Abstract
The pseuoduridine synthases (Ψ synthases) isomerize uridine (U) to pseudouridine (Ψ) in RNA, and they fall into five families that share very limited sequence similarity but have the same overall fold and active site architecture, including an essential Asp. The mechanism by which the Ψ synthases operate remains unknown, and mechanistic work has largely made use of RNA containing 5-fluorouridine (f5U) in place of U. The Ψ synthase TruA forms a covalent adduct with such RNA, and heat disruption of the adduct generates a hydrated product of f5U, which was reasonably concluded to result from the hydrolysis of an ester linkage between the essential Asp and f5U. In contrast, the Ψ synthase TruB, which is a member of a different family, does not form an adduct with f5U in RNA but catalyzes the rearrangement and hydration of the f5U, which labeling studies with [18O]water showed does not result from ester hydrolysis. To extend the line of mechanistic investigation to another family of Ψ synthases and an enzyme that makes an adduct with f5U in RNA, the behavior of RluA towards RNA containing f5U was examined. Stem-loop RNAs are shown to be good substrates for RluA. Heat denaturation of the adduct between RluA and RNA containing f5U produces a hydrated nucleoside product, and labeling studies show that hydration does not occur by ester hydrolysis. These results are interpreted in light of a consistent mechanistic scheme for the handling of f5U by Ψ synthases.
Pseudouridine (Ψ) is the most common modified nucleoside in RNA, and it is found in non-messenger RNA in all organisms (1). The pseudouridine synthases (Ψ synthases) generate Ψ by the post-transcriptional isomerization of U in RNA. These enzymes fall into five families that retain no statistically significant global sequence similarity (2–5) and are typically named for the first member identified in Escherichia coli: TruA1 (6), TruB (7), RluA (8), RsuA (9), and TruD (4). Crystal structures of members of all five families (10–21) reveal that they share the same fold with a core β-sheet and therefore likely arose by divergent evolution (22). An active site Asp is the sole amino acid residue conserved among all Ψ synthases (23), and it is essential for catalytic activity in all five families (4, 23–26).
The widespread occurrence of Ψ testifies to its importance, and several particular Ψ residues are physiologically essential. The lack of Ψ1911, Ψ1915, and Ψ1917 in 23S rRNA leads to a sharply decreased growth rate in E. coli (25). The loss of function by homologs of a nuclear Ψ synthase in yeast (Cbf5p), drosophila (mfl), rats (NAP57), and humans (dyskerin) leads to phenotypes ranging from severe physiological impairment to lethality (27–31). U2 and U6 snRNA require the presence of Ψ for the spliceosome to assemble and subsequently splice pre-mRNA in eukaryotes (32–34).
The experiments demonstrating the importance of Ψ in spliceosome assembly relied on inhibition of the responsible Ψ synthase(s) by in vitro transcripts of U2 and U6 snRNA that contained 5-fluorouridine (f5U) in place of U ([f5U]RNA). The use of [f5U]RNA was based on the reasonable assumption that Ψ synthases follow a mechanism involving the Michael addition of an enzymic nucleophile at C6 of U (Figure 1, panel A), which was derived from the similar mechanism of pyrimidine methylases typified by thymidylate synthase (35). In the methylases, the enzymic nucleophile is the thiol group of an active site Cys, but no Cys are conserved among the Ψ synthases, even within families (2, 3). In representatives of three families that have been examined, all Cys can be replaced with Ala without a significant effect on activity (36, 37). It was therefore proposed that the essential Asp acts as a nucleophile in the Michael addition (38) rather than attacking C1’ to form an acylal intermediate (Figure 1, panel B), which had been proposed earlier (23). The pH dependence of the Ψ synthase TruB (39) and consideration of the conserved acid/base groups in the Ψ synthase active sites (22) led us to propose that the catalytic Asp functions as a general base as well as a nucleophile. Recently reported site-directed mutagenesis studies led to an alternate proposal that Tyr-76 of TruB serves as the general base (40). We conclude that the results of those studies (40) and the pH dependence of TruB (39) are better accommodated by Tyr-76 serving as a general acid or hydrogen bond donor to assist glycosidic bond cleavage2, so the catalytic Asp remains depicted as the general base in Figure 1.
Figure 1.
Proposed mechanisms for the Ψ synthase reaction. A, the “Michael mechanism” in which the essential Asp nucleophilically adds to the pyrimidine ring. B, the “acylal” mechanism, in which the essential Asp nucleophilically adds to the ribose ring either by a concerted (upper path) or stepwise (lower path) process. The stereochemistry of the rearranged uridine is based on that observed in the cocrystal of TruB and [f5U]TSL (11).
Support for the participation of the catalytic Asp as a Michael nucleophile was provided by the inhibition of TruA upon the formation of a covalent adduct with [f5U]tRNA, as deduced from mobility shifts in denaturing gels (SDS-PAGE and urea-PAGE) and radiolabeling studies that indicated attachment of the enzyme to the pyrimidine rather than the ribose ring of f5U (38). After heat disruption of the adduct, mass spectrometry definitively revealed that f5U had been converted into a hydrated product (38), which was reasonably ascribed to ester hydrolysis of the Michael adduct between f5U and the catalytic Asp (Figure 2, panel A). In an attempt to capture the Michael adduct, cocrystals of TruB and T-arm stem-loop RNA3 containing f5U ([f5U]TSL) were grown and the structure determined (11). Surprisingly, a covalent adduct was not observed, and f5U had been rearranged to the C5-glycoside (as is U during its conversion to Ψ) and hydrated at C6. This product was quite reasonably ascribed to ester hydrolysis of the rearranged rather than the initial Michael adduct (Figure 2, panel B), a process that was assumed to occur slowly over the months required to grow crystals and acquire diffraction data.
Figure 2.
