Abstract
Background
The growth hormone secretagogue receptor type 1a gene (GHSR) encodes the cognate receptor of ghrelin, a gut hormone that regulates food intake and pituitary growth hormone secretion. Previous studies in US families and in a German population suggested GHSR to be a candidate quantitative locus for association with human obesity and growth.
Aim
To test common genetic variation in GHSR for association with body size in children and adults.
Methods
Sequencing was performed to systematically identify novel single nucleotide polymorphisms (SNPs) in GHSR. A set of three haplotype-tagging (ht)SNPs was identified which captured all the genetic variation in GHSR. These three htSNPs were then genotyped in three large population-based UK cohort studies (two adult and one childhood cohort) comprising 5,807 adults and 843 children.
Results
No significant genotype or haplotype associations were found with adult or childhood height, weight or BMI.
Conclusion
Common variation in GHSR is not associated with body size in UK adults or children.
Keywords: ghrelin, ghrelin receptor, body mass index, gene, growth, ALSPAC, Ely Study
Introduction
The growth hormone secretagogue receptor type 1a gene (GHSR) encodes the cognate receptor of ghrelin, a gut hormone that regulates energy homeostasis, food intake and the release of growth hormone by the anterior pituitary (1). The full-length receptor (type 1a) contains 366 amino-acids encoded by two exons on chromosome 3q25. A splice variant (type 1b) comprises only exon 1 and a short region of the intron. It is not clear if this variant is transcribed to protein in vivo but theoretically it would code for a 289 amino-acid protein, representing 5 transmembrane domains. The type 1b receptor is widely distributed (as opposed to the type 1a receptor), but has no biological activity in GH-releasing or calcium-related functional assays (2). In animal models genomic regions syntenic with the human GHSR gene are candidate quantitative trait loci for energy expenditure and body temperature regulation (3). In addition, Ghsr-null mice have a lean phenotype (4) and improvement of glycemic profiles in leptin deficient mice with concomitant suppression of Ghsr suggest a possible role of this gene in obesity and related co-morbidities (5). In humans the GHSR locus is in physical proximity to genomic linkage peaks for obesity (6, 7).
Rare deleterious mutations in GHSR have been associated with short stature in humans (8, 9). Carriers of an alanine 204 to glutamate amino-acid change were three standard deviations smaller than average for height and weight (8). The 204 glutamate variant lowers cell membrane receptor density, but does not alter receptor affinity for the agonist and increased signal transduction was recorded (8). Another non-synonymous GHSR variation at amino-acid 279 was detected in one heterozygous obese child with short normal stature, but its function is yet unknown (9).
Common polymorphisms in GHSR have been associated with obesity in both a cross-sectional and a family-based association study (10). Five SNPs in a single strong linkage disequilibrium (LD) block covering GHSR exon 1 and its 5′ adjacent region were all associated with BMI and obesity in the German population-based MONICA cohort (10). In a large US family study, the same susceptibility haplotype was more commonly transmitted to obese offspring (10).
The SNPs genotyped in the above study (10) were selected from a public database (dbSNP). In order to confirm those observations and to systemically test for association with common genetic variations we re-sequenced GHSR to identify the common haplotype tagging (ht)SNPs, and then examined these SNPs for association with BMI in three population-based UK cohort studies comprising 5,807 adults and 843 children.
Methods
Populations and measurements
The adult populations came from two distinct cohort studies: the MRC Ely Study (11) and EPIC-Norfolk (6, 12). The children were derived from one population, the Avon Longitudinal Study of Parents and Children (ALSPAC) (13, 14).
The Ely Study is a population-based study of the aetiology and pathogenesis of type 2 diabetes and related metabolic disorders (11). The study consists of an ethnically homogeneous Caucasian population, aged between 40 and 65 years at baseline. Body weight and height were measured at clinic visits using standard methods. This cohort was recruited from a population-sampling frame with a high response rate (74%), and is therefore representative of the eastern England population. Body weight was known for 811 Ely Study participants.
