Abstract
Background
Ethanolic fermentation of lignocellulosic biomass is a sustainable option for the production of bioethanol. This process would greatly benefit from recombinant Saccharomyces cerevisiae strains also able to ferment, besides the hexose sugar fraction, the pentose sugars, arabinose and xylose. Different pathways can be introduced in S. cerevisiae to provide arabinose and xylose utilisation. In this study, the bacterial arabinose isomerase pathway was combined with two different xylose utilisation pathways: the xylose reductase/xylitol dehydrogenase and xylose isomerase pathways, respectively, in genetically identical strains. The strains were compared with respect to aerobic growth in arabinose and xylose batch culture and in anaerobic batch fermentation of a mixture of glucose, arabinose and xylose.
Results
The specific aerobic arabinose growth rate was identical, 0.03 h-1, for the xylose reductase/xylitol dehydrogenase and xylose isomerase strain. The xylose reductase/xylitol dehydrogenase strain displayed higher aerobic growth rate on xylose, 0.14 h-1, and higher specific xylose consumption rate in anaerobic batch fermentation, 0.09 g (g cells)-1 h-1 than the xylose isomerase strain, which only reached 0.03 h-1 and 0.02 g (g cells)-1h-1, respectively. Whereas the xylose reductase/xylitol dehydrogenase strain produced higher ethanol yield on total sugars, 0.23 g g-1 compared with 0.18 g g-1 for the xylose isomerase strain, the xylose isomerase strain achieved higher ethanol yield on consumed sugars, 0.41 g g-1 compared with 0.32 g g-1 for the xylose reductase/xylitol dehydrogenase strain. Anaerobic fermentation of a mixture of glucose, arabinose and xylose resulted in higher final ethanol concentration, 14.7 g l-1 for the xylose reductase/xylitol dehydrogenase strain compared with 11.8 g l-1 for the xylose isomerase strain, and in higher specific ethanol productivity, 0.024 g (g cells)-1 h-1 compared with 0.01 g (g cells)-1 h-1 for the xylose reductase/xylitol dehydrogenase strain and the xylose isomerase strain, respectively.
Conclusion
The combination of the xylose reductase/xylitol dehydrogenase pathway and the bacterial arabinose isomerase pathway resulted in both higher pentose sugar uptake and higher overall ethanol production than the combination of the xylose isomerase pathway and the bacterial arabinose isomerase pathway. Moreover, the flux through the bacterial arabinose pathway did not increase when combined with the xylose isomerase pathway. This suggests that the low activity of the bacterial arabinose pathway cannot be ascribed to arabitol formation via the xylose reductase enzyme.
Background
Ethanol produced by fermentation of plant biomass is considered to be an environmentally friendly alternative to fossil fuels [1-3]. Cost-effective and sustainable production of ethanol as a transportation fuel entails the utilisation of microbial strains able to ferment completely all the sugars in lignocellulosic hydrolyzates [4-6]. Baker's yeast Saccharomyces cerevisiae, which has been used for ethanol production since the beginning of history [7], displays efficient ethanolic fermentation of sugar and starch-based raw materials. The selection process has also made S. cerevisiae a very robust organism, which tolerates high ethanol concentrations and is able to cope with harsh environments [8]. However, S. cerevisiae is unable to utilise arabinose and xylose, which in some raw materials such as agricultural residues and hardwoods, can account for more than 30% of total sugars [9], and which constitutes a significant barrier to the cost-effectiveness and sustainability of bioethanol production [6]. S. cerevisiae has been extensively engineered, developed and adapted to expand its substrate range to include the utilisation of the pentose sugars, arabinose and xylose, for growth and ethanol production [4].
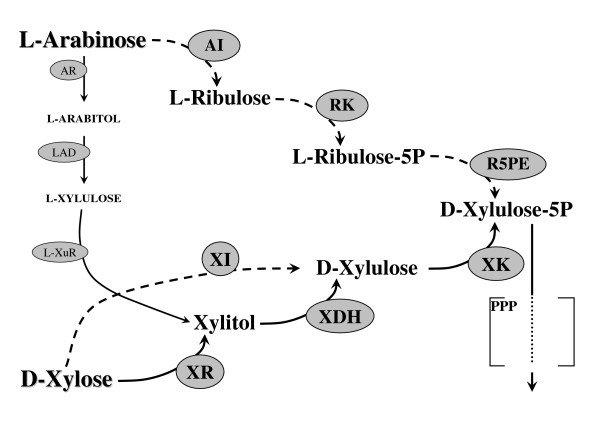
To enter the central carbon metabolism, arabinose and xylose must first be converted to xylulose 5-phosphate, an intermediate compound of the pentose phosphate pathway (PPP) (Figure 1). Essentially, two different pathways are available in nature for the conversion of pento-aldoses to xylulose: reduction/oxidation-based pathways and isomerisation-based pathways.
Figure 1.
Xylose and arabinose utilisation pathways. Solid lines: oxidation/reduction-based pathways; dashed lines: isomerisation-based pathways. PPP: pentose phosphate pathway. AI: arabinose isomerase; AR: arabinose reductase; LAD: arabitol dehydrogenase; L-XuR: L-xylulose reductase; R5PE: ribulose-phosphate-5-epimerase; RK: ribulokinase; XDH: xylitol dehydrogenase; XI: xylose isomerase; XK: xylulokinase; XR: xylose reductase.
In bacteria, L-arabinose is converted to L-ribulose, L-ribulose-5-P and finally D-xylulose-5-P via L-arabinose isomerase (AraA) [10-13], L-ribulokinase (AraB) [10,11,13,14] and L-ribulose-5-P 4-epimerase (AraD) [10,11,13,15], respectively (Figure 1). In fungi, L-arabinose is reduced to L-arabitol by arabinose reductase [16]. Arabitol is then re-oxidised by arabitol dehydrogenase to give L-xylulose [17], which is in turn converted to xylitol by L-xylulose reductase [17]. Xylitol is finally converted to xylulose by xylitol dehydrogenase (XDH) [18,19], whose activity is also part of xylose utilisation pathways (Figure 1).