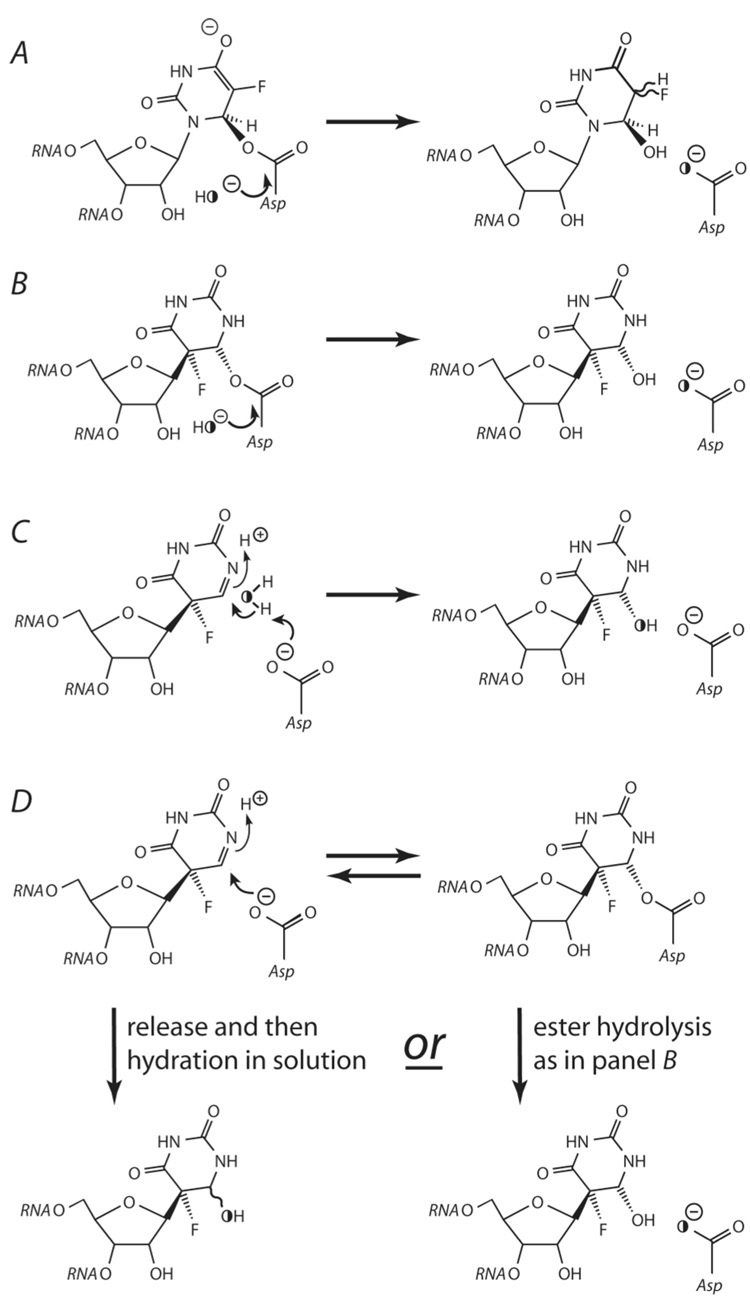
Possible reactions between Ψ synthases and f5U in RNA showing the predicted labeling in [18O]water (denoted by the half-filled O). A, the initial Michael adduct between f5U and the catalytic Asp undergoes ester hydrolysis. B, the ultimate Michael adduct between f5U and the catalytic Asp undergoes ester hydrolysis. C, the catalytic Asp acts as a general base in the direct hydration of rearranged f5U. D, the catalytic Asp adds to the rearranged f5U to make a covalent adduct, which can then suffer ester hydrolysis (downwards on the right) or elimination (reversal of the top reaction) followed by release into solution and spontaneous hydration (downwards on the left). Ester hydrolysis results in the labeling of the catalytic Asp (A, B, and right side of D), while direct hydration (C and left side of D) labels the nucleoside.
Subsequent biochemical investigations, however, demonstrated that [f5U]TSL does not potently inhibit TruB but is instead efficiently handled as a substrate to generate products of f5U that are hydrated and likely rearranged (41). Furthermore, labeling studies with [18O]water definitively showed that the oxygen atom added by hydration comes directly from solvent water rather than from the catalytic Asp as would be required by ester hydrolysis (42). Two schemes were offered to account for the direct hydration of rearranged f5U (Figure 2, panels C and D) by TruB, and it was proposed that the latter (Figure 2, panel D) scheme accounts for the disparate behavior of Ψ synthases towards [f5U]RNA: f5U is always rearranged by the same mechanism, either the “Michael mechanism” (Figure 1, panel A) or the “acylal mechanism” (Figure 1, panel B), and then the detailed geometry of the active site dictates whether or not the catalytic Asp lies near enough to the pyrimidine ring of the rearranged f5U to add at C6 and form a covalent adduct. An adduct forms in the case of TruA, indicating that the equilibrium lies in favor of the covalently bound rearranged f5U in Figure 2, panel D. Upon heat denaturation of the complex, the covalent adduct either undergoes ester hydrolysis (proceeding downwards on the right in Figure 2, panel D) or suffers elimination of Asp followed by spontaneous hydration in bulk solvent (proceeding to the left and then downwards in Figure 2, panel D). With TruB, either the Asp cannot efficiently reach C6 or the adduct is destabilized by some other interaction in the active site so that the equilibrium depicted in Figure 2, panel D strongly favors the noncovalent complex, which either undergoes hydration catalyzed by the Asp (Figure 2, panel C) or dissociates with spontaneous hydration of rearranged f5U in bulk solvent (proceeding to the left in Figure 2, panel D).
To examine the chemical fate of a covalent adduct between a Ψ synthase and [f5U]RNA, the 18O labeling studies have now been extended to RluA, which behaves very much like TruA with respect to [f5U]RNA (41). E. coli RluA isomerizes U32 in the anticodon loop of four tRNAs as well as U746 of 23S rRNA (Figure 3); the enzyme’s role as a ribosomal large subunit pseudouridine synthase gave rise to its name. The five physiological substrates differ in the stem region but have similar loop sequences, strongly suggesting that stem-loop RNAs will be good substrates for RluA. That prediction was borne out by the behavior of RluA towards anticodon stem-loop RNA containing f5U ([f5U]ASL): incubation of RluA with [f5U]ASL results in the stoichiometric inhibition of enzymatic activity and the formation of an adduct that appears to be covalent as judged by denaturing gel retardation analysis (41). The kinetic characterization of stem-loop RNA as a substrate for RluA and 18O labeling studies that probe the interaction of RluA with [f5U]ASL are presented here.
Figure 3.
Stem-loop structures in natural substrates for E. coli RluA that contain the isomerized uridine residue (underlined), U32 in the anticodon stem-loops (ASLs) of four tRNAs and U746 in a 23S rRNA stem-loop (RSL). The ASLs span residues 27–43 of tRNA, and RSL spans residues 737–760 of 23S rRNA.