The EPIC-Norfolk cohort (6, 12) is a prospective population-based study of 25,639 men and women aged between 40 and 79 years, resident in Norfolk, UK. Participants were recruited from age-sex registers of general practices in Norfolk as part of the 10-country collaborative EPIC study designed to investigate dietary and other determinants of cancer. The sub-cohort used for this study is a random sample of 5,000 participants (EPIC5000) who were free of disease (cancer, coronary heart disease and diabetes) at baseline, who had arrayed DNA samples available, and had height and weight measured at clinic visits using standard methods (15). Body weight was known for 4996 EPIC5000 participants.
ALSPAC is a geographically based birth cohort (14). The initial ALSPAC sample consists of 14,541 pregnancies. Children in the present study derived from a 10% sub-cohort (“Children in Focus”) who were chosen at random from the last 6 months of ALSPAC births and attended research clinics at various time intervals between 4 to 61 months of age (1432 families attended at least one clinic) (13). At age 7 years, body weight was measured using electronic scales, and standing height by stadiometer (Leicester height measure; Child Growth Foundation, London, UK). Using a topical anaesthetic, a non-fasting venous blood sample was collected. Samples were centrifuged and stored at −70°C. IGF-I levels were measured by direct ELISA (Diagnostic Systems Laboratories, Sinsheim, Germany) (16). Body weight at age 7 years was known for 843 of these children. Lymphocyte DNA was prepared as described previously (17).
All the studies were approved by the local research ethics committees, and informed consent was obtained from each participant or their parent.
SNP selection
The selection of SNPs to cover the genetic variation of the GHSR gene in the European population was based on DNA resequencing performed in 70 obese French children (BMI >97 percentile for age) with a strong component of early onset obesity in their family background (18). The SNPs rs495225 (T171C), rs2232169 (C447G) and rs572169 (G477A) were selected following haplotype-tagging identification methods suggested by Johnson et al (19).
Genotyping
Genotyping was performed sequentially; initially in the Ely Study and ALSPAC populations using the MassEXTEND (hME) assay on the SEQUENOM platform (Sequenom™, Sandiego, CA). Genotyping calling rate was 96% (95% CI: 0.95–0.99].
Subsequently EPIC5000 samples were genotyped using Custom TaqMan assays (Applied Biosystems, Warrington, UK) on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Genotyping call rate was 98% and repeated assays in 75 DNA samples showed 100% concordance. Details for all genotyping primers, probes and PCR conditions are available upon request from the corresponding author.
Statistical analysis
Each SNP was tested for Hardy-Weinberg equilibrium using the χ2 test. We used linear regression to test the association between each SNP (unilocus tests) and the continuous anthropometric outcomes (dependent variables) adjusting for age and sex. All SNPs were considered as linear factors (i.e. additive or co-dominant models). Results are displayed as p-value for the Wald test of the coefficients in the linear model, separately in each study, and then jointly for adult populations. These analyses were performed with SAS version 8.2 (SAS Institute Inc, Cary, NC) and SPSS v1 1 (SPPS, Chicago, IL).
Common haplotypes were inferred from the three htSNPs. As phase was unknown, assignment of haplotype probabilities was performed using the SNPHAP program (19). Tests for main haplotype effects were performed using a linear model weighted by haplotype probability, and clustered by the individual identification to obtain robust standard errors (STATA regression command xi:regres) (20). Results are displayed as p-value for main haplotype effects. Significance was taken at p<0.05.
Results
Details of the GHSR SNPs identified are shown in Figure 1. Genotype frequencies of the three GHSR htSNPs genotyped were in Hardy-Weinberg equilibrium (p>0.2) in each study population (see supplementary data Table 1). Genotype frequencies of T171C differed slightly between the EPIC5000 and Ely Study populations (p=0.044).
Figure 1.
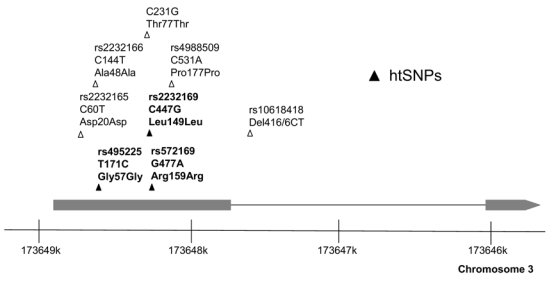
Schematic diagram of the GHSR gene demonstrating the common SNPs identified by sequencing of 70 French obese children. The three haplotype-tagging SNPs genotyped in the UK population studies are indicated by solid triangles and bold type. Only SNP C231G (ttccgcgagctgcgcaccacc/gaccaacctctacctgtccag) is not annotated in HAPMAP Build 35.