In pentose-growing yeasts, xylose is first reduced by xylose reductase (XR) to xylitol [20], which in turn is oxidised to xylulose by XDH (Figure 1) [18,19]. In bacteria and some anaerobic fungi, xylose isomerase (XI) is responsible for direct conversion of xylose to xylulose [21-23] (Figure 1). Xylulose is finally phosphorylated to xylulose-5-phosphate by xylulokinase (XK) [24]. In S. cerevisiae, pentose sugar fermentation has been achieved by introducing several different alternative pathways, recently reviewed in Hahn-Hägerdal et al [25].
In the present investigation, we compared two xylose and arabinose co-consuming S. cerevisiae strains, which expressed two different xylose utilisation pathways and were otherwise genetically identical. A strain expressing XR and XDH and harbouring a bacterial arabinose utilisation pathway was compared with an isogenic strain instead expressing the XI xylose utilisation pathway. Pentose utilisation was characterised with respect to aerobic arabinose or xylose growth. Substrate consumption and product formation during anaerobic co-fermentation of glucose, arabinose and xylose was also investigated. The XR/XDH strain displayed faster aerobic growth on xylose and outperformed the XI strain with respect to pentose sugar consumption and ethanol production.
Results
Construction of arabinose and xylose fermenting strains TMB3075 (XR/XDH strain) and TMB3076 (XI strain)
Two strains, harbouring a chromosomally integrated, bacterial arabinose utilisation pathway [26] that consists of L-arabinose isomerase (Bacillus subtilis AraA), L-ribulokinase (Escherichia coli AraB) and L-ribulose-5-phosphate 4-epimerase (E. coli AraD) [26,27], in combination with two different plasmid-borne xylose pathways, were constructed. The two strains, which contained either the XR/XDH pathway or the XI pathway (Figure 1), will be referred to as the 'XR/XDH strain' and 'XI strain', respectively (Table 1). The XR/XDH strain harbours the arabinose pathway in combination with P. stipitis XYL1 and XYL2 genes encoding XR and XDH, respectively [28]. For the XI strain, the same arabinose pathway is combined with the Piromyces XI gene (xylA) [29,30].
Table 1.
Plasmids and strains used in this study
| Plasmid | Features | Reference |
| prDNAAraA | pBluescript, NTS2::pHXT7tr-AraA(B. subtilis)-tCYC1 | [27] |
| prDNAAraD | pBluescript, NTS2::pHXT7tr-AraD (E. coli)-tCYC1 | [27] |
| pY7 | XYL1 (P. stipitis), XYL2 (P. stipitis), URA3 | [28] |
| YEplacHXT-XIp | pHXT7tr-XI (Pyromyces sp.)-tCYC1, URA3 | [29] |
| YIpAraB | KanMX, pHXT7tr-AraB (E. coli)-tCYC1, TRP1 | [27] |
| YIplac128 | LEU2 | [53] |
| Strain | Genotype | Reference |
| TMB3042 | CEN.PK 2-1C, Δgre3, his3:: p PGK1-XKS1- t PGK1, TAL1:: p PGK1-TAL1- t PGK1, TKL1:: p PGK1-TKL1- t PGK1, RKI1:: p PGK1-RKI1- t PGK1, RPE1:: p PGK1-RPE1- t PGK1, leu2, trp1, ura3 | [31] |
| TMB3070 | TMB3042, YIpAraB | This work |
| TMB3073 | TMB3070, pHXT7tr-AraA(B. subtilis)-tCYC1, NTS2::pHXT7tr-AraD (E. coli)-tCYC1 | This work |
| TMB3074 | TMB3073, YIplac128 | This work |
| TMB3075 (xylose reductase/xylitol dehydrogenase strain) | TMB3074, pY7 | This work |
| TMB3076 (xylose isomerase strain) | TMB3074, YEplacHXT-XIp | This work |
The starting strain TMB3042 (Table 1) harbours genetic modifications previously reported to be favourable for pentose fermentation, such as overexpression of PPP [31] and XK [32] (Figure 1) and deletion of GRE3, encoding an endogenous unspecific aldose reductase (AR) [33]. Gre3p, like XR, is able to reduce xylose to xylitol, but with the exclusive use of nicotinamide adenine dinucleotide phosphate (NADPH) as a cofactor [34]. Removal of the NADPH-dependent activity of Gre3p is therefore, beneficial for the cofactor balance of a strain expressing P. stipitis XR (XIL1), which can perform the reduction also using nicotinamide adenine dinucleotide (NADH) [31,35]. In addition, xylitol is a known inhibitor of XI [36] and deletion of GRE3 in an XI strain prevents the formation of this compound and has therefore, a beneficial effect on xylose fermentation [33,37].
First, the bacterial arabinose utilisation pathway (Figure 1) was integrated in TMB3042. The gene AraB from E. coli was integrated in a single copy [26,27], while B. subtilis AraA and E. coli AraD where targeted to the rDNA region of S. cerevisiae to allow multiple copy integration [27].
Transformants were selected for their ability to grow on arabinose on yeast extract peptone arabinose (YPA) (see Methods section) plates. The rich medium provided conditions for the cells to recover from transformation. After 3 days, visibly larger colonies emerged over a confluent background of minute colonies growing on the rich medium. No larger colonies were detected on negative control transformation plates, even after several days of incubation. Strain TMB3073 was purified from 16 independently picked clones, which were re-streaked on yeast nitrogen base/arabinose (YNBA) plates. The presence of the integrated constructs was verified by polymerase chain reaction (PCR) with specific primers for AraA, AraB and AraD genes. This is to the best of our knowledge the first plasmid-free, arabinose growing S. cerevisiae laboratory strain.