Materials and Methods
General
Synthetic RNA oligonucleotides with 2’-ACE protecting groups were purchased from Dharmacon, Inc. (Lafayette, CO) and deprotected by acid-catalyzed hydrolysis according to the manufacturer’s instructions. To remove any residual buffer, the deprotected RNA was dissolved in RNase-free water (1 mL) and concentrated in a Centricon-3 centrifugal ultrafiltration device (final V = 100–200 µL); the retentate was diluted with RNase-free water (to 1 mL) and reconcentrated (to 100–200 µL), and the dilution/reconcentration sequence was repeated once more. The RNA oligonucleotides corresponded to the anticodon stem-loop (ASL) of E. coli tRNAPhe (GGGGAUUGAAAAUCCCC), ASL containing 5-fluorouridine in place of the U that is converted to Ψ ([f5U]ASL, GGGGAf5UUGAAAAUCCCC), and C737 to G760 of E. coli 23S rRNA (CCGCUAAUGUUGAAAAAUUAGCGG). Concentrations were determined using A260 nm and the extinction coefficient provided by the manufacturer (170,500 M−1cm−1 for ASL and [f5U]ASL and 246,000 M−1cm−1 for the ribosomal stem-loop).
Protein overexpression and purification were achieved as described previously (24, 37) using plasmids based on pET15b (Novagen). Tris•HCl was purchased from EMD Biosciences (San Diego, CA). Trypsin (sequencing grade) was purchased from Roche Applied Science (Indianapolis, IN). S1 nuclease was purchased from Promega (Madison, WI). Prime RNase inhibitor was purchased from Eppendorf through Fisher Scientific (Pittsburgh, PA). All other chemicals were purchased from Fisher Scientific or its Acros Organics division (Pittsburgh, PA). HPLC was performed with either a Beckman System Gold HPLC instrument equipped with a System 125 solvent module, a Rheodyne 7725i injector, and a Beckman 168 diode array detector or with an Agilent 1100 system composed of a binary pump, micro vacuum degasser, variable wavelength detector, and thermostatted autosampler with an extended volume upgrade kit. In general, the Beckman system was used with the Clipeus C18 column (see below) and the Agilent system was used with the Zorbax C18 column (see below).
HPLC Separation of ASL from Product and Assay of RluA Activity
Initial tests of ASL as a substrate for RluA and subsequent activity assays were performed under conditions similar to those with full length tRNA transcript (24, 37). The reaction mixture (500 µL) was 50 mM HEPES buffer, pH 7.5, containing ammonium chloride (100 mM), DTT (5 mM), EDTA (1 mM), RNase inhibitor (30 units), and ASL (350 – 3500 nM). After pre-equilibration at 37 °C, reactions were initiated by the addition of RluA (typically 1 µL, 50 nM final concentration). Aliquots (75 µL) were removed at various times and immediately added to quench solution (25 µL), which was 0.5 M sodium phosphate buffer, pH 7.5, containing sodium chloride (0.5 M).
The quenched aliquots were passed over G-25 spin columns that had been pre-equilibrated with 125 mM sodium phosphate buffer, pH 7.5, containing sodium chloride (125 mM). The resulting sample was passed through a 0.2 µm filter and analyzed by HPLC over a C18 column (CLIPEUS C18 5 µm column, 150 × 4.6 mm, Higgins Analytical, Mountain View, CA). RNA was eluted with an acetonitrile gradient using the following program (the first number is the percentage of aqueous acetonitrile, 40% vol/vol, in 25 mM ammonium acetate buffer, pH 6.0; the second number is the elapsed time): 0, 0; 0, 3; 5, 8; 15, 18; 15, 23; 30, 26; 50, 27.5; 50, 29; 100, 30; 100, 31; 0, 32; 0, 36. A concave gradient was used from 0–5%; all other gradient steps were linear.
The peaks in the HPLC traces were integrated to determine the fraction of product and substrate RNA and hence their concentrations, since the total RNA concentration was known. Assays at each RNA concentration were completed in duplicate or triplicate, and the results varied less than 10% between determinations. KaleidaGraph 3.6 (Synergy Software, Reading, PA) was used to plot the variance in initial velocities with respect to the substrate concentration and fit the data to the Briggs-Haldane equation.
Digestion of RNA and Separation of Nucleosides
The digestion of RNA was accomplished by the method of Gehrke et al. (43) and the HPLC analysis of the resulting nucleotides by the method of Buck et al. (44) as described previously by us (45) with slight variation. RNA solutions that contained RluA were heated to 100 °C and cooled on ice; the protein precipitate was pelleted by centrifugation, and the supernatant was transferred to a fresh tube. RNA was diluted into 50 mM sodium acetate buffer, pH 4.5, containing sodium chloride (280 mM) and zinc chloride (4.5 mM). S1 nuclease4 (100 units) was added; after 1 h at 37 °C, the reaction mixture was heated to 100 °C for 5 min and then cooled on ice. Additional S1 nuclease (100 units) was added, and the reaction mixture was incubated for 1 h at 37 °C. For most ASL and [f5U]ASL digests, the reaction was diluted into 300 mM Tris buffer, pH 7.4, followed by addition of alkaline phosphatase (5 units) and incubated for 1 h at 37 °C to remove 3′ phosphate groups; to reduce the size of the void peak in HPLC runs, alkaline phosphatase treatments were sometimes performed without the addition of Tris, which had no effect on the efficacy of dephosphorylation in the incubation time.
Digested ASL was analyzed by HPLC over a C18 column (CLIPEUS C18 5 µm) using the same method described above for intact ASL. For digested [f5U]ASL, the HPLC analysis was performed using a Zorbax StableBond C18 column (150 × 4.6 mm, 5 µm; Agilent Technologies, Palo Alto, CA) using the same gradient of acetonitrile in 25 mM ammonium acetate buffer, pH 6.0, described for analysis of the intact stem-loop except that all gradient steps were linear.
Formation of the Adduct between RluA and [f5U]ASL
The adduct between RluA and [f5U]ASL was formed by incubating RluA (10 to 40 µM) with [f5U]ASL (20 to 80 µM) in the assay reaction buffer for 3 h at 37 °C. A time course was completed by incubating RluA (10 µM) with [f5U]ASL (50 µM) in the reaction buffer at 37 °C; aliquots (7.5 µL) were removed at various time points and quenched into an equal volume of 2× Laemmli sample buffer. SDS-PAGE analysis (10% gel) verified the presence of adduct, which was indicated by a shift to a higher apparent molecular weight. RNA recovered from the supernatant after heat disruption of the adduct was analyzed by HPLC over a C18 column (as described above for RluA activity assays with ASL) to confirm that [f5U]ASL had been modified.