The phenotypic characteristics of the study populations are summarised in Supplementary data Table 2. In ALSPAC children none of the three htSNPs were associated with height, weight, BMI or IGF-I levels (Table 1; all p>0.16). In adults from the Ely Study, borderline associations were seen between T171C and BMI (p=0.041, positive association with minor allele), and between G477A and IGF-I levels (p=0.015, positive association with minor allele). In contrast, in EPIC5000 adults there was only a weak inverse association between the G477A minor allele and height (p=0.035). None of the associations were concordant between the Ely and EPIC5000 studies, and in a combined analysis of these two adult populations no htSNP showed an association with height, weight or BMI (all p>0.11; Table 1).
Table 1.
P-values for the associations between each GHSR htSNP and body size in children (ALSPAC) and adults (Ely and EPIC5000 studies).
| GHSR htSNPs | ||||
|---|---|---|---|---|
| G477A | T171C | C447G | ||
| rs572169 | rs495225 | rs2232169 | ||
| Children | P-value | P-value | P-value | |
| ALSPAC (N=843) | Height | 0.66 | 0.22 | 0.62 |
| Weight | 0.29 | 0.88 | 0.62 | |
| BMI | 0.20 | 0.64 | 0.94 | |
| IGF-I | 0.16 | 0.64 | 0.40 | |
| Adult cohorts | P-value | P-value | P-value | |
| Ely Study (N=811) | Height | 0.36 | 0.20 | 0.51 |
| Weight | 0.52 | 0.014 (+ve) | 0.83 | |
| BMI | 0.67 | 0.041 (+ve) | 0.16 | |
| IGF-I | 0.015 (+ve) | 0.70 | 0.11 | |
| EPIC5000 (N=4996) | Height | 0.035 (-ve) | 0.97 | 0.47 |
| Weight | 0.15 | 0.83 | 0.51 | |
| BMI | 0.54 | 0.69 | 0.18 | |
| Combined adult cohorts | P-value | P-value | P-value | |
| EPIC5000 + Ely* (N=5,807) | Height | 0.11 | 0.69 | 0.63 |
| Weight | 0.30 | 0.26 | 0.83 | |
| BMI | 0.72 | 0.25 | 0.47 | |
Analyses are adjusted for age and gender.
Additionally adjusted for study population (EPIC5000 or Ely Study)
p-values are from co-dominant (additive) models.
Where p<0.05, “+ve” or “-ve” indicate the direction of association with minor allele copy number
Three major haplotypes (frequency >0.1) could be inferred from the three GHSR htSNPs in the adult populations (supplementary data Table 3). Similar to the individual SNP analyses, there was no association between common GHSR haplotypes and body size in adults from Ely and EPIC5000 combined (supplementary data Table 3; all p>0.20).
Discussion
In three large UK cohort studies we failed to identify any consistent association between common haplotypic variation in GHSR and body weight, height or BMI in adults or children.
Baessler et al. previously studied 2 SNPs within GHSR and a further 8 SNPs within the 40 to 50 kb up- and downstream adjacent regions (10) and found that an LD block of 5 SNPs was associated with BMI in 1,095 individuals from 178 pedigrees with multiple obese members. That LD block included the synonymous SNP G477A (rs572169); the minor allele (A) was transmitted more frequently than expected to obese cases, was more prevalent in obese cases than controls, and an additive genetic association with higher BMI was confirmed in a further 1,418 Caucasians (10). Of the 5 SNPs in that study, we included only G477A (rs572169), however this SNP would be expected to closely represent the other four obesity risk-associated SNPs due to high LD (r2 >0.75). The relatively large size of our study provided >98% power to replicate the association with obesity risk reported by Baessler et al. (10).
The reason for lack of consistency between the two reports is unknown. Baessler et al. observed a consistent association in Caucasian populations from US and Germany (10), and our three UK populations were also largely Caucasian in origin, with a minor allele frequency of the G477A SNP in our populations (MAF = 0.31) being similar to the other study. It is possible that the genetic association might be dependent on, or modified by some further environment, lifestyle or other genetic factor. However, the lack of further confirmatory studies published since that original report might suggest that the original association was a false positive finding. It is possible that genetic variations in the ghrelin gene, the ligand for the GHSR, may contribute to obesity risk directly, or by interaction with GHSR variants.
