Abstract
The human bcl-2 gene contains a GC-rich region upstream of the P1 promoter that has been shown to be critically involved in the regulation of bcl-2 gene expression. We have demonstrated that the guanine-rich strand of the DNA in this region can form any one of three distinct intramolecular G-quadruplex structures. Mutation and deletion analysis permitted isolation and identification of three overlapping DNA sequences within this element that formed the three individual G-quadruplexes. Each of these was characterized using nondenaturing gel analysis, DMS footprinting, and circular dichroism. The central G-quadruplex, which is the most stable, forms a mixed parallel/antiparallel structure consisting of three tetrads connected by loops of one, seven, and three bases. Three different G-quadruplex-interactive agents were found to further stabilize these structures, with individual selectivity toward one or more of these G-quadruplexes. Collectively, these results suggest that the multiple G-quadruplexes identified in the promoter region of the bcl-2 gene are likely to play a similar role to the G-quadruplexes in the c-myc promoter in that their formation could serve to modulate gene transcription. Last, we demonstrate that the complexity of the G-quadruplexes in the bcl-2 promoter extends beyond the ability to form any one of three separate G-quadruplexes to each having the capacity to form either three or six different loop isomers. These results are discussed in relation to the biological significance of this G-quadruplex-forming element in modulation of bcl-2 gene expression and the inherent complexity of the system where different G-quadruplexes and loop isomers are possible.
INTRODUCTION
The bcl-2 (B-cell CLL/lymphoma 2) gene product is a mitochondrial membrane protein that exists in delicate balance with other related proteins and is involved in the control of programmed cell death, functioning as an apoptosis inhibitor.1,2 The bcl-2 gene was originally discovered for its involvement in the t(14;18) chromosomal translocation frequently found in lymphomas.3 This translocation causes deregulated expression of the bcl-2 gene, leading to elevated levels of bcl-2 mRNA and protein.4 Since its initial discovery, the bcl-2 gene has been classified as a proto-oncogene because of its overexpression in a wide range of human cancers, including B-cell and T-cell lymphomas,5 breast,6 cervical,7 non-small cell lung,8 prostate,9 and colorectal.10 However, in contrast to other proto-oncogenes, the oncogenic potential of bcl-2 is not achieved by promoting cell proliferation, but by reducing the rate of cell death. In addition, bcl-2 overexpression has been found to interfere with the therapeutic action of current cancer treatment regimes, which act by inducing apoptosis in tumor cells.11 Its broad expression in a variety of tumors, together with its function in the resistance to chemotherapy-induced apoptosis, makes bcl-2 a rational target for anticancer therapy.
In contrast, bcl-2 has also emerged as an attractive target for neuroprotective therapy to prevent apoptotic neuronal cell death in neurodegenerative disorders of aging, such as Alzheimer’s disease, Parkinson’s disease, and stroke. For instance, in neuronal cells the overexpression of bcl-2 has been shown to prevent cell death induced by diverse apoptotic stimuli.12,13 Likewise, the overexpression of bcl-2 in transgenic mouse models results in an excess of neurons in selected regions of the brain, indicating that bcl-2 protects neurons from naturally occurring cell death during development.14,15 Conversely, bcl-2 knockout mice have significant degeneration of motor, sensory, and sympathetic neurons, suggesting that bcl-2 is necessary for the maintenance of neuronal populations.16 Thus far, targeted delivery treatments are being sought that increase intracellular levels and function of bcl-2 by employing the use of non-pathogenic viral vectors constructed to transport plasmids encoding bcl-2 inside neurons of affected brain regions.17,18 Although encouraging, the neuroprotection observed by means of targeted delivery of bcl-2 is limited in that they employ dosing regimens that may be unrealistic for human use.19
The human bcl-2 gene has two promoters, designated P1 and P2, that control its transcriptional initiation (Figure 1A). The major promoter, P1, located 1386 to 1423 base pairs upstream of the translation start site, is a TATA-less, GC-rich promoter containing multiple transcription initiation sites.20 The P2 promoter contains both a canonical TATA element and a CCAAT box, yet is only responsible for a minor fraction of the bcl-2 transcripts in most cell types.21 Thus far, the mechanisms involved in the transcriptional regulation of the bcl-2 gene are not well understood, although a number of trans-acting and cis-acting elements that take part have been identified. For example, one major contributor to P1 promoter activity is a cAMP response element, which is necessary for bcl-2 deregulation in t(14;18) cells, the activation of mature B cells, and the rescue of immature B cells from calcium-dependent apoptosis.22,23 In addition, NF-κB has been shown to act through interactions with the camp response element site and activate bcl-2 in t(14;18) lymphoma cells.24 The P1 promoter also contains WT1 bindings sites, which function to silence the normal bcl-2 allele in t(14;18) cells,25 as well as repress bcl-2 transcription in HeLa cells.26 C/EBPα and A-Myb have been demonstrated to act through a binding site for the homeodomain protein Cdx and activate P2 promoter activity in t(14;18) cells.27,28 Also described is the existence of a negative response element upstream of the P2 promoter through which p53 may either directly or indirectly transcriptionally downregulate bcl-2 expression.29
Figure 1.
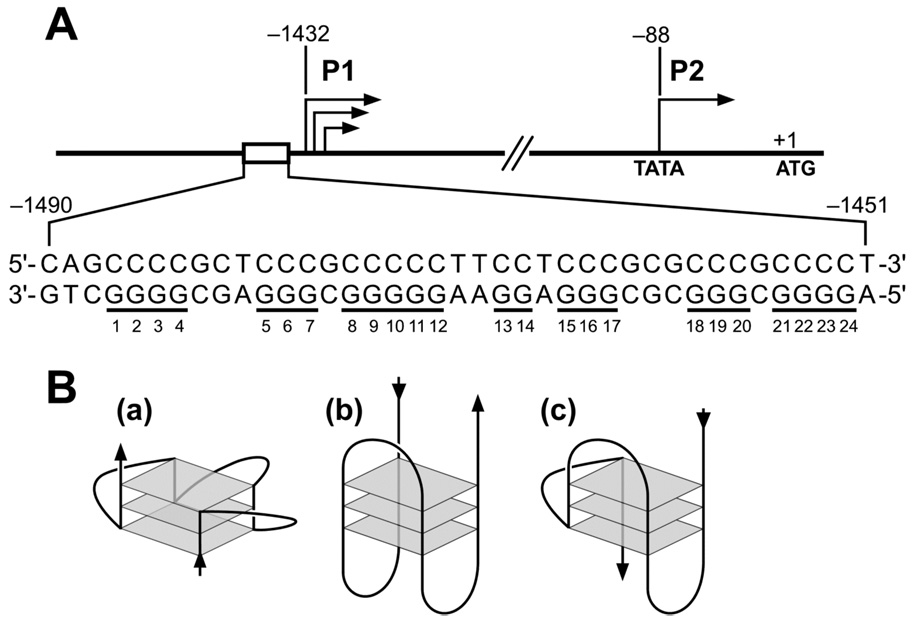
(A) Promoter structure of the human bcl-2 gene; shown in inset is the 39-mer sequence of the purine-rich strand upstream of the P1 promoter. Runs of guanines are underlined and numbered. (B) Examples of intramolecular G-quadruplex structures: (a) parallel, (b) antiparallel, (c) mixed parallel/antiparallel.
One region of particular interest is the GC-rich region located adjacent to the 5′-end of the bcl-2 P1 promoter (Figure 1A). Multiple transcription factors have been described to bind or regulate bcl-2 gene expression through this region, such as Sp1,20 WT1,25 E2F,30 and NGF (nerve growth factor).31 Additionally, deletion or mutation of this region has been shown to increase promoter activity by 2.1-fold or 2.6-fold compared with the full-length wild-type promoter, respectively.25 This GC-rich element is located in the proximity of a nuclease hypersensitive region,32 which has been shown to be dynamic in nature and capable of adopting paranemic structures or non-B-DNA conformations.33 In particular, guanine-rich sequences can adopt specific intramolecular DNA secondary structures termed G-quadruplexes (Figure 1B).34 Guanine-rich DNA sequences predisposed to the formation of G-quadruplexes have been identified in the telomeres of virtually all eukaryotic organisms,35 in immunoglobulin switch regions,36 and in the promoter regions of several growth-related genes.37,38 Interestingly, the guanine-rich DNA strand in the bcl-2 P1 promoter region contains seven runs of at least two contiguous guanines separated by two or more bases (Figure 1), which is consistent with the general motif capable of folding into at least one intramolecular G-quadruplex structure.