Subsequently, two different xylose utilisation pathways were independently introduced in TMB3074. The XR and XDH pathway was introduced by transformation with plasmid pY7 [28], harbouring XYL1 and XYL2 encoding genes form P. stipitis, while the XI pathway was introduced by transformation with plasmid YEplacHXT-XIp [29], carrying a synthetic Piromyces sp. XI gene [29,38]. Persistence of the integrated arabinose pathway was re-confirmed with PCR. The two resulting strains, named XR/XDH and XI strain, respectively, are prototrophic and genetically identical, except for the plasmid-borne xylose utilisation pathways.
Aerobic growth on defined pentose media
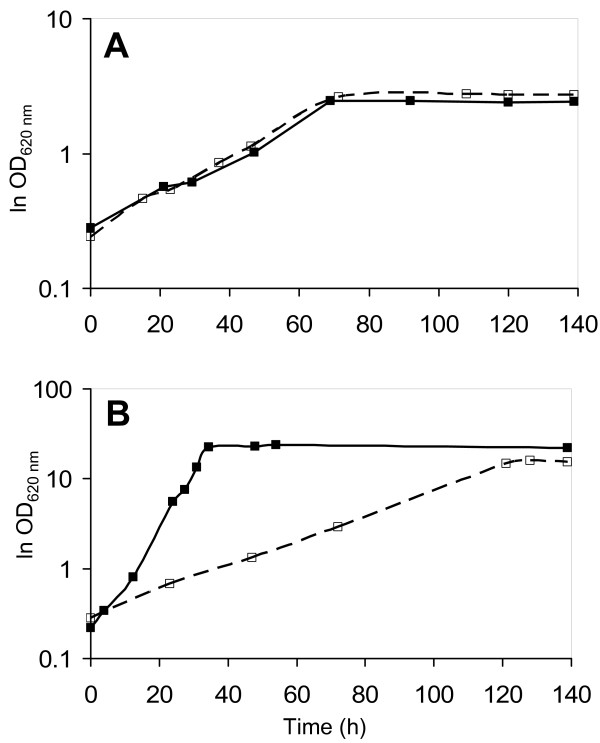
The reciprocal effect of the two combinations of pentose utilisation pathways was first assessed under aerobic conditions. The XR/XDH and XI strains were grown in YNBA and YNB/xylose (YNBX) media (see Methods section) (Figure 2). The two strains exhibited similar growth patterns in the arabinose medium (Figure 2A), with calculated maximum specific growth rates of 0.03 ± 0.004 h-1 for the XR/XDH strain and 0.03 ± 0.001 h-1 for the XI strain. When xylose was provided as the sole carbon source, the XR/XDH strain grew at higher growth rate than the XI strain, with calculated growth rates of 0.14 ± 0.06 h-1 for the XR/XDH strain and 0.03 ± 0.003 h-1 for the XI strain (Figure 2B), which is in agreement with previously reported results [29].
Figure 2.
Aerobic growth of Saccharomyces cerevisiae xylose reductase/xylitol dehydrogenase and xylose isomerase strains. (A) Yeast nitrogen base/arabinose medium and (B) yeast nitrogen base/xylose medium. Solid line, filled symbols: xylose reductase/xylitol dehydrogenase strain; dashed line, open symbols: xylose isomerase strain.
Anaerobic mixed sugar fermentation
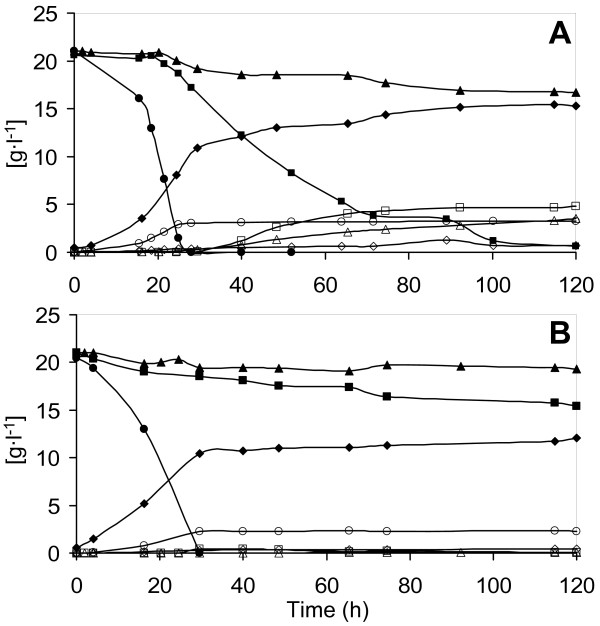
Next, two strains were compared in anaerobic batch fermentation with a mixture of glucose, arabinose and xylose as a carbon source (Figure 3) (Table 2). Glucose was consumed first and completely exhausted at around 28–30 hours (Figure 3). Arabinose and xylose were co-consumed by both strains in the pentose phase, that is, the fermentation phase subsequent to glucose depletion, with xylose being consumed at a higher rate than arabinose by both strains. Biomass formation ceased after glucose depletion. Specific substrate consumption and product formation rates (Table 2) were calculated only for the pentose phase, when biomass was constant.
Figure 3.
Substrate and product concentration during anaerobic batch fermentation of defined mineral medium containing glucose, arabinose and xylose. (A) Xylose reductase/xylitol dehydrogenase strain; (B) xylose isomerase strain. ●: glucose; ■: xylose; ▲: arabinose; ◆: ethanol; ◇: acetate; ○: glycerol; □: xylitol; △: arabitol.
Table 2.