Mass Spectrometry
Solutions of RluA or the adduct between RluA and [f5U]ASL were incubated for 5 min at 100 °C to denature the RluA. The protein precipitate was pelleted by centrifugation, and the supernatant was found to contain <1% of the protein as determined by absorbance at 280 nm. The protein pellet was suspended in a solution (100 µL) of trypsin (0.02 mg/mL; 1:50 trypsin:RluA by weight) in 25 mM ammonium bicarbonate buffer, pH 8. The tube containing the digestion mixture was taped flat on the face of a drum and slowly rotated overnight at 37 °C, during which time all of the precipitate dissolved. The tryptic peptides were cleaned and concentrated by loading the digestion mixture onto a ZipTipC18 pipetter tip (Millipore, Bedford, MA) and washing (10 × 10 µL) with aqueous trifluoroacetic acid (0.1% vol/vol). Samples were spotted directly on the MALDI plate by eluting the peptides with matrix solution, which is saturated α-cyano-4-hydroxycinnamic acid in 50% aqueous acetonitrile containing trifluoroacetic acid (0.1%). ESI MS/MS sequencing was performed as described previously (39, 42). RNA (5 µM, 20 µL) was digested by adding RNase A (0.4 µg) or RNase T1 (100 units) followed by incubation for 1 h at 37 °C. For experiments with [18O]water, alkaline phosphatase (2 units) was added after the RNase digestion, and the sample was incubated an additional 1 h at 37 °C. The RNA digestion products were cleaned and concentrated on a ZipTipC18 using a protocol designed for oligonucleotides provided by the manufacturer. Briefly, the crude sample was loaded on the tip and washed (5 × 10 µL) first with 0.1 M triethylammonium acetate buffer, pH 7.0, and then with RNase free water (5 × 10 µL). Samples were eluted and spotted directly on the MALDI plate with matrix solution, which was saturated 3-hydroxypicolinic acid in 70% aqueous acetonitrile containing trifluoroacetic acid (0.1%). Mass spectra were acquired with a Bruker Omniflex or Biflex III MALDI-TOF mass spectrometer as described previously (41, 42). The complete mass spectrum of RluA gave 66% sequence coverage, and all of the expected products larger than dinucleotides from the RNase digestions of stem-loop RNA were observed.
Controls for Oxygen Exchange with Solvent
A number of controls were run to insure that oxygen atoms did not exchange between solvent and either Asp-64 or the hydrated product of f5U at any time during the work-up and analysis. To test for exchange between Asp-64 and solvent in native RluA, a sample (30 µM, 60 µL) of RluA was incubated in storage buffer (in which RluA is fully active) containing [18O]water (50%); after 3 h at 37 °C, the incubation mixture was heated to 100 °C for 5 min to precipitate the protein. The precipitated RluA was pelleted by centrifugation and gently washed with 25 mM ammonium bicarbonate buffer, pH 8.0, to remove any residual buffer containing [18O]water and pelleted again by centrifugation. The RluA pellet was resuspended in 25 mM ammonium bicarbonate buffer (50 µL), pH 8.0, digested overnight with trypsin (1 µg), and analyzed by MALDI-MS. To test for incorporation of 18O into the tryptic peptides of RluA, they were incubated in buffer containing [18O]water (50%) at 37 °C for 3 h and analyzed by MALDI-MS. To test for exchange of 18O into the hydrated product of f5U, the oligonucleotides from the digestion of modified [f5U]ASL with RNase T1 or RNase A were incubated in buffer containing [18O]water (50%) and allowed to incubate for 2 h at 22 °C and analyzed by MALDI-MS To test for exchange of 18O into Asp-64 or the hydrated product of f5U during the preparation or analysis of MALDI-MS samples, matrix solutions made with [18O]water (50%) were used. To test for exchange between solvent and the hydrated product of f5U during the RNase digestions, modified [f5U]ASL was digested with RNase and treated with alkaline phosphatase in buffer containing [18O]water (50%). To show that 18O incorporation into the tryptic peptide from RluA could be observed, RluA was digested with trypsin as described above except the buffer contained [18O]water (50%); the peptides were then analyzed by MALDI-MS.
Test for Ester Hydrolysis using [18O]water
An adduct formation reaction (200 µL) was run with and under the same conditions described above, except that the buffer contained [18O]water (50%). To allow exchange of any unlabeled bound water, RluA (40 µM) was pre-incubated for 5 min before [f5U]ASL (to 20 µM) was added to initiate adduct formation; after 3 h at 37 °C, adduct formation was confirmed by SDS-PAGE analysis of an aliquot (5 µL) of the reaction mixture. The adduct was disrupted by heating the remainder of the sample to 100 °C for 5 min, and precipitated RluA was removed by centrifugation. The complete modification of [f5U]ASL was verified by HPLC analysis of an aliquot (20 µL) of the supernatant. To insure the complete removal of residual [18O]water, the pelleted RluA was gently washed with 25 mM ammonium bicarbonate buffer, pH 8.0, and re-pelleted. The supernatant was removed, the RluA digested with trypsin, and the resulting peptides analyzed by MALDI-MS. Protein was phenol-extracted from the original supernatant containing the modified [f5U]ASL, and the aqueous phase was washed with chloroform:isoamyl alcohol (24:1); the modified [f5U]ASL was precipitated by the addition of 0.1 volume of 3 M sodium acetate buffer, pH 5.2, and then 3 volumes of absolute ethanol. After at least 2 h at −20 °C, the RNA was pelleted by centrifugation and washed twice with cold aqueous ethanol (70%). The pellet was dissolved in RNase free water, the product [f5U]ASL digested with RNase T1, and the resulting oligonucleotides analyzed by MALDI-MS.