In the present study, we demonstrate that the guanine-rich DNA strand of the GC-rich region, −58 to −19 base pairs upstream of the bcl-2 P1 promoter, has the ability to form multiple G-quadruplex structures in vitro. We also isolate and characterize three separate DNA sequences within this region that are consistent with the formation of the three individual G-quadruplex structures. In addition, we establish that G-quadruplex-interactive agents, such as TMPyP4, Se2SAP, and telomestatin (Figure 2), can further stabilize these structures with a type of individual selectivity to one or more of these G-quadruplexes.
Figure 2.
Structures of the G-quadruplex-interactive compounds TMPyP4, telomestatin, and Se2SAP, and the control compound TMPyP2.
EXPERIMENTAL PROCEDURES
Enzymes and Drugs
T4 kinase and Taq DNA polymerase were purchased from Promega. TMPyP2 (four conformational isomers), TMPyP4, and Se2SAP were synthesized in our laboratory. Telomestatin was kindly provided by Dr. Kazuo Shin-ya (University of Tokyo, Japan).
DNA Oligonucleotides
All synthetic DNA oligonucleotides were supplied by Operon. Oligonucleotide extinction coefficients were calculated using the nearest neighbor method.39 Since this calculation produces a theoretical value for single stranded DNA, the resulting nearest neighbor extinction coefficients were optimized for higher order structural hyperchromicity using UV/Vis thermal denaturation techniques.40 Oligonucleotides were heated to 20 °C above their respective Tm and the absorbance at λmax was extrapolated back to 25 °C to correct for structural hyperchromicity. The structural hyperchromicity of the quadruplexes was nominally 25%. The nearest neighbor extinction coefficient was corrected using the following relation: ε260(Quad) = ε260(NN) / (1 + (%Hyperchromicity / 100)). The corrected ε260 were as follows: Pu39WT, 309040 M−1 cm−1, 5′G4, 196480 M−1 cm−1, MidG4, 207840 M−1 cm−1, and 3′G4, 230400 M−1 cm−1.
Preparation and End-Labeling of Oligonucleotides
The 5′-end-labeled single-strand DNA oligonucleotides were obtained by incubating the oligomer with T4 polynucleotide kinase and [γ-32P] ATP for 1 h at 37 °C. Labeled DNA was purified with a Bio-Spin 6 chromatography column (BioRad) after inactivation of the kinase by heating for 5 min at 95 °C. Labeled oligonucleotides were then purified by denaturing gel electrophoresis (16% PAGE).
Polymerase Stop Assay
The 5′-end [γ-32P]-labeled primer d[TAATACGACTCACTATAGC AATTGCGTG] was annealed to a cassette DNA, d[TCCAACTATGTATAC(G-quadruplex-forming insert)TTAGCGGCACGCAATTGCTATAGTGAGTCGTATTA], containing the G-quadruplex-forming sequence of interest. DNA formed by annealing the primer to the template sequence was purified using gel electrophoresis on an 8% native polyacrylamide gel. The assays were carried out as described previously.41 Briefly, the purified DNA was mixed with a reaction buffer (50 mM Tris-HCl, pH 7.6,10 mM MgCl2, 0.5 mM DTT, 0.1 mM EDTA, 1.5 g/L BSA), 0.2 mM deoxynucleotide triphosphates, and KCl (as specified in the figures). The reaction was incubated for 1 h at 25 °C. Taq DNA polymerase was then added (5 U/reaction) and allowed to extend the primer for 30 min at 37 °C. The reactions were stopped by adding an equal volume of stop buffer (95% formamide, 10 mM EDTA, 10 mM NaOH, 0.1% xylene cyanol, 0.1% bromphenol blue) and loaded on a 16% sequencing PAGE gel. For the drug stabilization experiments, drugs were added and incubated an additional 1 h at 25 °C before primer extension by Taq DNA polymerase.
Dimethyl Sulfate Footprinting
The 5′-end [γ-32P]-labeled DNA oligonucleotides were denatured by heating to 95 °C for 5 min and then slowly cooled to 4 °C in 10 mM Tris-HCl with or without 100 mM KCl. The reactions were then subjected to preparative gel electrophoresis (16% polyacrylamide in 1 × TBE and 10 mM KCl for 15 h at 4 °C). Briefly, each band of interest was excised and the DNA species eluted by soaking in running buffer (1 ×TBE and 10 mM KCl) overnight at 4 °C. DNA solutions were then used to perform the DMS footprinting, as outlined previously.37 In brief, following the addition of 1 µg of calf thymus DNA, the DNA solutions were subjected to dimethyl sulfate (0.5% DMS in 50% ethanol) for 10 min. Each reaction was quenched by adding 18 µL of stop buffer. After ethanol precipitation and treatment with piperidine, the cleaved products were separated on a 16% sequencing PAGE gel.
Circular Dichroism
CD spectra were recorded on a Jasco-810 spectropolarimeter (Jasco, Easton, MD), using a quartz cell of 1 mm optical path length, an instrument scanning speed of 100 nm/min, with a response time of 1 s, and over a wavelength range of 200–320 nm. All DNA samples, TBA d[GGTTGGTGTGGTTGG], Tetrahymena d[GGGTTGGGTTGGGTTGGGTT], c-myc Pu27-mer d[TGGGGAGGGTGGGGAGGGTGGGGAAGG], and bcl-2 Pu39WT (sequence shown in Table 1), were dissolved in Tris-HCl buffer (50 mM, pH 7.6) containing 100 mM KCl or 100 mM NaCl to a strand concentration of 5 µM. The DNA oligonucleotides were then denatured by heating to 95 °C for 5 min and slowly cooled to 25 °C over several hours. The CD spectra herein are representations of three averaged scans taken at 25 °C and are baseline corrected for signal contributions due to the buffer.
Table 1.
| Name | Sequence (5′–3′) |
|---|---|
| Pu39WT | AGGGGCGGGCGCGGGAGGAAGGGGGCGGGAGCGGGGCTG |
| 5′G4 | AGGGGCGGGCGCGGGAGGAAGGGGGC |
| MidG4 | CGGGCGCGGGAGGAAGGGGGCGGGAGC |
| 3′G4 | CGCGGGAGGAAGGGGGCGGGAGCGGGGCTG |
| MidG4-mut1 | CGGGCGCGGGAGGAATTGGGCGGGAGC |
| MidG4-mut2 | CGGGCGCGGGAGGAAGGGTTCGGGAGC |
| MidG4-mut3 | CGGGCGCGGGAGGAATGGGTCGGGAGC |
Imaging and Quantification
The dried gels were exposed on a phosphor screen. Imaging and quantification were performed using a Storm 820 PhosphorImager and ImageQuant 5.1 software (Molecular Dynamics).
RESULTS
Formation of G-Quadruplex Structures by the Guanine-Rich Strand in the bcl-2 P1 Promoter Region
To initially test our hypothesis of the occurrence of intramolecular G-quadruplex structures within the guanine-rich strand of the bcl-2 P1 promoter region, a DNA polymerase stop assay was used. This assay provides a straightforward approach to identifying guanine-rich DNA sequences that have the ability to form stable G-quadruplex structures in vitro, based on the assumption that a DNA polymerase is incapable of traversing a stable DNA secondary structure.42,43 A cassette DNA sequence containing the G-quadruplex-forming sequence of interest, or in this case, the 39-mer guanine-rich strand (Pu39WT, Table 1) from the bcl-2 P1 promoter region, was annealed with a [γ-32P]-labeled primer, which subsequently was extended with Taq DNA polymerase in the presence of increasing concentrations of KCl. A full-length product was synthesized in the absence of KCl (Figure 3A, lane 3); however, upon the addition of KCl to the reaction, three premature primer arrest sites were produced (Figure 3A, lanes 4–6). The DNA polymerase arrests sites (1, 2, and 3) occurred directly upstream of G1, G5, and G8, respectively (see numbering in Figure 1). All three arrest sites increased in a K+-dependent manner. In addition, the G-quadruplex-interactive agent TMPyP4, a cationic porphyrin, had the ability to stabilize the three G-quadruplex structures formed in the Pu39WT sequence in the presence of minimal K+ (20 mM), whereas TMPyP2 (Figure 2), a structural isomer of TMPyP4, which lacks the ability to interact with G-quadruplex structures,44 had a negligible effect on the arrest products (Figure 3B). Taken together these results suggest that the Pu39WT sequence has the capacity to form three separate intramolecular G-quadruplex structures, which are stabilized by K+ or TMPyP4.