Substrate consumption and product formation parameters during anaerobic batch fermentation of a mixture of glucose, arabinose and xylose in defined mineral medium
| TMB3075 (xylose reductase/xylitol dehydrogenase) | TMB3076 (xylose isomerase) | |
| Consumed arabinose g l-1 | 4.04 ± 0.33 | 1.65 ± 0.01 |
| Consumed xylose g l-1 | 20.1 ± 0.2 | 4.9 ± 0.7 |
| q arabinose* (g arabinose) (g cells)-1 h-1 | 0.014 ± 0.001 | 0.005 ± 0.001 |
| q xylose* (g xylose) (g cells)-1 h-1 | 0.09 ± 0.001 | 0.02 ± 0.001 |
| Final ethanol titre g l-1 | 14.7 ± 0.5 | 11.8 ± 0.3 |
| q ethanol* (g ethanol) (g cells)-1 h-1 | 0.024 ± 0.001 | 0.010 ± 0.001 |
| Y ethanol, on total added sugars (g ethanol) (g sugar)-1 | 0.23 ± 0.01 | 0.18 ± 0.01 |
| Y ethanol, on consumed sugars (g ethanol) (g sugar)-1 | 0.32 ± 0.01 | 0.41 ± 0.01 |
| Y xylitol (g xylitol) (g consumed xylose)-1 | 0.27 ± 0.04 | 0.04 ± 0.02 |
| Y arabitol (g arabitol) (g consumed arabinose)-1 | 0.87 ± 0.03 | ND |
|
Y biomass, on total added sugars (g dry cell weight) (g sugar) -1 |
0.03 ± 0.01 |
0.03 ± 0.01 |
|
Y biomass, on consumed sugars (g dry cell weight) (g sugar) -1 |
0.04 ± 0.01 |
0.07 ± 0.01 |
| Y glycerol (g acetate) (g consumed arabinose)-1 | 0.08 ± 0.001 | 0.09 ± 0.01 |
| Y acetate (g acetate) (g consumed arabinose)-1 | 0.020 ± 0.002 | 0.016 ± 0.001 |
Values are the calculated average of two biological replicates. *Calculated after glucose depletion.
ND.: not determined. q: specific productivity, Y: yield.
The XR/XDH strain consumed 4.04 g l-1 arabinose compared with 1.65 g l-1 for the XI strain. The XR/XDH strain consumed nearly all xylose with an overall rate of 0.09 g (g cells)-1 h-1. In contrast, only 8% of the xylose was consumed by the XI strain, at an overall rate of 0.02 g (g cells)-1 h-1.
The XR/XDH strain produced a final ethanol concentration of 14.7 g l-1, with a yield based on total sugars of 0.23 g (g sugar)-1, while the XI strain reached an ethanol concentration of 11.8 g l-1, with a yield on total sugars equal to 0.18 g (g sugar)-1 (Table 2). The specific ethanol productivity from pentose sugars for the XR/XDH strain was 0.024 g (g cells)-1 h-1, versus 0.010 g (g cells)-1h-1 for the XI strain (Table 2).
Almost 90% of the arabinose consumed by the XR/XDH-strain was converted to arabitol, whereas arabitol formation in the XI strain was essentially negligible. As a result, the final arabitol concentration was 3.51 g l-1 for the XR/XDH strain, whereas it was below detection for the XI strain. At the same time, the XR/XDH strain converted part of the consumed xylose to xylitol, with a yield of 0.27 g xylitol per gramme of consumed xylose. The lower by-product formation gave the XI strain a higher ethanol yield on consumed sugars, 0.41 (g ethanol) (g consumed sugar)-1, compared with 0.32 (g ethanol) (g consumed sugar)-1 for the XR/XDH strain.
Biomass was formed with a yield of 0.032 g g-1 calculated on total sugars, with a final dry cell weight of 1.95 g l-1 for both strains. Acetate is usually generated by S. cerevisiae in response to a demand for NADPH. This can be associated with the need of this cofactor for biosynthetic reactions. In the XR/XDH strain, NADPH demand might be higher due to the utilisation of this cofactor by the XR NADPH requirement. Consistently, acetate yield was slightly but significantly higher in the XR/XDH strain, while the opposite was observed for glycerol yield.
Discussion
To the best of our knowledge, arabinose and xylose co-metabolism in recombinant S. cerevisiae strains has previously only been reported for one recombinant industrial strain of S. cerevisiae [27]. In this strain, the XR/XDH pathway was combined with a functional bacterial arabinose utilising pathway [26]. While arabinose and xylose were co-consumed, only xylose was further metabolised to ethanol. Arabinose was instead converted to arabitol, which was believed to be catalysed by the overexpressed heterologous P. stipitis XR enzyme. In fact, this enzyme has a lower Km for arabinose than for xylose [39]. Arabitol is a known inhibitor of arabinose isomerase [40] and the results suggested that arabinose metabolism in this recombinant S. cerevisiae strain was limited by arabitol inhibition of the first enzyme in the pathway.
The current investigation aimed to explore whether arabitol formation during co-utilisation of arabinose and xylose could be avoided or minimised when xylose metabolism was instead governed by an isomerase pathway (Figure 1). This would allow co-utilisation of arabinose and xylose under conditions where arabitol formation and inhibition of arabinose isomerase was reduced or absent. In fact, arabitol formation was completely abolished when the two isomerase pathways were combined in one strain, TMB3076 (Table 2), confirming that arabitol formation was caused by overexpression of the heterologous XR. However, the absence of arabitol formation was the only benefit observed during arabinose and xylose co-utilisation by the isomerase strain. The XR/XDH strain was superior in all other aspects, that is, sugar uptake rate, ethanol concentration, ethanol yield on total sugars and ethanol productivity.