Results
Stem-loop RNA as a Substrate of RluA
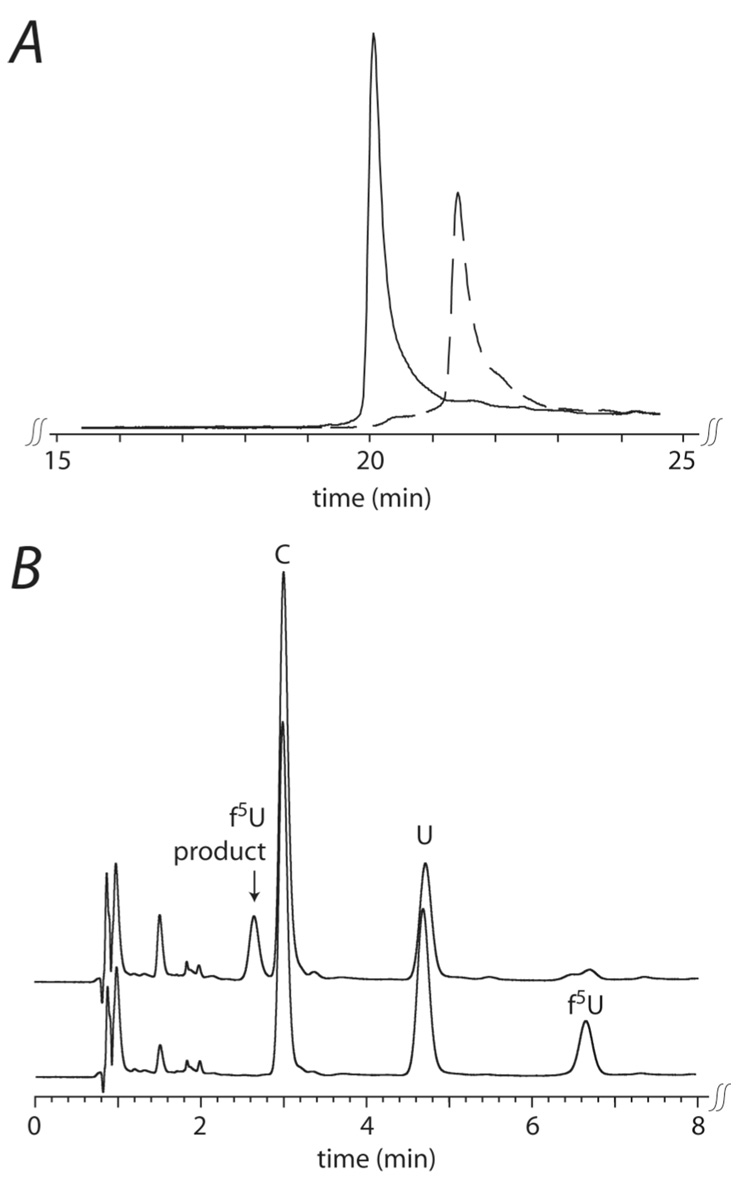
In order to verify that RluA isomerizes the U in ASL corresponding to U32 in tRNA (Figure 3), ASL was incubated with RluA and analyzed by reverse phase HPLC, which revealed a shift to shorter retention time after incubation with RluA (Figure 4). To ensure that this shift was due to formation of Ψ, ASL was digested to free nucleosides and analyzed by reverse phase HPLC both before and after incubation with RluA. As expected based on preliminary reports (46, 47) and our results with [f5U]ASL (41), Ψ was generated as a result of RluA action (Figure 4). Similarly, the stem-loop corresponding to C737 to G760 (Figure 3) of E. coli 23S ribosomal RNA was handled by RluA to generate Ψ (data not shown).
Figure 4.
HPLC analysis of ASL before (solid) and after (dashed) incubation with RluA. A, partial HPLC trace of intact ASL. The major peaks are substrate ASL (with U) at 15 min and product ASL (with Ψ) at 14 min. A small amount of substrate ASL remains in the product trace; the small peak at 11 minutes is a component of the reaction buffer. B, partial HPLC trace of the nucleosides from the complete digestion of ASL. The substrate trace shows U (5.00 min) at the expected ratio to C (3.05 min); the product trace shows diminished U and the appearance of Ψ (2.08 min) at the expected ratio relative to the other nucleosides. The G and A peaks (not shown) are not changed in intensity upon incubation with RluA, as expected.
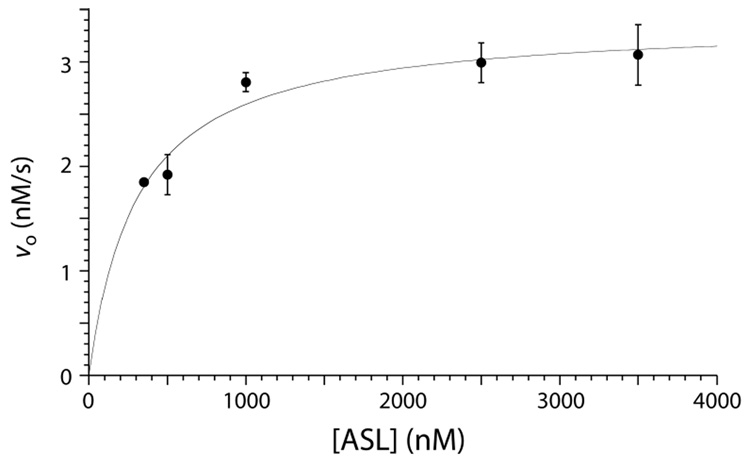
RluA activity assays were run with ASL to compare it to full-length tRNA as a substrate. When testing different buffer systems for optimal storage conditions, we discovered that sodium phosphate buffers inhibited RluA. After testing several different buffer concentrations, it was determined that 125 mM sodium phosphate buffer, pH 7.5, containing sodium chloride (125 mM) completely (and reversibly) inhibited RluA; neither sodium phosphate nor sodium chloride alone effected complete inhibition. Although the physical basis of the inhibition remains unclear, these mild quench conditions allowed for activity assays based on reverse phase HPLC separation of substrate ASL (with U) and product ASL (with Ψ). Activity assays were run at several concentrations of ASL, and the data was fit to the Briggs–Haldane equation (Figure 5). The kinetic parameters indicated that ASL is only a very slightly worse substrate than full-length tRNA (Table 5.1), with kcat = 0.068 s−1 (1.5-fold lower than the value with full-length tRNA) and Km = 308 nM (2.9-fold higher than with tRNA) so that kcat/Km for RluA with ASL as the substrate is 2.2 × 105 M−1s−1 (4.2-fold lower than with tRNA). The difference between the two substrates is slight and largely due to the increased Km, which is reasonable given the smaller net charge of ASL to make non-specific ionic interactions with RluA.
Figure 5.
Briggs-Haldane plot of the kinetic data for RluA with ASL as substrate. The enzyme concentration was 50 nM. Each point is the average of at least two independent determinations; error bars are omitted when the error is smaller than the size of data point.
Confirmation of an Adduct between RluA and [f5U]ASL
The adduct between RluA and [f5U]ASL was formed under conditions similar to those of the activity assays, as described previously (41), and adduct formation was confirmed by SDS-PAGE analysis. The adduction of [f5U]ASL and RluA causes a gel shift; heating disrupts the adduct and returns the normal migration of RluA (Figure 6, panel A). To determine if adduct formation with RluA required the essential Asp-64, D64A RluA (24) was tested, and no adduct formed after more than 3 h (Figure 6, panel A), whereas detectable adduct formed in less than 1 min with wild-type enzyme (Figure 6, panel B).