Figure 3.
Determination of the ability of the Pu39WT sequence (Table 1) to form multiple G-quadruplex structures. (A) DNA polymerase stop assay showing three K+-dependent DNA polymerase arrests. Lane 1 shows a guanine sequencing reaction, lane 2 is a negative control containing no Taq DNA polymerase, and lanes 3–6 contain 0, 25, 50, and 100 mM KCl, respectively. Arrows point to the positions of the full-length product of DNA synthesis, the DNA polymerase arrest sites (1, 2, and 3), and the free primer. (B) Concentration-dependent inhibition of Taq polymerase DNA synthesis by stabilization of the bcl-2 G-quadruplex structures with TMPyP4 (0.5, 1, 2.5, 5 µM) compared to TMPyP2 (0.5, 1, 2.5, 5 µM). All lanes contain 20 mM KCl. (C) Nondenaturing gel analysis of Pu39WT sequence preincubated in the absence or presence of 100 mM KCl. (D) DMS footprinting of band 1 (lane 2) and band 2 (lane 3) from (C). Lane 1 shows a purine sequencing reaction. The protected guanines from DMS are indicated with filled circles, partial protection with half-filled circles, no protection with open circles, and hypersensitive guanines with arrowheads. (E) Comparative CD spectra of three known G-quadruplex-forming sequences with the unknown bcl-2 promoter Pu39WT sequence. Line colors: green = c-myc promoter sequence (parallel G-quadruplex in 100 mM KCl); blue = thrombin binding aptamer (TBA) sequence (antiparallel G-quadruplex in 100 mM KCl); red = Tetrahymena telomeric repeat sequence (mixed parallel/antiparallel G-quadruplex in 100 NaCl); black = Tetrahymena telomeric repeat sequence (100 mM KCl); orange = bcl-2 promoter Pu39WT sequence (100 mM KCl). All CD data were obtained with a 5 µM strand concentration at 25 °C.
Further evidence for the presence of multiple intramolecular G-quadruplex structures in the bcl-2 P1 promoter region was obtained from native polyacrylamide gel electrophoresis of the 5′-end-labeled Pu39WT in the presence of 100 mM KCl. This gel showed a more rapidly moving band than the expected electrophoretic mobility of an unstructured oligonucleotide (band 2 in Figure 3C). Several minor bands of slower electorophetic mobility were also seen, likely corresponding to two- or four-stranded higher-order G-quadruplex structures. The broadness of the high mobility band is likely attributable to the potential of this sequence to form an equilibrium mixture of multiple intramolecular G-quadruplex structures, as observed previously in the DNA polymerase stop assay (Figure 3A). The guanine bases involved in the formation of the G-tetrads can be deduced by dimethyl sulfate (DMS) footprinting, which probes the accessibility of the N7 position of guanines to alkylation by DMS. Since the N7 position of guanine is involved in Hoogsteen bonding to form the G-tetrad of a G-quadruplex structure, they are inaccessible to DMS methylation.37 Therefore, bands 1 and 2 corresponding to the species formed without and with KCl were isolated and subjected to DMS treatment and piperidine cleavage (Figure 3). The N7 guanine methylation pattern produced by band 1 was consistent with a single-stranded, unstructured form of DNA (Figure 3D, lane 2). For band 2, several guanine bases within the G-quadruplex forming region were either partially or fully protected from DMS methylation; however, the cleavage pattern was fairly indefinite in that no specific structure prevailed (Figure 3D, lane 3). It is significant to note that the run of two guanines (G13 and G14) in the middle of Pu39WT were hypersensitive to methylation, suggesting that they are in an external loop region and not involved in the G-tetrads. In addition, the run of five guanines (G8–G12) provides some evidence for sequence ambiguity in that only partial DMS protection is observed in the two guanines at the 5′-end of this guanine tract, compared to full protection of the remaining three guanine bases.
CD provides additional support for the existence of intramolecular G-quadruplex structures, also specifically assisting in the determination of DNA strand orientation and the conformation of the guanine glycosyl bonds within a given G-quadruplex structure. However, caution is advised when analyzing CD spectra since oversimplification of structure–spectrum relationships may lead to misinterpretation of G-quadruplex CD spectra.45 Signature CD spectra have been determined for several novel G-quadruplex structures in the presence of monovalent cations.46,47 In general, antiparallel-type G-quadruplexes containing alternating syn- and anti-glycosyl bond conformations have been shown to have a CD spectrum characterized by a positive ellipticity maximum at 295 nm and a negative minimum at 265 nm, whereas, parallel-type G-quadruplexes composed of all anti-glycosyl bond conformations have a positive maximum at 264 nm and a negative minimum at 240 nm.47 Comparative CD analysis was performed between different types of known G-quadruplex structures, the c-myc promoter NHE III1 region (“a” in Figure 1B), the thrombin binding DNA aptamer (“b” in Figure 1B), the telomeric repeat of Tetrahymena (“c” in Figure 1B), and the unknown Pu39WT sequence in the presence of 100 mM KCl or 100 mM NaCl. The CD spectrum for the Pu39WT resembles most closely that of the Tetrahymena sequence by displaying an absorption maximum at 264 and a shoulder at 295 nm. These results infer the formation of a mixed parallel/antiparallel G-quadruplex structure in the bcl-2 P1 promoter sequence (Figure 3E); however, based upon the results from the electophoretic mobility analysis, DMS footprinting, and the polymerase stop assay, the appearance of these two undefined peaks most likely occurs because there is a mixture of more than one G-quadruplex structure in solution.
Mutational Analysis of the Seven Guanine Tracts Identifies the Sequences Involved in the Formation of Each of the Three G-Quadruplex Structures
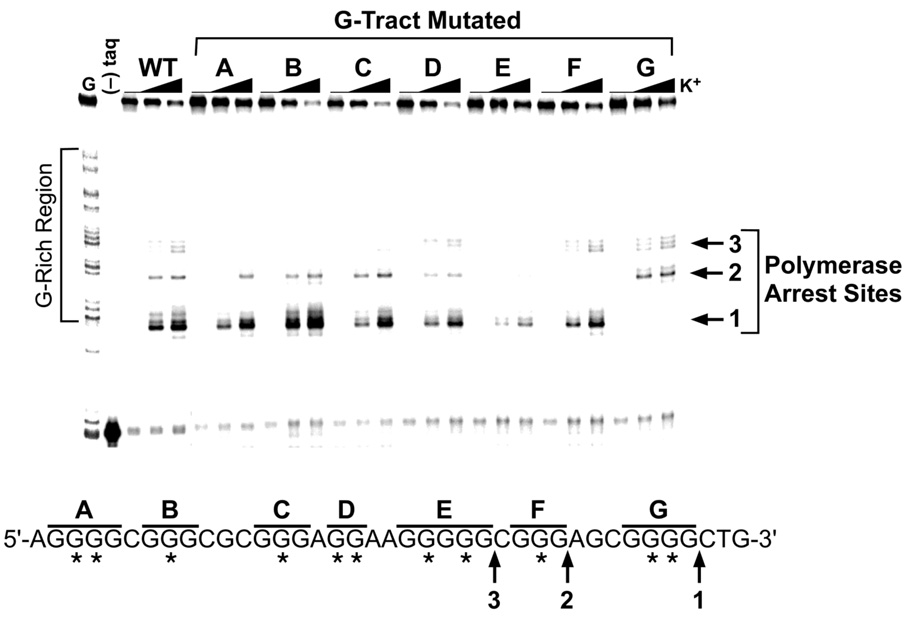
To extend the study and evaluate which of the seven G-tracts are responsible for the formation of the three different G-quadruplex structures within the bcl-2 P1 promoter region, oligomers containing mutations within each of the seven G-tracts were designed and synthesized. The actual mutations selected were devised with the intention of knocking-out each tract of guanines independently and thus preventing their participation in G-quadruplex formation. For this reason, single or double G-to-T base mutations were made to the central guanine base(s) of each G-tract of Pu39WT (see asterisks in Figure 4) based on the assumption that the central guanine base(s) participate(s) in the formation of the critical core G-tetrad of any G-quadruplex produced. Use of these G-tract knockout sequences in the DNA polymerase stop assay resulted in reduced or complete disappearance of one or more of the three polymerase arrest sites (Figure 4). Mutation of any of the three 5′-end G-tracts (A, B, and C) inhibited the formation of the 5′-end G-quadruplex structure at arrest site 3. Arrest sites 1 and 2 were only eliminated upon mutation of the two 3′-end G-tracts (G and F, respectively). Knocking-out the run of five guanines (E) was detrimental to all three of the G-quadruplex structures, while mutation of the central G-tract (D) consisting of a run of only two guanines produced a small but comparable reduction of all three of the DNA polymerase arrest sites. This result is in agreement with the hypersensitivity of these two guanines to DMS methylation and an indication of their peripheral involvement in all three G-quadruplex structures formed. Overall the mutational analysis strengthens the hypothesis that three intramolecular G-quadruplex structures can form in the guanine-rich region of the bcl-2 P1 promoter. In addition, it provides insight for a more focused deletion study in order to isolate and identify the three individual G-quadruplex-forming sequences within the bcl-2 P1 promoter region.