In addition to G477A (rs572169), we genotyped two other SNPs to further cover the common variation in GHSR exons as indicated by our resequencing of 70 obese children. Inspection of the HapMap Build 35 CEU population data (http://www.hapmap.org/), which were released following the genotyping of this study, showed that in addition to the three SNPs that we genotyped, only one further intronic SNP (A216G, rs2948694) would be needed to fully cover the GHSR gene including the introns and 1 kbp up- and downstream; however that additional SNP is relatively rare (MAF = 0.07).
A limitation of our study is the slight discrepancies we obtained between the different study populations. Slight differences in genotype frequencies between the Ely Study and EPIC5000 populations could reflect differences in selection of these cohorts. Furthermore, borderline associations in the Ely Study with adult body weight and BMI were not confirmed in larger EPIC5000 study. We checked that the first results were not driven by a few individuals (data not shown), and conclude that they might have arisen by chance. Associations with circulating IGF-1 levels in the Ely Study could not be tested in EPIC5000 due to lack of data, but were not supported by associations with body size.
In conclusion, in a systematic study of common GHSR variation in three large population-based UK cohort studies comprising 5807 adults and 843 children, we found no association with body weight, height or BMI.
Acknowledgments
The UK Medical Research Council, the Wellcome Trust and the University of Bristol provide core support for ALSPAC. The Ely Study was funded by the Medical Research Council and Diabetes UK. EPIC-Norfolk is supported by programme grants from the UK Medical Research Council UK and Cancer Research UK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We are extremely grateful to the families and volunteers in the ALSPAC, EPIC-Norfolk and Ely Studies who gave their time to take part in these studies.
Footnotes
Author Disclosure Summary The authors have nothing to declare
References
- 1.Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 1999;402:656–660. doi: 10.1038/45230. [DOI] [PubMed] [Google Scholar]
- 2.Gnanapavan S, Kola B, Bustin SA, Morris DG, McGee P, Fairclough P, Bhattacharya S, Carpenter R, Grossman AB, Korbonits M. The tissue distribution of the mRNA of ghrelin and subtypes of its receptor, GHS-R, in humans. J Clin Endocrinol Metab. 2002;87:2988. doi: 10.1210/jcem.87.6.8739. [DOI] [PubMed] [Google Scholar]
- 3.Ueda H, Ikegami H, Kawaguchi Y, Fujisawa T, Yamato E, Shibata M, Ogihara T. Genetic analysis of late-onset type 2 diabetes in a mouse model of human complex trait. Diabetes. 1999;48:1168–1174. doi: 10.2337/diabetes.48.5.1168. [DOI] [PubMed] [Google Scholar]
- 4.Zigman JM, Nakano Y, Coppari R, Balthasar N, Marcus JN, Lee CE, Jones JE, Deysher AE, Waxman AR, White RD, Williams TD, Lachey JL, Seeley RJ, Lowell BB, Elmquist JK. Mice lacking ghrelin receptors resist the development of diet-induced obesity. J Clin Invest. 2005;115:3564–3572. doi: 10.1172/JCI26002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, Asnicar M, Saha PK, Chan L, Smith RG. Ablation of ghrelin improves the diabetic but not obese phenotype of ob/ob mice. Cell Metab. 2006;3:379–386. doi: 10.1016/j.cmet.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Rice T, Chagnon YC, Perusse L, Borecki IB, Ukkola O, Rankinen T, Gagnon J, Leon AS, Skinner JS, Wilmore JH, Bouchard C, Rao DC. A genomewide linkage scan for abdominal subcutaneous and visceral fat in black and white families: The HERITAGE Family Study. Diabetes. 2002;51:848–855. doi: 10.2337/diabetes.51.3.848. [DOI] [PubMed] [Google Scholar]