Heterologous expression of xylose and later, arabinose isomerase in S. cerevisiae have been very tedious undertakings. Already suggested in the late 1970s, it required another 16 years before the first XI was functionally expressed in S. cerevisiae [28]. Furthermore, it was only when XI expression from multi-copy plasmids was combined with extensive adaptation protocols that a functional xylose fermenting recombinant S. cerevisiae strain was obtained [41]. Similarly, the first arabinose fermenting recombinant S. cerevisiae strain had also undergone extensive adaptation in addition to being transformed with genes for the bacterial arabinose pathway [26]. The same results were later obtained with an arabinose pathway based on other bacterial genes [42]. While the beneficial effects of these extensive adaptation protocols remain to be clarified, it is tempting to speculate that heterologous isomerases may not be able to express their full catalytic potential, in terms of substrate conversion rate, in the S. cerevisiae intracellular environment.
In addition to adaptation, codon optimisation has been shown to improve the performance of heterologous bacterial isomerase pathways in S. cerevisiae [43]. Both ethanol yield and specific ethanol productivity were significantly increased when the codon usage of the bacterial genes was adapted to the usage of glycolytic yeast genes.
In S. cerevisiae, pentose sugars are reduced to the corresponding pentitols by the unspecific AR encoded by GRE3 [33,34] as well as by a number of uncharacterised open reading frames [44]. This was well illustrated by an arabinose-adapted recombinant strain of S. cerevisiae carrying a GRE3 deletion [42]. This strain reduced xylose to xylitol most likely through one of its unspecific reductases, while arabinose reduction to arabitol formation was negligible. However, despite the presence of a highly expressed xylA gene encoding XI, the strain was incapable of xylose growth.
In the XR/XDH strain, arabitol formation represents a dead end in the metabolism, since yeast lacks the enzymatic activity to further convert arabitol. In addition, arabitol has been extensively reported to be a potent inhibitor of arabinose isomerase [36,40,45-48], which was illustrated by the observation that the highest arabinose consumption rate occurred between 25 and 40 hours (Figure 3A), when glucose was depleted and prior to arabitol build-up. Thus, the XR/XDH strain displays a lower yield on consumed sugars, but the higher consumption rates of pentose sugars compensate for this, resulting eventually in higher specific ethanol productivity.
Conclusion
The combination of the XR/XDH pathway and the bacterial arabinose isomerase pathway resulted in both higher pentose sugar uptake and higher overall ethanol production than the combination of the XI pathway and bacterial arabinose isomerase pathway. Moreover, the flux through the bacterial arabinose pathway did not increase when combined with the XI pathway. This suggests that the low activity of the bacterial arabinose pathway cannot be ascribed to arabitol formation via the XR enzyme.
Methods
Strains and media
Yeast strains and plasmid utilised for this work are summarised in Table 1. E. coli DH5α (Life Technologies, Rockville, MD, US) was used as an intermediate host for cloning steps and plasmid amplification and was routinely grown in lysogeny broth medium [49] containing 100 mg l-1 ampicillin (Shelton Scientific, Shelton, CT, US). S. cerevisiae strains were grown aerobically in YNB (Difco Laboratories-Becton, Dickinson and Co., Sparks, NV, US), buffered at pH 5.5 with 50 mM potassium hydrogen phthalate (Merck, Darmstadt, Germany) [50] and formulated as it follows: YNB/glucose 20 g l-1 glucose, 6.7 g l-1 YNB; YNBA 50 g l-1 arabinose, 13.4 g l-1 YNB; YNBX 50 g l-1 xylose, 13.4 g l-1 YNB. According to strain requirements, the medium was supplemented with uracil and/or leucine at concentrations of 40 mg l-1 and 240 mg l-1, respectively. YPA medium for initial selection of arabinose-growing strains was composed of 10 g l-1 yeast extract (Merck, Darmstadt, Germany), 20 g l-1 peptone (Merck, Darmstadt, Germany), 50 g l-1 arabinose (Sigma-Aldrich, St Louis, MO, US). Solid media were obtained by addition of 20 g l-1 agar (Merck, Darmstadt, Germany).
Anaerobic mixed sugar batch fermentation was performed in defined mineral medium [51] supplemented with 0.4 g l-1 Tween 80 (Sigma-Aldrich, St Louis, MO, US), 0.01 g l-1 ergosterol (Alfa Aesar, Karlsruhe, Germany), 20 g l-1 glucose (VWR International, Poole, UK), 20 g l-1 xylose (Acros Organics, Geel, Belgium) and 20 g l-1 arabinose (Sigma-Aldrich, St Louis, MO, US).
Nucleic acid manipulation
Standard molecular biology techniques were used [49]. The lithium acetate/dimethyl sulphoxide protocol was used for yeast transformation [52]. Yeast chromosomal DNA was extracted with a bead-beater (Biospecs Products, Bartlesville, OK, US) and phenol/chloroform [49]. Plasmid DNA was purified from E. coli with Gene JET plasmid miniprep kit (Fermentas, St Leon-Rot, Germany). Linear integration cassettes were PCR-amplified from plasmid prDNAAraA and prDNAAraD [27] with PWO DNA polymerase (Fermentas, St Leon-Rot, Germany) using the following thermal cycler programme: 94°C 5 min; 25 cycles of 94°C 30 s, 46.5°C 30 s, 72°C 3 min; final extension 72°C 7 min. The two PCR reactions were treated with DpnI endonuclease (Fermentas, St Leon-Rot, Germany) for the removal of template plasmid DNA, pooled, precipitated with ethanol and resuspended in 15 μl of TE buffer (10 mM Tris, 1 mM ethylenediaminetetraacetic acid, pH 8.0), which was used for yeast transformation. Analytical PCR was performed with Taq DNA polymerase (Fermentas, St Leon-Rot, Germany).
Cultivation conditions
S. cerevisiae was grown aerobically in 1 litre baffled flasks containing 0.1 litre medium, incubated at 30°C in a rotary shake-incubator (INR-200 Shake Incubator, Gallenkamp, Leicester, UK) at 200 rpm. Cultures were inoculated at an initial optical density (OD)620 nmof 0.2 ± 0.02 with sterile H2O-washed cells from a late-exponential YNBG pre-culture. The maximum specific growth rate, μ, was calculated from exponential fitting of growth curves from at least two biological duplicates.