Figure 4.
DNA polymerase stop assay to analyze the effect on stability of the three individual G-quadruplex structures formed by the Pu39WT sequence by knocking-out each G-tract with mutations. In the sequence below the gel, letters (A–G) indicate which G-tract contains the mutation(s) and asterisks indicate G-to-T mutations. The three DNA polymerase arrest sites (1, 2, and 3) are identified with arrows. KCl concentrations are 0, 50, and 100 mM.
Isolation and Identification of the Guanine-Tracts Involved in the Formation of Each of the Three Different G-Quadruplex Structures and Comparison of Their CD Signatures
Isolation of three individual G-quadruplex-forming sequences
In general, an intramolecular G-quadruplex structure can arise from a DNA sequence that contains four runs of at least two or more contiguous guanines separated by one or more bases ([G≥2Nn]4). In the case of the Pu39WT sequence, there are a total of six runs of three or more uninterrupted guanines, each with an equal likelihood to participate in G-quadruplex formation. The utilization of basic combination probability analysis reveals the possibility of fifteen different G-quadruplex structures consisting of any four G-tracts without regard to their position in the Pu39WT sequence. However, the majority of these hypothetical G-quadruplex structures are formed from four nonconsecutive G-tracts, which would generate an unstable structure in view of the existence of lengthy external loops.48 If the locality of the G-tracts is taken into consideration, given that only arrangements of four successive G-tracts are allowed, then the number of potential G-quadruplex structures is decreased to three, two external and one internal located at the 5′-end, 3′-end, and middle of the Pu39WT sequence, respectively.
As a result of the mutational and sequence examination, three truncated sequences were designed (5′G4, MidG4, and 3′G4; Table 1) consisting of each of the four consecutive G-tracts existing in the Pu39WT sequence and placed in the primer extension cassette sequence for analysis using the DNA polymerase stop assay. Figure 5, A′–C′, shows the results of Taq DNA polymerase primer extension using these substrates in the presence of increasing concentrations of KCl. Each of the three substrates gave rise to a single premature primer arrest site in the presence of K+. The individual arrest sites in the three truncated sequences, 5′G4, MidG4, and 3′G4, occurred at G8, G5, and G1, respectively, which is in full agreement with the three arrest sites observed using the Pu39WT sequence (Figure 3A). In addition, the internal four G-tracts (MidG4) produces a G-quadruplex structure that is remarkably more stable then either the 3′ or 5′ G-quadruplexes, based on the minimal amount of K+ required to produce a DNA polymerase arrest, suggesting that it could be the predominant structure within the bcl-2 P1 promoter region (Figure 5B″).
Figure 5.
Characterization of the three individual G-quadruplex-forming sequences. (A′–C′) DNA polymerase stop assays utilizing the 5′G4, MidG4, and 3′G4 sequences (see Table 1). Arrows point to the positions of the K+-dependent DNA polymerase arrest sites (G8, G5, and G1). KCl concentrations are 0, 25, 50, and 100 mM. (A″–C″) DMS footprinting and graphical representation of autoradiograms of isolated bands from the nondenaturing gels shown in the inset. Nondenaturing gel analysis of 5′G4, MidG4, and 3′G4 sequences preincubated under the conditions specified in the figure are shown in panel insets. The guanines protected from DMS are indicated with filled circles, partial protection with half-filled circles, no protection with open circles, and hypersensitive guanines with arrowheads. (D) The top panel shows comparative CD spectra of the bcl-2 promoter full-length and truncated sequences in the presence of 100 mM KCl. Line colors: orange = Pu39WT; red = 5′G4; blue = MidG4; black = 3′G4. The bottom panel compares the Tetrahymena telomeric repeat sequence with the bcl-2 MidG4 sequence. Line colors: red = Tetrahymena telomeric repeat sequence (mixed parallel-antiparallel G-quadruplex in 100 NaCl); black = Tetrahymena telomeric repeat sequence (100 mM KCl); blue = MidG4 (100 mM KCl); orange = bcl-2 promoter Pu39WT sequence (100 mM KCl). All CD data were obtained with a 5 µM strand concentration at 25° C.
DMS footprinting identifies the guanines involved in each G-quadruplex structure
To determine the guanines involved in each of the G-tetrads of these three isolated G-quadruplex structures, electorphoretic mobility shift and DMS footprinting methods were used. The three oligonucleotides 5′G4, MidG4, and 3′G4 were denatured at 95 °C for 5 min and slowly cooled to 4 °C over several hours in the presence or absence of 100 mM KCl. The samples were then subjected to native polyacryamide gel electrophoresis to separate the intramolecular G-quadruplexes from unstructured single-stranded DNA. Incubation of the oligonucleotides 3′G4 and MidG4 in the absence of K+ resulted in the formation of one primary band (band 1 in insets of Figure 5, B″ and C″); however, two discrete bands were produced in the presence of K+, one major band (band 2 in insets of Figure 5, B″ and C″) with a faster mobility than that of band 1 and one minor band that ran similar to the K+-deficient DNA, suggesting that not all the DNA was converted to the higher mobility species. In the case of the 5′G4 oligonucleotide, two bands were seen in the absence (bands 1 and 2 in inset of Figure 5A″) and the presence of K+ (bands 3 and 4 in inset of Figure 5A″), with band 4 shifting to a more rapid mobility than all the other bands. All the numbered bands in the insets of Figure 5, A″–C″, were excised, subjected to DMS treatment and piperidine cleavage, then separated on sequencing gels. The results of DMS footprinting are shown in Figure 5 (A″–C″), as well as graphical illustrations of the relative band intensities for each lane. The internal MidG4 sequence, which is the most stable, based on low K+-dependence, was the only sequence to conform to a specific intramolecular G-quadruplex structure derived from its cleavage pattern, in which four runs of three guanines were partially or fully protected from DMS methylation (Figure 5B″). Derivations of the G-tetrads formed in sequences 5′G4 and 3′G4 are not so straightforward. The 3′G4 N7 methylation pattern showed protection of at least three guanines in the three 3′-end G-tracts; however, the fourth G-tract only displayed protection of two guanines (Figure 5C″). This suggests that the adjacent adenine or the central run of two guanines, which did show some degree of DMS protection, may contribute to the formation of a unique intramolecular G-quadruplex structure in the 3′G4 sequence. For the 5′G4 sequence, the footprinting patterns of bands 2 and 4 were compared, since a mobility shift was observed, whereas no appreciable shift occurred between bands 1 and 3. Several guanine bases within the G-tracts were either partially or fully protected from DMS methylation in the 5′G4 sequence (Figure 5A″); however, the unclear N7 methylation pattern did not allow for determination of specific G-tracts involved in G-quadruplex formation. Significantly, this is the only sequence to show full protection of the run of two guanines, which may be attributable to another unique G-quadruplex structure reminiscent of the 3′G4 sequence.
