L, Azzi A. Aldose reductase induction: A novel response to oxidative stress of smooth muscle cells. FASEB J. 1997;11:181–188. doi: 10.1096/fasebj.11.2.9039961. [DOI] [PubMed] [Google Scholar]
- 6.Rittner HL, Hafner V, Klimiuk PA, Szweda LI, Goronzy JJ, Weyand CM. Aldose reduc-tase functions as a detoxification system for lipid peroxidation products in vasculitis. J Clin Invest. 1999;103:1007–1013. doi: 10.1172/JCI4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomlinson DR, Stevens EJ, Diemel LT. Aldose reductase inhibitors and their potential for the treatment of diabetic complications. Trends Pharmacol Sci. 1994;15:293–297. doi: 10.1016/0165-6147(94)90010-8. [DOI] [PubMed] [Google Scholar]
- 8.Lo AC, Cheung AK, Hung VK, Yeung CM, He QY, Chiu JF, Chung SS, Chung SK. Deletion of aldose reductase leads to protection against cerebral ischemic injury. J Cereb Blood Flow Metab. 2007;27:1496–1509. doi: 10.1038/sj.jcbfm.9600452. [DOI] [PubMed] [Google Scholar]
- 9.Petrash JM. All in the family: Aldose reductase and closely related aldo-keto reductases. Cell Mol Life Sci. 2004;61:737–749. doi: 10.1007/s00018-003-3402-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kyselova Z, Stefek M, Bauer V. Pharmacological prevention of diabetic cataract. J Diabetes Complications. 2004;18:129–140. doi: 10.1016/S1056-8727(03)00009-6. [DOI] [PubMed] [Google Scholar]
- 11.Larkins RG, Dunlop ME. The link between hyperglycaemia and diabetic nephropathy. Diabetologia. 1992;35:499–504. doi: 10.1007/BF00400475. [DOI] [PubMed] [Google Scholar]
- 12.McHugh JM, McHugh WB. Diabetes and peripheral sensory neurons: What we don’t know and how it can hurt us. AACN Clin Issues. 2004;15:136–149. doi: 10.1097/00044067-200401000-00012. [DOI] [PubMed] [Google Scholar]
- 13.Hotta N, Akanuma Y, Kawamori R, Matsuoka K, Oka Y, Shichiri M, Toyota T, Naka-shima M, Yoshimura I, Sakamoto N, Shigeta Y. Long-term clinical effects of epalrestat, an aldose reductase inhibitor, on diabetic peripheral neuropathy: The 3-year, multicenter, comparative aldose reductase inhibitor-diabetes complications trial. Diabetes Care. 2006;29:1538–1544. doi: 10.2337/dc05-2370. [DOI] [PubMed] [Google Scholar]
- 14.Vikramadithyan RK, Hu Y, Noh HL, Liang CP, Hallam K, Tall AR, Ramasamy R, Gold-berg IJ. Human aldose reductase expression accelerates diabetic atherosclerosis in trans-genic mice. J Clin Invest. 2005;115:2434–2443. doi: 10.1172/JCI24819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho HJ, Shashkin P, Gleissner CA, Dunson D, Jain N, Lee JK, Miller Y, Ley K. Induction of dendritic cell-like phenotype in macrophages during foam cell formation. Physiol Genomics. 2007;29:149–160. doi: 10.1152/physiolgenomics.00051.2006. [DOI] [PubMed] [Google Scholar]
- 16.Kapinsky M, Torzewski M, Buchler C, Duong CQ, Rothe G, Schmitz G. Enzymatically degraded LDL preferentially binds to cd14(high) cd16(+) monocytes and induces foam cell formation mediated only in part by the class B scavenger-receptor CD36. Arterioscler Thromb Vasc Biol. 2001;21:1004–1010. doi: 10.1161/01.atv.21.6.1004. [DOI] [PubMed] [Google Scholar]
- 17.Reutershan J, Morris MA, Burcin TL, Smith DF, Chang D, Saprito MS, Ley K. Critical role of endothelial CXCR2 in LPS-induced neutrophil migration into the lung. J Clin Invest. 2006;116:695–702. doi: 10.1172/JCI27009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wermuth B, von Wartburg JP. Aldose reductase from human tissues. Methods Enzymol. 1982;89:181–186. doi: 10.1016/s0076-6879(82)89032-0. [DOI] [PubMed] [Google Scholar]
- 20.Mizuno K, Suzuki T, Tanaka T, Taniko K. Inhibitory effects of fidarestat on aldose reductase and aldehyde reductase activity evaluated by a new method using hplc with post-column spectrophotometric detection. Biol Pharm Bull. 2000;23:244–248. doi: 10.1248/bpb.23.244. [DOI] [PubMed] [Google Scholar]
- 21.Ishii A, Kotani T, Nagaki Y, Shibayama Y, Toyomaki Y, Okukado N, Ienaga K, Okamoto K. Highly selective aldose reductase inhibitors. 1. 3-(arylalkyl)-2,4,5-trioxoimidazolidine-1-acetic acids. J Med Chem. 1996;39:1924–1927. doi: 10.1021/jm9508393. [DOI] [PubMed] [Google Scholar]