Figure 6.
SDS-PAGE analysis of the formation of the adduct between RluA and [f5U]ASL. A, the gel shift of wild-type RluA upon incubation with [f5U]ASL indicates adduct formation (lanes d and e). Heating disrupts the adduct (lanes b and c), which does not form with D64A RluA (lane h). B, The time course for the formation of the adduct between wild-type RluA and [f5U]ASL. Adduct formation was monitored in a reaction containing a five fold excess of [f5U]ASL over RluA. The adduct is detected at 0.5 min, and the reaction is half complete by 4 min.
Characterization of the Product of f5U
The product of f5U from the action of RluA on [f5U]ASL was characterized by reverse phase HPLC and mass spectrometry to see if its features matched those reported for the products of the action of TruA and TruB on [f5U]RNA. RluA and [f5U]ASL were incubated together, and after adduct formation was confirmed by SDS-PAGE, a portion of the reaction mixture was heated to disrupt the adduct. The protein precipitated upon heating and was pelleted by centrifugation; the product RNA in the supernatant was then analyzed. The RluA-modified [f5U]ASL shifted to a greater retention time on reverse phase HPLC, and peak integration indicated that >95% of the [f5U]ASL was modified (Figure 7, panel A). Since RluA action on f5U is expected to convert it into more polar product(s), the increased retention time was unanticipated. To insure that the change in [f5U]ASL was indeed at f5U, substrate and product [f5U]ASL were digested to free nucleosides and analyzed by HPLC, which confirmed that f5U had been changed into a more polar product (Figure 7, panel B).
Figure 7.
HPLC analysis of [f5U]ASL before (solid) and after (dashed) incubation with RluA and heat denaturation of the adduct. A, intact stem-loop showing an increased retention time for product [f5U]ASL. B, after digestion to free nucleosides; f5U elutes at 6.67 min, and the modified f5U product elutes at 2.64 min.
The f5U product was further characterized by MALDI-MS to determine if the product was hydrated, as was true for f5U after incubation of [f5U]RNA with TruA (48) and TruB (41). The [f5U]ASL signal (5465.8 m/zpred) was not intense, highly susceptible to forming salt adducts, and often not isotopically resolved. RNase A and RNase T1 were therefore used to digest [f5U]ASL and its product into smaller fragments and so afford better mass spectra and localize the site of modification. RNase T1 digestion of [f5U]ASL generated the expected tetranucleotide containing f5U (Af5UUG; 1323.2 m/zpred, 1324.0 m/zobs). The same analysis of [f5U]ASL that had been incubated with RluA yielded the corresponding tetranucleotide at 1341.7 m/z (data not shown), a gain of 18 m/z that demonstrates the hydration of f5U; the mass spectrum also shows a smaller unshifted peak, which is likely a combination of unmodified [f5U]ASL and the tetranucleotide from modified [f5U]ASL with a 2′,3′-cyclic phosphoric diester end rather than the 3′-phosphate end that results from complete hydrolysis. Digestion with RNase A yielded similar results, with a peak for the hexanucleotide (GGGGAf5U) that contains f5U (2052.3 m/zpred, 2052.8 m/zobs) and a peak at 2069.7 m/z for the hydrated product after incubation with RluA (data not shown).
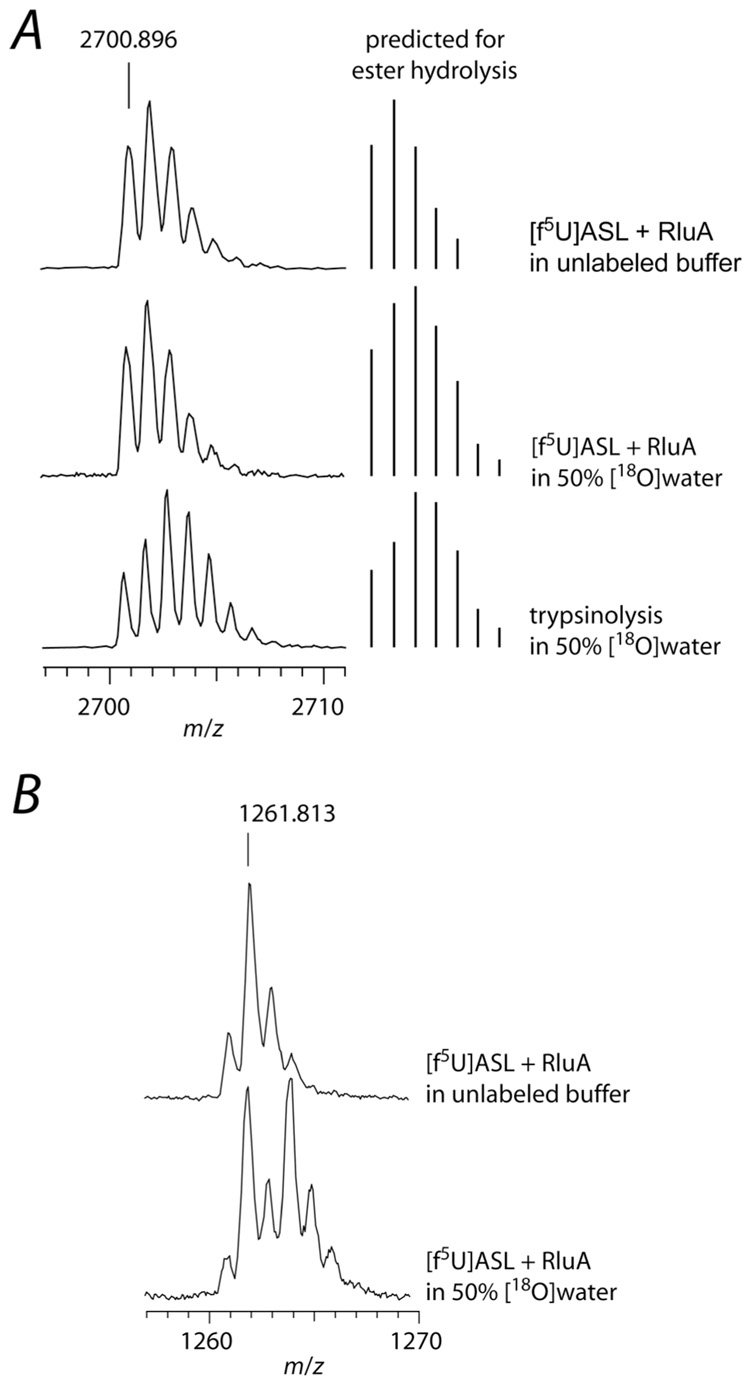
The heat-precipitated RluA from the adduct formation reactions was digested with trypsin and analyzed by MALDI-MS to verify that tryptic peptides showing Asp-64 would be observable and amenable to experiments with [18O]water to test for ester hydrolysis of a Michael adduct. The tryptic peptide containing Asp-64 (63Leu-Lys77) was detected but very small, presumably because of poor ionization due to the lower basicity of C-terminal Lys relative to Arg that generally biases against Lys-terminated peptides in MALDI-MS (49). Fortunately, a second peptide (53Asp-Lys77) that contains a single missed cleavage at Arg-62 (2700.4 m/zpred, 2700.9 m/zobs) was prominent and isotopically resolved (Figure 8, panel A). This peptide afforded an appropriate window and adequate resolution to observe the +2 m/z shift that would result from ester hydrolysis in [18O]water. Even allowing up to 3 missed cleavages, no other peptides from the tryptic digestion of RluA or trypsin were expected in this region, and the identity of the peptide was confirmed by ESI MS/MS sequencing (data not shown).