Circular dichroism analysis shows the three G-quadruplexes are mixed parallel/antiparallel structures
Comparative circular dichroism studies were also carried out on the three truncated sequences and the full-length Pu39WT sequence. As shown in Figure 5D (top panel), all the sequences had similar CD signatures, each comprising a major and minor positive peak at 264 and 295 nm, respectively. Based on the intensity and sharpness of the peaks at 264 nm and 295 nm, the 5′G4 and 3′G4 sequences had more parallel character with less defined antiparallel character. In contrast, a well-defined peak at 295 nm was observed in the CD spectrum of the MidG4 sequence, which is quite analogous to that of the Na+- and K+-stabilized Tetrahymena telomeric sequence (Figure 5D, bottom panel). However, of these two apparently similar forms (at least by CD), only the structure of that form in Na+ is known.49 This implies that the G-quadruplex formed in the MidG4 sequence has a similar folding topology to the Tetrahymena G-quadruplex structure in Na+, which consists of two lateral loops and one double-chain reversal. This was subsequently determined by NMR.50 Overall, these results are consistent with the presence of mixed parallel/antiparallel G-quadruplex structures for all the sequences; however, the most convincing evidence is seen with the MidG4 sequence.
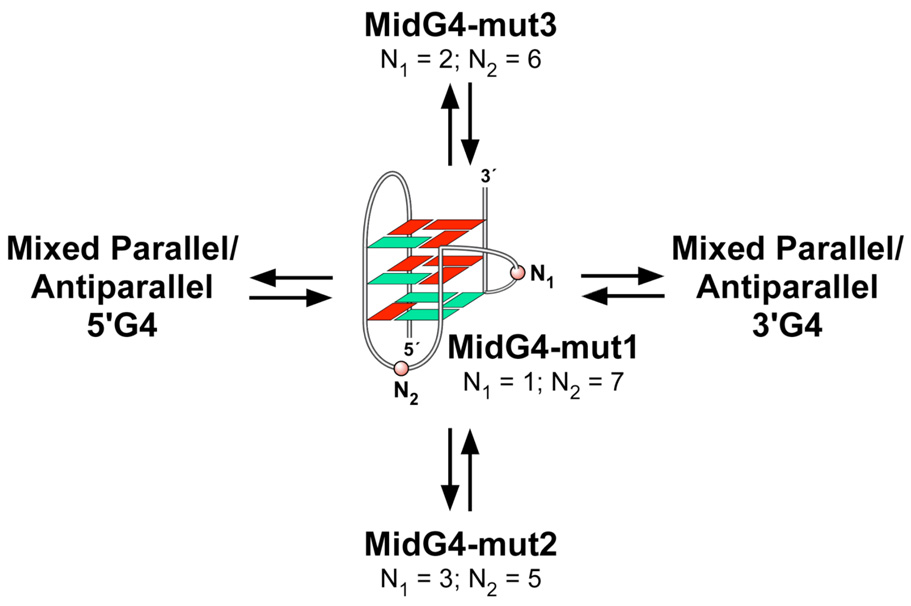
Comparison of Stability of the Three Possible Loop Isomers in the MidG4 Sequence
Even though three specific G-quadruplex-forming sequences were isolated, it is possible for each of these sequences to have the ability to form alternative loop isomers. For example, because the MidG4 sequence has three G-tracts, each consisting of only three guanines, this limits the G-quadruplex to having three G-tetrads; therefore, only three guanines are needed from the run of five guanines to participate in the formation of the three G-tetrads, and the remaining two guanines are forced into loop regions. There are three possible loop isomers due to the different combinations of three adjacent guanines in the run of five guanines. To test our hypothesis of loop isomers within the MidG4 sequence, dual G-to-T mutations were designed within the run of five guanines (Table 1; MidG4-mut1, -mut2, and -mut3) to determine the selection of guanines involved in the G-tetrads. These unambiguous mutated sequences were then compared for stability to the wild-type sequence using the DNA polymerase stop assay. The results revealed that the mutant sequences were less stable than the wild-type sequence based on arrest site intensities (Figure 6). Moreover, utilization of the three guanines at either the 5′-end or 3′-end (mut1 and mut2) of the run of five guanines in G-quadruplex formation appeared to be more favorable than the central three guanines (mut3). Mutation of the two 5′-end guanines generated the most stable structure based on the minimal amount of K+ required to produce a DNA polymerase arrest. These results indicate that loop isomerization can occur within the MidG4 sequence, and since this run of five guanines, as well as an additional run of four guanines, is also present in the 5′G4 and 3′G4 sequences, this phenomenon is likely to be even more exaggerated in these G-quadruplexes.
Figure 6.
DNA polymerase stop assay showing the difference in stability of the three possible loop isomers in the MidG4 sequence in comparison to the wild-type sequence. The wild-type MidG4 sequence is shown below the gel. MidG4-mut1, MidG4-mut2, and MidG4-mut3 contain dual G-to-T mutations within G5 (see Table 1). KCl concentrations are 0, 25, 50, and 100 mM.
The Three Different G-Quadruplex-Forming Sequences Bind G-Quadruplex-Interactive Drugs with Different Affinities
We have demonstrated that three individual G-quadruplex structures can form in the G-rich strand of the bcl-2 P1 promoter region. However, the long-term goal of this study is to identify small molecules with the ability to modulate bcl-2 gene expression through the stabilization and/or modification of these unique G-quadruplex structures. Therefore, we next examined the ability of a number of well-known G-quadruplex-interactive agents (TMPyP4, Se2SAP, and telomestatin) to selectively interact and stabilize the three individual G-quadruplex structures characterized in our preceding studies. While TMPyP4 is able to bind to and stabilize a variety of G-quadruplex structures, TMPyP2 lacks the ability to interact with these structures44 and serves as our negative control compound. Se2SAP is a synthetic, core-modified, expanded porphyrin that was first synthesized in our laboratory with the specific aim of targeting a subset of G-quadruplex structures.41 Telomestatin is a natural product,51 which has been shown to inhibit telomerase through its strong G-quadruplex interaction with the human telomeric sequence.52 DNA polymerase stop assays were preformed with this series of compounds together with the three individual G-quadruplex-forming sequences (5′G4, MidG4, and 3′G4) to test whether selective stabilization of specific G-quadruplexes was achievable (Figure 7). Surprisingly, the results revealed that two of the compounds (TMPyP4 and Se2SAP) did have a sequence or structural selectiveness for the different G-quadruplexes. The negative control, TMPyP2, was unable to stabilize any of the three G-quadruplex structures, whereas telomestatin had the ability to interact quite strongly with all sequences, increasing the arrest sites 4- to 6-fold above the no-drug control at the maximum concentration tested. In addition, telomestatin induced the formation of another arrest site (2 in lower-panel gel of Figure 7) or G-quadruplex structure in the 3′G4 sequence. TMPyP4 (Figure 3B) and Se2SAP (data not shown) intensified all three arrest sites when employing the Pu39WT sequence, yet when examining their binding to the individual G-quadruplexes they were selective to the 3′G4 and MidG4 sequences, respectively. These results suggest that it is possible to selectively target with different G-quadruplex-interactive molecules the three constitutive G-quadruplexes within the bcl-2 promoter, which could lead to different biological outcomes.
Figure 7.
Stabilization of the three individual G-quadruplex structures formed by the 5′G4, MidG4, and 3′G4 sequences with the addition of increasing concentrations of TMPyP2 (0.1, 0.25, 0.5, 1, and 2.5 µM), TMPyP4 (0.1, 0.25, 0.5, 1, and 2.5 µM), Se2SAP (0.05, 0.1, 0.25, 0.5, and 1 µM), and telomestatin (0.05, 0.1, 0.25, 0.5, and 1 µM) in the presence of 20 mM KCl. Quantification of autoradiograms is shown in right panels.