- 22.Rajagopalan S, Meng XP, Ramasamy S, Harrison DG, Galis ZS. Reactive oxygen species produced by macrophage-derived foam cells regulate the activity of vascular matrix met-alloproteinases in vitro. Implications for atherosclerotic plaque stability. J Clin Invest. 1996;98:2572–2579. doi: 10.1172/JCI119076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore KJ, Freeman MW. Scavenger receptors in atherosclerosis: Beyond lipid uptake. Arterioscler Thromb Vasc Biol. 2006;26:1702–1711. doi: 10.1161/01.ATV.0000229218.97976.43. [DOI] [PubMed] [Google Scholar]
- 24.Ramana KV, Srivastava SK. Mediation of aldose reductase in lipopolysaccharide-induced inflammatory signals in mouse peritoneal macrophages. Cytokine. 2006;36:115–122. doi: 10.1016/j.cyto.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer B, von Knethen A, Brune B. Dualism of oxidized lipoproteins in provoking and attenuating the oxidative burst in macrophages: Role of peroxisome proliferator-activated receptor-γ. J Immunol. 2002;168:2828–2834. doi: 10.4049/jimmunol.168.6.2828. [DOI] [PubMed] [Google Scholar]
- 26.Ruef J, Liu SQ, Bode C, Tocchi M, Srivastava S, Runge MS, Bhatnagar A. Involvement of aldose reductase in vascular smooth muscle cell growth and lesion formation after arterial injury. Arterioscler Thromb Vasc Biol. 2000;20:1745–1752. doi: 10.1161/01.atv.20.7.1745. [DOI] [PubMed] [Google Scholar]
- 27.Funk CD. The molecular biology of mammalian lipoxygenases and the quest for eicosa-noid functions using lipoxygenase-deficient mice. Biochim Biophys Acta. 1996;1304:65–84. doi: 10.1016/s0005-2760(96)00107-5. [DOI] [PubMed] [Google Scholar]
- 28.Markus HB, Raducha M, Harris H. Tissue distribution of mammalian aldose reductase and related enzymes. Biochem Med. 1983;29:31–45. doi: 10.1016/0006-2944(83)90051-0. [DOI] [PubMed] [Google Scholar]
- 29.Sun W, Oates PJ, Coutcher JB, Gerhardinger C, Lorenzi M. A selective aldose reductase inhibitor of a new structural class prevents or reverses early retinal abnormalities in experimental diabetic retinopathy. Diabetes. 2006;55:2757–2762. doi: 10.2337/db06-0138. [DOI] [PubMed] [Google Scholar]
- 30.Nishikawa T, Edelstein D, Du XL, Yamagishi S, Matsumura T, Kaneda Y, Yorek MA, Beebe D, Oates PJ, Hammes HP, Giardino I, Brownlee M. Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature. 2000;404:787–790. doi: 10.1038/35008121. [DOI] [PubMed] [Google Scholar]
- 31.Kuzumoto Y, Kusunoki S, Kato N, Kihara M, Low PA. Effect of the aldose reductase inhibitor fidarestat on experimental diabetic neuropathy in the rat. Diabetologia. 2006;49:3085–3093. doi: 10.1007/s00125-006-0400-7. [DOI] [PubMed] [Google Scholar]
- 32.Ramana KV, Bhatnagar A, Srivastava SK. Inhibition of aldose reductase attenuates TNF-α-induced expression of adhesion molecules in endothelial cells. FASEB J. 2004;18:1209–1218. doi: 10.1096/fj.04-1650com. [DOI] [PubMed] [Google Scholar]
- 33.Shashkin P, Dragulev B, Ley K. Macrophage differentiation to foam cells. Curr Pharm Des. 2005;11:3061–3072. doi: 10.2174/1381612054865064. [DOI] [PubMed] [Google Scholar]
- 34.Griendling KK. Novel NAD(P)H oxidases in the cardiovascular system. Heart. 2004;90:491–493. doi: 10.1136/hrt.2003.029397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danesh FR, Wada J, Wallner EI, Sahai A, Srivastava SK, Kanwar YS. Gene regulation of aldose-, aldehyde- and a renal specific oxido reductase (RSOR) in the pathobiology of diabetes mellitus. Curr Med Chem. 2003;10:1399–1406. doi: 10.2174/0929867033457368. [DOI] [PubMed] [Google Scholar]
- 36.Dent MT, Tebbs SE, Gonzalez AM, Ward JD, Wilson RM. Neutrophil aldose reductase activity and its association with established diabetic microvascular complications. Diabet Med. 1991;8:439–442. doi: 10.1111/j.1464-5491.1991.tb01628.x. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi Y, Tachikawa T, Ito T, Takayama S, Omori Y, Iwamoto Y. Erythrocyte aldose reductase protein: A clue to elucidate risk factors for diabetic neuropathies independent of glycemic control. Diabetes Res Clin Pract. 1998;42:101–107. doi: 10.1016/s0168-8227(98)00102-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.