Figure 8.
Labeling studies to probe for ester hydrolysis during the decomposition of the adduct between RluA and [f5U]ASL. The adduct was formed with a 2-fold excess of RluA and then heat-disrupted, and the protein and RNA were separately digested and subjected to MALDI-MS analysis. A, Partial mass spectra of the tryptic digest of RluA showing the tryptic peptide 53Leu-Lys77 (2700.4 m/z predicted) containing Asp-64 (left) and the predicted mass distribution for ester hydrolysis of a Michael adduct (right); top, RluA incubated in unlabeled buffer with [f5U]ASL; middle, RluA incubated with [f5U]ASL in buffer containing [18O]water (50%), showing no 18O incorporation into the peptide, which does not match the prediction for ester hydrolysis; bottom, RluA incubated in unlabeled buffer with [f5U]ASL with trypsinolysis in buffer containing [18O]water (50%). B, Partial mass spectra of [f5U]ASL after incubation with RluA and digestion with RNase T1; top, incubation in unlabeled buffer; bottom, incubation in buffer containing [18O]water (50%), showing 18O incorporation into the hydrated [f5U]ASL (50% of the product contains 16O and 50% contains 18O and is therefore shifted +2 m/z). The small [M]+ peak observed could be due to amination rather than hydration since ammonium chloride (100 mM) was present in the reaction mixture; however, the [M]+ was still present when ammonium chloride was replaced with sodium chloride (data not shown), so it more likely arises from the high laser power necessary to obtain the mass spectra of the oligonucleotides.
Labeling experiments with [18O]water
With the observability of the key RNA and enzyme fragments established, it was possible to probe for ester hydrolysis at Asp-64 by 18O labeling experiments. To simplify the analysis of RNA fragments, alkaline phosphatase was added after the RNase digestions to remove the 3′ phosphate group, which would incorporate 18O from the solvent upon RNase digestion. Controls were performed to verify that the 18O labeling experiments would be uncompromised by oxygen exchange between solvent and Asp-64 in native RluA or its tryptic peptides or between solvent and the product of f5U in intact modified [f5U]ASL or its product oligonucleotides. These various species were incubated in buffer containing [18O]water (50%), and in no case did Asp-64 or the hydrated product of f5U increase in mass, indicating the absence of oxygen exchange with solvent. Since trypsinolysis is the hydrolysis of peptide bonds, each new C-terminal carboxylate group contains one oxygen atom from solvent; trypsinolysis in buffer containing [18O]water therefore provides a positive control for the detection of 18O incorporation into the peptides. RluA was digested with trypsin in buffer containing [18O]water (50%), and the expected isotopic pattern for each peptide containing 18O was observed (Figure 8, panel A).
With the controls in place, [f5U]ASL was incubated with RluA in buffer containing [18O]water (50%); the reaction was terminated by heating, and the complete conversion of [f5U]ASL was confirmed by HPLC analysis. RluA was digested overnight with trypsin, and the modified [f5U]ASL was digested with either RNase A or RNase T1 and alkaline phosphatase, and the digestion products were analyzed by MALDI-MS. No shift was observed in the tryptic peptides from RluA after an adduct formation reaction in labeled buffer, indicating that 18O was not incorporated into the protein (Figure 8, panel A). The digestion products of [f5U]ASL from the same incubation with RluA in labeled buffer showed the pattern with the +2 m/z peak shifts expected for direct hydration by solvent (Figure 8, panel B). These results eliminate ester hydrolysis of a Michael adduct as the source of the hydrated product of f5U.
Discussion
It was initially thought that all Ψ synthases were potently inhibited by [f5U]RNA, based largely on experiments with TruA that indicated a covalent complex formed between TruA and [f5U]RNA to inactivate the enzyme irreversibly (23, 38). Unexpectedly, the crystal structure of TruB and [f5U]RNA did not show a covalent adduct but a rearranged and hydrated product of f5U (11). The product was reasonably ascribed to progression along the Michael mechanism to the rearranged adduct and then slow ester hydrolysis over the months required for crystal growth and analysis (Figure 2, panel B) (11). However, more recent work with TruB showed that it is not inhibited by [f5U]RNA and instead rearranges and hydrates f5U with no evidence of a covalent intermediate (41). Labeling studies with [18O]water demonstrated that the products of f5U arise by direct hydration of the nucleobase rather than ester hydrolysis (42).
Four possible scenarios can account for all the proposals for the handling of f5U by different Ψ synthases (Figure 2). As presented above, we have proposed that all Ψ synthases rearrange f5U by a common reaction mechanism but that the fate of the rearranged f5U differs based on the idiosyncratic geometry of a particular active site, which accounts for the differences in adduct formation (Figure 2, panel D). In the case of TruB, an adduct is not observed and direct hydration of rearranged f5U occurs. The investigations with RluA now test the generality of the proposed scheme by extending the examination to a Ψ synthase that forms an adduct with [f5U]RNA.
The analysis of the f5U product is technically easier starting with ASL (a 17-mer) rather than full-length tRNAPhe (a 76-mer), but it was possible that RluA would behave differently towards f5U in the context of a stem-loop RNA than in the context of a full-length physiological substrate. The kinetic parameters reveal that the ASL is an excellent substrate for RluA, with kcat/Km down by only 4-fold relative to full-length tRNAPhe (Table 1). Based on these results, the use of [f5U]ASL rather than [f5U]tRNA seems highly unlikely to have introduced changes in enzyme-substrate interactions that significantly perturb the handling of f5U in the context of the smaller substrate.