DISCUSSION
Deregulation of the bcl-2 proto-oncogene is responsible for neoplastic cell growth in many types of cancer. In addition, overproduction of the bcl-2 protein has been associated with a poor response to chemotherapy in several tumors, due to its role in blocking drug-induced apoptosis.53 As a consequence of these properties, the interest in blocking bcl-2 activity has strongly increased in recent years, leading to multiple approaches to discover selective bcl-2 inhibitors. Two current strategies include the use of antisense oligonucleotides54 or peptidomimetics55 and small molecules, both of which recognize the hydrophobic surface pocket of the bcl-2 protein and interfere with its protein–protein interactions.56,57 Recently, ABT-737, a small molecule inhibitor of bcl-2 protein–protein interactions was identified using NMR-based screening and structure-based design, which exhibits activity alone and in combination with chemotherapy or radiation in vivo.57
There is accumulating in vitro evidence of the importance of G-quadruplex structures in promoter regions; however, direct confirmation for the existence of these G-quadruplexes in vivo is still to be obtained. While a bioinformatics search of the human genome for the simplest G-quadruplex-forming motif reveals an impressive number of possible G-quadruplex structures,58 a search for the more complex motifs actually found in gene promoter regions has not been performed. To date, the presence of G-quadruplex structures functioning as transcriptional regulators has been implicated in at least three genes. The most well-studied example is the specific intramolecular G-quadruplex structure formed in the G-rich region upstream of the human c-myc gene promoter, which has been shown to be stabilized by G-quadruplex-interactive agents and to act as a c-myc transcriptional repressor.37,38 In contrast, the presence of G-quadruplex structures within the insulin-linked polymorphic region of the human insulin gene has been associated with high gene transcription, specifically functioning as enhancer elements.59,60 Finally, and most recently, the promoter and enhancer regions of several muscle-specific genes have been shown to adopt multiple DNA secondary structures in vitro, including bimolecular G-quadruplexes.61,62 This sequence motif is not only exclusive to the examples mentioned, but has been found in the upstream promoter regions of several other cellular genes (see ref. 38 for a partial list). With the exception of telomeres, all of these sequence motifs are in duplex regions of the genome. Thus, a major limitation to the formation of G-quadruplexes is the stability of duplex DNA. However, unwinding of DNA and the formation of single-stranded tracts during transcription or replication could provide the necessary conditions for the formation of G-quadruplexes.63–65
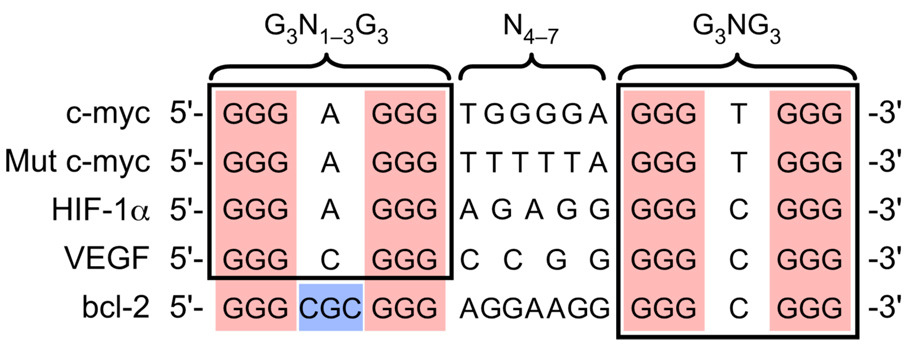
In this study we explored a candidate G-quadruplex-forming sequence located in the proximity of a nuclease hypersensitive element within the promoter region of the human bcl-2 gene that may possibly function as a regulator of bcl-2 transcription. We demonstrate that the G-rich strand within this region, which contains seven runs of two or more contiguous guanines (Figure 1), has the propensity to form three individual intramolecular G-quadruplex structures (Figure 3 and Figure 5). Isolation of the three individual G-quadruplex-forming sequences within this G-rich region revealed that the structure formed from the central four runs of guanines was the most stable (Figure 5B′). Comparison of this bcl-2 core sequence with an NMR-determined G-quadruplex structure of a mutant c-myc promoter sequence,66 as well as with the wild-type c-myc promoter sequence and two other promoter sequences with the potential to form G-quadruplexes, HIF-1α and VEGF (papers submitted for publication), revealed an apparent sequence similarity (Figure 8). However, one notable difference is observed in the 5′-end loop region, wherein the bcl-2 sequence contains three nucleotides instead of one. The mutant c-myc sequence (Figure 8) has been shown to produce a parallel G-quadruplex structure containing three double-chain-reversal loops of 1, 6, and 1 nucleotide in length (“a” in Figure 1B).66 DMS footprinting (Figure 5B″) and CD spectroscopy (Figure 5D) suggests that the core bcl-2 sequence forms a mixed parallel/antiparallel G-quadruplex similar to that of the Na+-stabilized Tetrahymena telomeric sequence, consisting of three G-tetrads, one double-chain-reversal loop of one nucleotide in size, and two lateral loops comprising three and seven nucleotides (“c” in Figure 1B). This difference in folding pattern between the bcl-2 sequence and the c-myc mutant sequence has been determined by NMR50 and is likely caused by the variation in the number of nucleotides present in the 5′-end loop region. In addition, it is important to note that the 3′-ends of all five sequences in Figure 8 are composed of an identical motif (G3NG3) capable of forming a single nucleotide double-chain-reversal loop, which may serve as a stable foundation for the formation of each G-quadruplex structure. The commonality of the G3NG3 motif extends also to two of the three constitutive G-quadruplexes in the bcl-2 promoter (Table 1). In the MidG4, the most stable mutant (MidG4-mut1) has a G5NG3 at the 3′-end, which is presumably common to the 3′G4 (see DMS footprinting in Figure 5C″). The 5′G4 quadruplex is the least stable of the three constituent bcl-2 promoter structures, and although it contains this motif at the 5′-end, DMS footprinting (Figure 5A″) shows incomplete protection of the appropriate guanines.
Figure 8.
Comparison of truncated G-quadruplex-forming sequences within selected gene promoters.
Comparison of putative G-quadruplex-forming sequences reveals variable degrees of complexity based on sequence composition. Both the bcl-2 and c-myc G-quadruplex-forming sequences are composed of more than four G-tracts and are capable of forming multiple G-quadruplex structures, based on the assumption that each G-tract has an equal probability of participating in G-quadruplex formation. Specifically, the wild-type c-myc promoter sequence is composed of five G-tracts and theoretically has the potential to form five different G-quadruplex structures, based on the number of combinations of four G-tracts; however, nondenaturing gel analysis shows only two intramolecular species.37 These are most likely two distinct parallel intramolecular G-quadruplex structures that can exist within the c-myc promoter sequence. The first predominant structure determined by NMR involves three G-tetrads, two single nucleotide double-chain-reversal loops, and a central double-chain-reversal loop containing two nucleotides utilizing the four 3′-end G-tracts.66 Even though the minor species, which utilizes the two 5′- and 3′-end G-tracts, has not been shown by NMR to be a similar all-parallel structure, a mutant (Mut c-myc in Figure 8) in which the middle G-tract is substituted by a T-tract does form a parallel structure with three double-chain reversal loops.66 When an additional G-tract is present, such as in the bcl-2 promoter sequence, the number of combinations of four G-tracts significantly increases to fifteen and continues to rapidly increase with more G-tracts. This concept of multiple combinations of four G-tracts within a given sequence represents the first level of complexity within G-quadruplex-forming sequences. A second level of complexity occurs when a G-quadruplex-forming sequence is composed of G-tracts containing unequal numbers of guanines. This is represented by the bcl-2 MidG4 sequence in Figure 6, in which an individual G-quadruplex structure has the ability to form three different loop isomers. Similarly, the G-quadruplex structure formed by the four 3′-end G-tracts in the c-myc promoter sequence has been shown to exist as a mixture of four different loop isomers.67 Therefore, besides the choice of G-tracts, there is a option among the guanines within a given G-tract that ultimately leads to the complexity found in most G-quadruplex-forming sequences. These layers of complexity for the G-quadruplex-forming regions in the bcl-2 promoter are illustrated in Figure 9. This unexpected degree of complexity is probably important from at least two perspectives. First, it gives rise to an enhanced degree of stability, and second, it provides alternative G-quadruplex structures and duplex regions for differential binding of proteins and drugs. Intriguingly, in both the c-myc and bcl-2 promoter regions in which multiple runs of guanines occur, there is only one predominant G-quadruplex species in each case, i.e. the parallel 1:2:1 loop isomer for c-MYC and the MidG4-mut1 sequence for bcl-2.