- 7.Vionnet N, Hani EH, Dupont S, Gallina S, Francke S, Dotte S, De Matos F, Durand E, Lepretre F, Lecoeur C, Gallina P, Zekiri L, Dina C, Froguel P. Genomewide search for type 2 diabetes-susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2-diabetes locus on chromosome Iq21-q24. Am J Hum Genet. 2000;67:1470–1480. doi: 10.1086/316887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pantel J, Legendre M, Cabrol S, Hilal L, Hajaji Y, Morisset S, Nivot S, Vie-Luton MP, Grouselle D, de Kerdanet M, Kadiri A, Epelbaum J, Le Bouc Y, Amselem S. Loss of constitutive activity of the growth hormone secretagogue receptor in familial short stature. J Clin Invest. 2006;116:760–768. doi: 10.1172/JCI25303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang HJ, Geller F, Dempfle A, Schauble N, Friedel S, Lichtner P, Fontenla-Horro F, Wudy S, Hagemann S, Gortner L, Huse K, Remschmidt H, Bettecken T, Meitinger T, Schafer H, Hebebrand J, Hinney A. Ghrelin receptor gene: identification of several sequence variants in extremely obese children and adolescents, healthy normal-weight and underweight students, and children with short normal stature. J Clin Endocrinol Metab. 2004;89:157–162. doi: 10.1210/jc.2003-031395. [DOI] [PubMed] [Google Scholar]
- 10.Baessler A, Hasinoff MJ, Fischer M, Reinhard W, Sonnenberg GE, Olivier M, Erdmann J, Schunkert H, Doering A, Jacob HJ, Comuzzie AG, Kissebah AH, Kwitek AE. Genetic linkage and association of the growth hormone secretagogue receptor (ghrelin receptor) gene in human obesity. Diabetes. 2005;54:259–267. doi: 10.2337/diabetes.54.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wareham NJ, Hennings SJ, Byrne CD, Hales CN, Prentice AM, Day NE. A quantitative analysis of the relationship between habitual energy expenditure, fitness and the metabolic cardiovascular syndrome. Br J Nutr. 1998;80:235–241. doi: 10.1017/s0007114598001287. [DOI] [PubMed] [Google Scholar]
- 12.Day N, Oakes S, Luben R, Khaw KT, Bingham S, Welch A, Wareham N. EPIC-Norfolk: study design and characteristics of the cohort. European Prospective Investigation of Cancer. Br J Cancer. 1999;80(Suppl 1):95–103. [PubMed] [Google Scholar]
- 13.Ong KK, Ahmed ML, Emmett PM, Preece MA, Dunger DB. Association between postnatal catch-up growth and obesity in childhood: prospective cohort study. Bmj. 2000;320:967–971. doi: 10.1136/bmj.320.7240.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golding J, Pembrey M, Jones R. ALSPAC-the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatr Perinat Epidemiol. 2001;15:74–87. doi: 10.1046/j.1365-3016.2001.00325.x. [DOI] [PubMed] [Google Scholar]
- 15.Sandhu MS, Heude B, Young EH, Luben R, Luan J, Khaw KT, Todd J, Wareham NJ. INS VNTR class genotype and indexes of body size and obesity: population-based studies of 7,999 middle-aged men and women. Diabetes. 2005;54:2812–2815. doi: 10.2337/diabetes.54.9.2812. [DOI] [PubMed] [Google Scholar]
- 16.Ong K, Kratzsch J, Kiess W, Dunger D. Circulating IGF-I levels in childhood are related to both current body composition and early postnatal growth rate. J Clin Endocrinol Metab. 2002;87:1041–1044. doi: 10.1210/jcem.87.3.8342. [DOI] [PubMed] [Google Scholar]
- 17.Jones RW, Ring S, Tyfield L, Hamvas R, Simmons H, Pembrey M, Golding J. A new human genetic resource: a DNA bank established as part of the Avon longitudinal study of pregnancy and childhood (ALSPAC) Eur J Hum Genet. 2000;8:653–660. doi: 10.1038/sj.ejhg.5200502. [DOI] [PubMed] [Google Scholar]
- 18.Gueorguiev MLC, Mein CA, Meyre D, Benzinou M, Weill J, Grossman AB, Froguel P, Korbonits M. Ghrelin receptor gene: involvement in eating behaviour. ENEA O3.4 2004 [Google Scholar]
- 19.Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- 20.Mander A. QHAPIPF:Stata module to perform analysis of quantitative traits using refression and log-linear modelling when PHASE is unknown. Economics BCDo ed 2003 [Google Scholar]