Anaerobic mixed-sugar batch fermentation was performed in 1.5 litre working volume bioreactors (Applikon Biotechnology, Schiedam, The Netherlands), for 120 hours, at 30°C, 200 rpm at pH 5.5 automatically controlled by the addition of 3 M potassium hydroxide. Anaerobic conditions were established prior to inoculation by sparging the medium for at least 3 hours with nitrogen (< 5 ppm oxygen) (AGA, Malmö, Sweden) at 0.2 litre/min flow rate. Nitrogen was sparged throughout the fermentation at the same flow rate. Cells were pre-grown aerobically in shake flasks in YNBG medium, harvested by centrifugation, resuspended in about 10 ml sterile medium and inoculated in the fermentor at an initial OD620 nm of 0.2. Fermentation experiments were performed in duplicate.
Analyses
Samples were drawn from the fermentors after discharging the sample tubing dead-volume, cells were quickly separated by centrifugation and the supernatant was filtered through 0.20 μm membrane filters (Toyo Roshi Kaish, Tokyo, Japan) and stored at 4°C until further analysis.
Concentrations of glucose, xylose, acetate, glycerol and ethanol were determined by high performance liquid chromatography (HPLC) (Beckman Instruments, Fullerton, CA, US). The compounds were separated with three Aminex HPX-87H resin-based columns (Bio-Rad, Hercules, CA, US) connected in series and preceded by a Micro-Guard Cation-H guard column (Bio-Rad, Hercules, CA, US). Separation was performed at 45°C, with 5 mM sulphuric acid at a flow rate of 0.6 ml/min as mobile phase. Owing to evaporation, ethanol concentrations were calculated from the degree of reduction balance of the overall carbon stoichiometry of the fermentation.
Concentrations of arabinose, arabitol and xylitol were determined by HPLC (Waters, Milford, MA, US) using an HPX-87P resin-based column (Bio-Rad, Hercules, CA, US) preceded by a Micro-Guard Carbo-P guard column (Bio-Rad, Hercules, CA, US). Separation was performed at 85°C, with water at a flow rate of 0.5 ml/min as mobile phase. All compounds were quantified by refractive index detection (Shimadzu, Kyoto, Japan). For each HPLC run, a seven-point calibration curve was made for each compound to calculate concentrations. Each sample was analysed at least in duplicate and a maximum of 10% difference between replicate analyses was accepted.
For each fermentation experiment, dry weight measurements were made in three points at least, in triplicate for each point. The end point of the fermentation (t = 120 h) was always included. For dry weight determination, a known volume of cell culture was filtered through dry pre-weighed 0.45 μm nitrocellulose filters, which were subsequently dried in a microwave oven and weighed.
Abbreviations
AR: aldose reductase; HPLC: high performance liquid chromatography; NADH: nicotinamide adenine dinucleotide; NADPH: nicotinamide adenine dinucleotide phosphate; OD: optical density; PCR: polymerase chain reaction; PPP: pentose phosphate pathway; XDH: xylitol dehydrogenase; XI: xylose isomerase; XK: xylulokinase; XR: xylose reductase; YNB: yeast nitrogen base; YNBA: yeast nitrogen base/arabinose; YNBG: yeast nitrogen base/glucose; YNBX: yeast nitrogen base/xylose; YPA: yeast extract peptone arabinose
Authors' contributions
MB participated in the design of the study, performed the experimental work and wrote the manuscript. BHH participated in the design of the study and commented on the manuscript. MFGG participated in the design of the study and commented on the manuscript. All the authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
MB is recipient of a Marie Curie Intra European Post Doctoral fellowship (EIF), project No. 039998-DESYRE 'Designed yeast for renewable bioethanol production'; project coordinator: BHH.
Contributor Information
Maurizio Bettiga, Email: maurizio.bettiga@tmb.lth.se.
Bärbel Hahn-Hägerdal, Email: barbel.hahn-hagerdal@tmb.lth.se.
Marie F Gorwa-Grauslund, Email: marie-francoise.gorwa@tmb.lth.se.