Figure 9.
Proposed equilibrium between the multiple G-quadruplex structures formed within the bcl-2 promoter region (red = anti conformation; green = syn conformation). The G-quadruplex folding pattern is as determined by an NMR study.40
The inability to selectively target a specific G-quadruplex structure was one of the initial criticisms of G-quadruplexes as drug targets. However, a number of studies have established that small molecules can be designed that have varying degrees of selectively to different G-quadruplex structures.41,68–70 For example, cationic porphyrins and sapphyrins have been shown to bind to and stabilize a number of different types of G-quadruplexes.41,44,71,72 Specifically, the cationic porphyrin and sapphyrin used in this study, TMPyP4 and Se2SAP, have been shown to have variable selectivity for G-quadruplex structures. TMPyP4 is less discriminating compared to Se2SAP, which has been shown to specifically stabilize one of the c-myc G-quadruplexes,41 as well as the K+ form of the human telomeric G-quadruplex.71 In addition, TMPyP4 and Se2SAP at low salt concentrations have been shown to convert the c-myc parallel G-quadruplex to a double- or single-loop hybrid G-quadruplex structure, respectively. In this study these two cationic ligands demonstrated selective stabilization of two of the three G-quadruplex structures formed within the bcl-2 promoter (Figure 7); however, whether similar drug-induced structural conversion occurs has yet to be determined.
Although the biological relevance of this G-quadruplex-forming region upstream of the bcl-2 P1 promoter is unknown, we speculate that it may function to modulate bcl-2 transcription. Depending on which of the three G-quadruplex structures is present in vivo, this will provide different binding sites for transcriptional factors. Targeting these preexisting paranemic secondary structures separately with selective small molecules may result in variable biological consequences. Studies are ongoing to investigate the effect of G-quadruplex-interactive agents on bcl-2 gene transcription.
Overall, we provide in vitro evidence for the formation of three separate intramolecular G-quadruplex structures in the proximal P1 promoter region of the human bcl-2 gene, which are selectively stabilized by different G-quadruplex-interactive agents. Our results also reveal another proto-oncogene, besides c-myc, that may possibly be regulated by the presence of G-quadruplex structures within its promoter. This study strengthens the idea that G-quadruplexes may play a role in biological processes and that selective targeting of individual G-quadruplex structures is achievable.
ACKNOWLEDGEMENTS
Research was supported by the National Institutes of Health (CA94166 and CA09213). We thank Kazuo Shin-ya (University of Tokyo) for providing the telomestatin for this study and Edwin Lewis and his lab for oligonucleotide extinction coefficient calculations. We are grateful to David Bishop for preparing, proofreading, and editing the final version of the manuscript and figures.
REFERENCES
- 1.Chao DT, Korsmeyer SJ. Annu. Rev. Immunol. 1998;16:395–419. doi: 10.1146/annurev.immunol.16.1.395. [DOI] [PubMed] [Google Scholar]
- 2.Adams JM, Cory S. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 3.Yunis JJ. Science. 1983;221:227–236. doi: 10.1126/science.6336310. [DOI] [PubMed] [Google Scholar]
- 4.Cleary ML, Smith SD, Sklar J. Cell. 1986;47:19–28. doi: 10.1016/0092-8674(86)90362-4. [DOI] [PubMed] [Google Scholar]
- 5.Akagi T, Kondo E, Yoshino T. Leuk. Lymphoma. 1994;13:81–87. doi: 10.3109/10428199409051655. [DOI] [PubMed] [Google Scholar]
- 6.Joensuu H, Pylkkanen L, Toikkanen S. Am. J. Pathol. 1994;145:1191–1198. [PMC free article] [PubMed] [Google Scholar]
- 7.Tjalma W, De Cuyper E, Weyler J, Van Marck E, De Pooter C, Albertyn G, van Dam P. Am. J. Obstet. Gynecol. 1998;178:113–117. doi: 10.1016/s0002-9378(98)70636-2. [DOI] [PubMed] [Google Scholar]
- 8.Pezzella F, Turley H, Kuzu I, Tungekar MF, Dunnill MS, Pierce CB, Harris A, Gatter KC, Mason DY. N. Engl. J. Med. 1993;329:690–694. doi: 10.1056/NEJM199309023291003. [DOI] [PubMed] [Google Scholar]
- 9.McDonnell TJ, Troncoso P, Brisbay SM, Logothetis C, Chung LW, Hsieh JT, Tu SM, Campbell ML. Cancer Res. 1992;52:6940–6944. [PubMed] [Google Scholar]
- 10.Baretton GB, Diebold J, Christoforis G, Vogt M, Müller C, Dopfer K, Schneiderbanger K, Schmidt M, Löhrs U. Cancer. 1996;77:255–264. doi: 10.1002/(SICI)1097-0142(19960115)77:2<255::AID-CNCR6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 11.Reed JC, Kitada S, Takayama S, Miyashita T. Ann. Oncol. 1994;5 Suppl. 1:61–65. doi: 10.1093/annonc/5.suppl_1.s61. [DOI] [PubMed] [Google Scholar]
- 12.Middleton G, Nunez G, Davies AM. Development. 1996;122:695–701. doi: 10.1242/dev.122.2.695. [DOI] [PubMed] [Google Scholar]
- 13.Allsopp TE, Wyatt S, Paterson HF, Davies AM. Cell. 1993;73:295–307. doi: 10.1016/0092-8674(93)90230-n. [DOI] [PubMed] [Google Scholar]
- 14.Farlie PG, Dringen R, Rees SM, Kannourakis G, Bernard O. Proc Natl Acad Sci U S A. 1995;92:4397–4401. doi: 10.1073/pnas.92.10.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubois-Dauphin M, Frankowski H, Tsujimoto Y, Huarte J, Martinou JC. Proc Natl Acad Sci U S A. 1994;91:3309–3313. doi: 10.1073/pnas.91.8.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michaelidis TM, Sendtner M, Cooper JD, Airaksinen MS, Holtmann B, Meyer M, Thoenen H. Neuron. 1996;17:75–89. doi: 10.1016/s0896-6273(00)80282-2. [DOI] [PubMed] [Google Scholar]
- 17.Linnik MD, Zahos P, Geschwind MD, Federoff HJ. Stroke. 1995;26:1670–1674. doi: 10.1161/01.str.26.9.1670. discussion 1675. [DOI] [PubMed] [Google Scholar]
- 18.Shimazaki K, Urabe M, Monahan J, Ozawa K, Kawai N. Gene Ther. 2000;7:1244–1249. doi: 10.1038/sj.gt.3301211. [DOI] [PubMed] [Google Scholar]
- 19.Shacka JJ, Roth KA. Curr Drug Targets CNS Neurol Disord. 2005;4:25–39. doi: 10.2174/1568007053005127. [DOI] [PubMed] [Google Scholar]
- 20.Seto M, Jaeger U, Hockett RD, Graninger W, Bennett S, Goldman P, Korsmeyer SJ. EMBO J. 1988;7:123–131. doi: 10.1002/j.1460-2075.1988.tb02791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsujimoto Y, Croce CM. Proc. Natl. Acad. Sci. USA. 1986;83:5214–5218. doi: 10.1073/pnas.83.14.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson BE, Mochon E, Boxer LM. Mol. Cell. Biol. 1996;16:5546–5556. doi: 10.1128/mcb.16.10.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji L, Mochon E, Arcinas M, Boxer LM. J. Biol. Chem. 1996;271:22687–22691. doi: 10.1074/jbc.271.37.22687. [DOI] [PubMed] [Google Scholar]
- 24.Heckman CA, Mehew JW, Boxer LM. Oncogene. 2002;21:3898–3908. doi: 10.1038/sj.onc.1205483. [DOI] [PubMed] [Google Scholar]