References
- Mabee WE. Policy options to support biofuel production. Adv Biochem Eng Biotechnol. 2007;108:329–357. doi: 10.1007/10_2007_059. [DOI] [PubMed] [Google Scholar]
- Hahn-Hägerdal B, Galbe M, Gorwa-Grauslund MF, Lidén G, Zacchi G. Bio-ethanol – the fuel of tomorrow from the residues of today. Trends Biotechnol. 2006;24:549–556. doi: 10.1016/j.tibtech.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Farrell AE, Plevin RJ, Turner BT, Jones AD, O'Hare M, Kammen DM. Ethanol can contribute to energy and environmental goals. Science. 2006;311:506–508. doi: 10.1126/science.1121416. [DOI] [PubMed] [Google Scholar]
- Hahn-Hägerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF. Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol. 2007;74:937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- Galbe M, Sassner P, Wingren A, Zacchi G. Process engineering economics of bioethanol production. Adv Biochem Eng Biotechnol. 2007;108:303–327. doi: 10.1007/10_2007_063. [DOI] [PubMed] [Google Scholar]
- Sassner P, Mårtensson C-G, Galbe M, Zacchi G. Steam pretreatment of H2SO4-impregnated Salix for the production of bioethanol. Bioresour Technol. 2008;99:137–145. doi: 10.1016/j.biortech.2006.11.039. [DOI] [PubMed] [Google Scholar]
- Piskur J, Rozpedowska E, Polakova S, Merico A, Compagno C. How did Saccharomyces evolve to become a good brewer? Trends Genet. 2006;22:183–186. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Almeida JR, Modig T, Petersson A, Hahn-Hägerdal B, Lidén G, Gorwa-Grauslund MF. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biotechnol. 2007;82:340–349. http://www3.interscience.wiley.com/cgi-bin/fulltext/114206806/PDFSTART [Google Scholar]
- Olofsson K, Bertilsson M, Lidén G. A short review on SSF – an interesting process option for ethanol production from lignocellulosic feedstocks. Biotechnol Biofuels. 2008;1:7. doi: 10.1186/1754-6834-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englesberg E. Enzymatic characterization of 17 l-arabinose negative mutants of Escherichia coli. J Bacteriol. 1961;81:996–1006. doi: 10.1128/jb.81.6.996-1006.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englesberg E, Squires C, Meronk F. The L-arabinose operon in Escherichia coli B/r: a genetic demonstration of two functional states of the product of a regulator gene. Proc Natl Acad Sci USA. 1969;62:1100–1107. doi: 10.1073/pnas.62.4.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath EC, Horecker BL, Smyrniotis PZ, Takagi Y. Pentose fermentation by Lactobacillus plantarum. II. L-arabinose isomerase. J Biol Chem. 1958;231:1031–1037. [PubMed] [Google Scholar]
- Sa-Nogueira I, de Lencastre H. Cloning and characterization of araA, araB, and araD, the structural genes for L-arabinose utilization in Bacillus subtilis. J Bacteriol. 1989;171:4088–4091. doi: 10.1128/jb.171.7.4088-4091.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma DP, Horecker BL. Pentose fermentation by Lactobacillus plantarum. III. Ribulokinase. J Biol Chem. 1958;231:1039–1051. [PubMed] [Google Scholar]
- Burma DP, Horecker BL. Pentose fermentation by Lactobacillus plantarum. IV. L-Ribulose-5-phosphate 4-epimerase. J Biol Chem. 1958;231:1053–1064. [PubMed] [Google Scholar]
- Chiang C, Knight SG. A new pathway of pentose metabolism. Biochem Biophys Res Commun. 1960;3:554–559. doi: 10.1016/0006-291x(60)90174-1. [DOI] [PubMed] [Google Scholar]
- Chiang C, Knight SG. L-arabinose metabolism by cell-free extracts of Penicillium chrysogenum. Biochim Biophys Acta. 1961;46:271–278. doi: 10.1016/0006-3002(61)90750-8. [DOI] [PubMed] [Google Scholar]
- Chiang C, Knight SG. D-xylose metabolism by cell-free extracts of Penicillum chrisogenum. Biochim Biophys Acta. 1959;35:454–463. doi: 10.1016/0006-3002(59)90395-6. [DOI] [PubMed] [Google Scholar]
- Veiga L. Polyol dehydrogenases in Candida albicans 2. Xylitol oxydation to D-xylulose. J Gen Appl Microbiol. 1968;14:79–87. [Google Scholar]
- Veiga L. Polyol dehydrogenases in Candida albicans 1.Reduction of D-xylose to xylitol. J Gen Appl Microbiol. 1968;14:65–78. [Google Scholar]
- Mitsuhashi S, Lampen JO. Conversion of D-xylose to D-xylulose in extracts of Lactobacillus pentosus. J Biol Chem. 1953;204:1011–1018. [PubMed] [Google Scholar]
- Harhangi HR, Akhmanova AS, Emmens R, Drift C van der, de Laat WTAM, van Dijken JP, Jetten MSM, Pronk JT, Op den Camp HJM. Xylose metabolism in the anaerobic fungus Piromyces sp. strain E2 follows the bacterial pathway. Arch Microbiol. 2003;180:134–141. doi: 10.1007/s00203-003-0565-0. [DOI] [PubMed] [Google Scholar]
- Chen W. Glucose isomerase (a review) Process Biochem. 1980;15:36–41. [Google Scholar]
- Chang SF, Ho NW. Cloning the yeast xylulokinase gene for the improvement of xylose fermentation. Appl Biochem Biotechnol. 1988;17:313–318. doi: 10.1007/BF02779165. [DOI] [PubMed] [Google Scholar]
- Hahn-Hägerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund MF. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. In: Olsson L, editor. Adv Biochem Eng Biotechnol. Vol. 108. Berlin/Heidelberg: Springer; 2007. pp. 147–177. Advances in Biochemical Engineering/Biotechnology. [DOI] [PubMed] [Google Scholar]
- Becker J, Boles E. A modified Saccharomyces cerevisiae strain that consumes L-Arabinose and produces ethanol. Appl Environ Microbiol. 2003;69:4144–4150. doi: 10.1128/AEM.69.7.4144-4150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhumaa K, Wiedemann B, Hahn-Hägerdal B, Boles E, Gorwa-Grauslund MF. Co-utilization of L-arabinose and D-xylose by laboratory and industrial Saccharomyces cerevisiae strains. Microb Cell Fact. 2006;5:18. doi: 10.1186/1475-2859-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walfridsson M, Anderlund M, Bao X, Hahn-Hägerdal B. Expression of different levels of enzymes from the Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effects on product formation during xylose utilisation. Appl Microbiol Biotechnol. 1997;48:218–224. doi: 10.1007/s002530051041. [DOI] [PubMed] [Google Scholar]
- Karhumaa K, Sanchez R, Hahn-Hägerdal B, F G-GM. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb Cell Fact. 2007;6:5. doi: 10.1186/1475-2859-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyper M, Hartog MMP, Toirkens MJ, Almering MJH, Winkler AA, van Dijken JP, Pronk JT. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 2005;5:399–409. doi: 10.1016/j.femsyr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Karhumaa K, Hahn-Hägerdal B, Gorwa-Grauslund MF. Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast. 2005;22:359–368. doi: 10.1002/yea.1216. [DOI] [PubMed] [Google Scholar]
- Ho NW, Chen Z, Brainard AP. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Environ Microbiol. 1998;64:1852–1859. doi: 10.1128/aem.64.5.1852-1859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Träff KL, Otero Cordero RR, van Zyl WH, Hahn-Hagerdal B. Deletion of the GRE3 aldose reductase gene and its influence on xylose metabolism in recombinant strains of Saccharomyces cerevisiae expressing the xylA and XKS1 genes. Appl Environ Microbiol. 2001;67:5668–5674. doi: 10.1128/AEM.67.12.5668-5674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, van Zyl C, van Tonder A, Prior BA. Purification and partial characterization of an aldo-keto reductase from Saccharomyces cerevisiae. Appl Environ Microbiol. 1995;61:1580–1585. doi: 10.1128/aem.61.4.1580-1585.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzi M, Harwart K, Erlemann P, Bui-Thahn NA, Dellweg H. Purification and properties of the NAD+-xylitol-dehydrogenase from the yeast Pichia stipitis. J Ferment Bioeng. 1989;67:20–24. http://www.sciencedirect.com/science?_ob=MImg&_imagekey=B6T8G-47FX6V8-4D-1&_cdi=5086&_user=745831&_orig=search&_coverDate=12%2F31%2F1989&_sk=999329998&view=c&wchp=dGLbVtz-zSkWA&md5=22919af8be718161eceb3c796ca7264d&ie=/sdarticle.pdf [Google Scholar]
- Yamanaka K. Inhibition of D-xylose isomerase by pentitols and D-lyxose. Arch Biochem Biophys. 1969;131:502–506. doi: 10.1016/0003-9861(69)90422-6. [DOI] [PubMed] [Google Scholar]
- Lönn A, Träff-Bjerre KL, Cordero Otero RR, van Zyl WH, Hahn-Hägerdal B. Xylose isomerase activity influences xylose fermentation with recombinant Saccharomyces cerevisiae strains expressing mutated xylA from Thermus thermophilus. Enzyme Microb Technol. 2003;32:567–573. http://www.sciencedirect.com/science?_ob=MImg&_imagekey=B6TG1-480CSJC-1-2&_cdi=5241&_user=745831&_orig=search&_coverDate=04%2F08%2F2003&_sk=999679994&view=c&wchp=dGLbVzb-zSkzS&md5=19c05841d5f1112f8003729301324655&ie=/sdarticle.pdf [Google Scholar]
- Kuyper M, Harhangi HR, Stave AK, Winkler AA, Jetten MSM, Laat WTAM, Ridder JJJ, Op den Camp HJM, Dijken JP, Pronk JT. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res. 2003;4:69–78. doi: 10.1016/S1567-1356(03)00141-7. [DOI] [PubMed] [Google Scholar]
- Rizzi M, Elrlemann P, Bui-Thahn NA, Dellweg H. Xylose fermentation by yeasts. 4. Purification and kinetic studies of xylose reductase from Pichia stipitis. Appl Microbiol Biotechnol. 1988;29:148–154. http://www.springerlink.com/content/wlg43687wu643532/fulltext.pdf [Google Scholar]
- Patrick JW, Lee N. Purification and properties of an L-arabinose isomerase from Escherichia coli. J Biol Chem. 1968;243:4312–4318. [PubMed] [Google Scholar]
- Kuyper M, Winkler AA, Van Dijken JP, Pronk JT. Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Res. 2004;4:655–664. doi: 10.1016/j.femsyr.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Wisselink HW, Toirkens MJ, del Rosario Franco Berriel M, Winkler AA, van Dijken JP, Pronk JT, van Maris AJA. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of L-arabinose. Appl Environ Microbiol. 2007;73:4881–4891. doi: 10.1128/AEM.00177-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann B, Boles E. Codon-optimized bacterial genes improve L-Arabinose fermentation in recombinant Saccharomyces cerevisiae. Appl Environ Microbiol. 2008;74:2043–2050. doi: 10.1128/AEM.02395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Träff KL, Jönsson LJ, Hahn-Hägerdal B. Putative xylose and arabinose reductases in Saccharomyces cerevisiae. Yeast. 2002;19:1233–1241. doi: 10.1002/yea.913. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Wood WA. Methods in Enzymology. Vol. 41. London: Academic Press; 1975. L-arabinose isomerase from Lactobacillus gayonii; pp. 458–461. [DOI] [PubMed] [Google Scholar]
- Patrick J, Lee N, Wood WA. Methods in Enzymology. Vol. 41. London: Academic Press; 1975. L-arabinose isomerase; pp. 453–458. [DOI] [PubMed] [Google Scholar]
- Nakamatu T, Yamanaka K. Crystallization and properties of -arabinose isomerase from Lactobacillus gayonii. Biochim Biophys Acta. 1969;178:156–165. doi: 10.1016/0005-2744(69)90142-9. [DOI] [PubMed] [Google Scholar]
- Izumori K, Yamanaka K. Selective inhibition of Klebsiella aerogenes growth on pentoses by pentitols. J Bacteriol. 1978;134:713–717. doi: 10.1128/jb.134.3.713-717.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Hahn-Hägerdal B, Karhumaa K, Larsson CU, Gorwa-Grauslund M, Gorgens J, van Zyl WH. Role of cultivation media in the development of yeast strains for large scale industrial use. Microb Cell Fact. 2005;4:31. doi: 10.1186/1475-2859-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppsson M, Bengtsson O, Franke K, Lee H, Hahn-Hägerdal B, Gorwa-Grauslund MF. The expression of a Pichia stipitis xylose reductase mutant with higher K(M) for NADPH increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Biotechnol Bioeng. 2006;93:665–673. doi: 10.1002/bit.20737. [DOI] [PubMed] [Google Scholar]
- Hill J, Donald KAIG, Griffiths DE. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791–5796. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]