- 25.Heckman C, Mochon E, Arcinas M, Boxer LM. J. Biol. Chem. 1997;272:19609–19614. doi: 10.1074/jbc.272.31.19609. [DOI] [PubMed] [Google Scholar]
- 26.Hewitt SM, Hamada S, McDonnell TJ, Rauscher FJ, 3rd, Saunders GF. Cancer Res. 1995;55:5386–5389. [PubMed] [Google Scholar]
- 27.Heckman CA, Wheeler MA, Boxer LM. Oncogene. 2003;22:7891–7899. doi: 10.1038/sj.onc.1206639. [DOI] [PubMed] [Google Scholar]
- 28.Heckman CA, Mehew JW, Ying GG, Introna M, Golay J, Boxer LM. J. Biol. Chem. 2000;275:6499–6508. doi: 10.1074/jbc.275.9.6499. [DOI] [PubMed] [Google Scholar]
- 29.Miyashita T, Harigai M, Hanada M, Reed JC. Cancer Res. 1994;54:3131–3135. [PubMed] [Google Scholar]
- 30.Gomez-Manzano C, Mitlianga P, Fueyo J, Lee H-Y, Hu M, Spurgers KB, Glass TL, Koul D, Liu T-J, McDonnell TJ, Yung WK. Cancer Res. 2001;61:6693–6697. [PubMed] [Google Scholar]
- 31.Liu YZ, Boxer LM, Latchman DS. Nucleic Acids Res. 1999;27:2086–2090. doi: 10.1093/nar/27.10.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young RL, Korsmeyer SJ. Mol. Cell. Biol. 1993;13:3686–3697. doi: 10.1128/mcb.13.6.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watson JD, Crick FH. Cold Spring Harb. Symp. Quant. Biol. 1953;18:123–131. doi: 10.1101/sqb.1953.018.01.020. [DOI] [PubMed] [Google Scholar]
- 34.Hurley LH, Wheelhouse RT, Sun D, Kerwin SM, Salazar M, Fedoroff OYu, Han FX, Han H, Izbicka E, Von Hoff DD. Pharmacol. Ther. 2000;85:141–158. doi: 10.1016/s0163-7258(99)00068-6. [DOI] [PubMed] [Google Scholar]
- 35.Blackburn EH. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 36.Sen D, Gilbert W. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 37.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Proc. Natl. Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simonsson T, Pecinka P, Kubista M. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cantor CR, Warshaw MM, Shapiro H. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 40.Sun XG, Cao EH, He YJ, Qin JF. J. Biomol. Struct. Dyn. 1999;16:863–872. doi: 10.1080/07391102.1999.10508298. [DOI] [PubMed] [Google Scholar]
- 41.Seenisamy J, Bashyam S, Gokhale V, Vankayalapati H, Sun D, Siddiqui-Jain A, Streiner N, Shin-ya K, White E, Wilson WD, Hurley LH. J. Am. Chem. Soc. 2005;127:2944–2959. doi: 10.1021/ja0444482. [DOI] [PubMed] [Google Scholar]
- 42.Weitzmann MN, Woodford KJ, Usdin K. J. Biol. Chem. 1996;271:20958–20964. doi: 10.1074/jbc.271.34.20958. [DOI] [PubMed] [Google Scholar]
- 43.Han H, Hurley LH, Salazar M. Nucleic Acids Res. 1999;27:537–542. doi: 10.1093/nar/27.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han H, Langley DR, Rangan A, Hurley LH. J. Am. Chem. Soc. 2001;123:8902–8913. doi: 10.1021/ja002179j. [DOI] [PubMed] [Google Scholar]
- 45.Dapic V, Abdomerovic V, Marrington R, Peberdy J, Rodger A, Trent JO, Bates PJ. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petraccone L, Erra E, Esposito V, Randazzo A, Mayol L, Nasti L, Barone G, Giancola C. Biochemistry. 2004;43:4877–4884. doi: 10.1021/bi0300985. [DOI] [PubMed] [Google Scholar]
- 47.Miyoshi D, Nakao A, Sugimoto N. Nucleic Acids Res. 2003;31:1156–1163. doi: 10.1093/nar/gkg211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hazel P, Huppert J, Balasubramanian S, Neidle S. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Patel DJ. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 50.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. J. Am. Chem. Soc. 2005;127:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin-ya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, Hayakawa Y, Seto H. J. Am. Chem. Soc. 2001;123:1262–1263. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 52.Kim M-Y, Vankayalapati H, Shin-ya K, Wierzba K, Hurley LH. J. Am. Chem. Soc. 2002;124:2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 53.Desoize B. Anticancer Res. 1994;14:2291–2294. [PubMed] [Google Scholar]
- 54.Klasa RJ, Gillum AM, Klem RE, Frankel SR. Antisense Nucleic Acid Drug Dev. 2002;12:193–213. doi: 10.1089/108729002760220798. [DOI] [PubMed] [Google Scholar]
- 55.Tzung SP, Kim KM, Basanez G, Giedt CD, Simon J, Zimmerberg J, Zhang KY, Hockenbery DM. Nat. Cell. Biol. 2001;3:183–191. doi: 10.1038/35055095. [DOI] [PubMed] [Google Scholar]
- 56.Enyedy IJ, Ling Y, Nacro K, Tomita Y, Wu X, Cao Y, Guo R, Li B, Zhu X, Huang Y, Long YQ, Roller PP, Yang D, Wang S. J. Med. Chem. 2001;44:4313–4324. doi: 10.1021/jm010016f. [DOI] [PubMed] [Google Scholar]
- 57.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM, O’Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 58.Todd AK, Johnston M, Neidle S. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hammond-Kosack MC, Dobrinski B, Lurz R, Docherty K, Kilpatrick MW. Nucleic Acids Res. 1992;20:231–236. doi: 10.1093/nar/20.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lew A, Rutter WJ, Kennedy GC. Proc. Natl. Acad. Sci. USA. 2000;97:12508–12512. doi: 10.1073/pnas.97.23.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yafe A, Etzioni S, Weisman-Shomer P, Fry M. Nucleic Acids Res. 2005;33:2887–2900. doi: 10.1093/nar/gki606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Etzioni S, Yafe A, Khateb S, Weisman-Shomer P, Bengal E, Fry M. J. Biol. Chem. 2005;29:26805–26812. doi: 10.1074/jbc.M500820200. [DOI] [PubMed] [Google Scholar]
- 63.Leonard MW, Patient RK. Mol. Cell Biol. 1991;11:6128–6138. doi: 10.1128/mcb.11.12.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pan G, Greenblatt J. J. Biol. Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 65.Liang CP, Garrard WT. Mol. Cell Biol. 1997;17:2825–2834. doi: 10.1128/mcb.17.5.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Phan AT, Modi YS, Patel DJ. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seenisamy J, Rezler EM, Powell TJ, Tye D, Gokhale V, Joshi CS, Siddiqui-Jain A, Hurley LH. J. Am. Chem. Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 68.Teulade-Fichou MP, Carrasco C, Guittat L, Bailly C, Alberti P, Mergny JL, David A, Lehn JM, Wilson WD. J. Am. Chem. Soc. 2003;125:4732–4740. doi: 10.1021/ja021299j. [DOI] [PubMed] [Google Scholar]
- 69.Harrison RJ, Cuesta J, Chessari G, Read MA, Basra SK, Reszka AP, Morrell J, Gowan SM, Incles CM, Tanious FA, Wilson WD, Kelland LR, Neidle S. J. Med. Chem. 2003;46:4463–4476. doi: 10.1021/jm0308693. [DOI] [PubMed] [Google Scholar]
- 70.Perry PJ, Jenkins TC. Mini Rev. Med. Chem. 2001;1:31–41. doi: 10.2174/1389557013407304. [DOI] [PubMed] [Google Scholar]
- 71.Rezler EM, Seenisamy J, Bashyam S, Kim M-Y, White E, Wilson WD, Hurley LH. J. Am. Chem. Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- 72.Dixon IM, Lopez F, Esteve JP, Tejera AM, Blasco MA, Pratviel G, Meunier B. ChemBioChem. 2005;6:123–132. doi: 10.1002/cbic.200400113. [DOI] [PubMed] [Google Scholar]