Table 1.
Kinetic Parameters for RluA with tRNA and ASL
| substrate | kcat (s−1) | Km (nM) | kcat/ Km (M−1s−1) |
|---|---|---|---|
| tRNAa | 0.099 ± 0.003 | 108 ± 20 | 9.2 × 105 |
| ASL | 0.068 ± 0.003 | 308 ± 66 | 2.2 × 105 |
From Ramamurthy et al. (37)
We have confirmed the formation of a heat-sensitive adduct between RluA and [f5U]ASL, demonstrated that it forms quickly, and showed that the product of f5U is hydrated. No adduct is observed with catalytically inactive D64A RluA. The behavior of RluA towards [f5U]RNA, then, closely matches the behavior of TruA (23, 38) in forming an adduct that is dependent on the active site Asp and is disrupted by heating to generate a hydrated product of f5U.
In order to determine if the product forms by ester hydrolysis or direct hydration, the adduct forming reaction was run in buffer containing [18O]water. No 18O was incorporated into Asp-64 (or anywhere else in RluA) whereas the RNase digestion products of [f5U]ASL do show 18O incorporation (Figure 8). These results indicate that ester hydrolysis does not occur and that, instead, a water molecule is added directly to the pyrimidine ring. Although the labeling results alone cannot rule out the Michael mechanism, a hydrated product of f5U can no longer be considered strong evidence in support of the conclusion that the Michael mechanism operates since that conclusion rested on the assumed hydrolysis of an ester adduct between the active site Asp and [f5U]RNA as the source of the hydrated nucleoside.
The labeling results eliminate the schemes involving ester hydrolysis during the handling of [f5U]RNA by RluA (Figure 2), leaving two schemes to account for the direct hydration of f5U. In the first (Figure 2, panel C), the conserved Asp serves as a general base to catalyze hydration of rearranged f5U. This scheme is reasonable for TruB, but it cannot account for the formation of an apparently covalent adduct between RluA and [f5U]ASL. Alternatively, the complex between RluA and [f5U]ASL may be a very tight but noncovalent one that resists denaturation by both urea (7 M) and SDS (0.1%) unless heated (41). Although not physically unreasonable in principle, it remains unclear why RluA would form such a tight noncovalent complex but TruB does not and why D64A RluA would not form such a tight noncovalent adduct. Experiments that more thoroughly address the covalent nature of the adduct are underway.
We are left to discuss the scheme that we proposed to account for the handling of [f5U]RNA by all Ψ synthases (Figure 2, panel D). The scheme features an equilibrium between rearranged f5U and the Michael adduct resulting from the addition of the catalytic Asp. All of our results are consistent with the proposal that RluA follows the same reaction course as TruB to rearranged f5U; at that point, fine differences in the active site allow Asp-64 of RluA, but not Asp-48 of TruB, to reach the nucleobase and make a stable covalent adduct. For RluA (and TruA), the depicted equilibrium must lie far to the right to account for the observed adduct (Figure 2, panel D). Heating both disrupts the tertiary structure of the complex and leads to the collapse of the adduct by elimination of Asp-64 to generate the rearranged f5U that then becomes hydrated (proceeding to the left in Figure 2, panel D). Based on the HPLC data (Figure 7, panel B) and preliminary 1D 19F NMR data (C. Hamilton, unpublished observations), it appears likely that only a single product of f5U is generated by the action of RluA. A single hydrated product implies that access by bulk solvent is restricted to only one face of the pyrimidine ring, which suggests that the enzyme catalyzes the process (Figure 2, panel C) or the melting protein retains enough structure to shield one face of the pyrimidine ring as hydration occurs following the elimination of Asp-64. Although it seems unlikely, it also remains possible that RluA may form a Michael adduct with f5U (unrearranged), which suffers elimination upon heating (the reversal of the reaction shown in Figure 2, panel A) and then enzyme-catalyzed hydration of f5U (similar to Figure 2, panel C) before complete enzyme denaturation releases the hydrated product. A full chemical characterization of the products of f5U from the action of TruB and RluA to resolve these possibilities is underway.
Regardless of the number of hydrated products of f5U and the nature of the adduct between RluA and [f5U]ASL as covalent or noncovalent, the main results presented here remain definitive: f5U is hydrated upon heat disruption of the adduct, and hydration does not occur as a result of ester hydrolysis. The 18O labeling studies have now revealed the same outcome (direct hydration rather than ester hydrolysis) for two Ψ synthases that differ in family membership and the formation of a stable adduct with [f5U]RNA. Although several more Ψ synthases must be similarly examined to extend the comparison to members of the same family that differ in adduct formation and members of different families that share adduction behavior, our current observations are well accommodated by the proposal that all Ψ synthases rearrange f5U by a common route and apparent discrepancies in their handling of f5U arise from fine differences in the position of the active site Asp relative to rearranged f5U.
abbreviations
- Ψ
pseudouridine
- f5U
5-fluorouridine
- ASL
anticodon stem-loop of E. coli tRNAPhe
- [f5U]ASL
ASL containing f5U in the position corresponding to U33 (the residue isomerized by RluA)
- TSL
T-arm stem-loop
- [f5U]TSL
TSL containing f5U in the position corresponding to U55 in tRNA
Footnotes
This work was supported by the National Institutes of Health (GM59636 to E.G.M.) and the National Science Foundation (DBI0096578 to M.V.J.). C.A.V. was supported by United States Public Health Service Grant T32 GM08550. T.M.G. was supported by HHMI Undergraduate Science Education Grant 52002062 to the University of Delaware.
TruA was originally named Ψ synthase I.
Briefly, when Phannachet et al. incubated stem-loop substrate with Y76F TruB, they detected no conversion of U to Ψ (40). However, when f5U replaced the isomerized U in the stem-loop, the f5U was rearranged and hydrated just as with wild-type TruB (vide infra). In the cocrystal of wild-type TruB (11, 13), the phenol group of Tyr-76 makes a hydrogen bond with the fluorine atom in the product of f5U, and Phannachet et al. therefore concluded that Tyr-76 conducts the final deprotonation to convert U to Ψ, a step that does not occur with f5U since a base cannot remove the fluorine as it does a proton.
However, it remains unclear to us whether or not the nucleoside conformation in the cocrystal is one that reflects the case with the natural substrate (U) or results from the interactions required for binding of the unnatural product of f5U, in which case the observed conformation does not reflect events along the physiological reaction pathway. The observed conformation of the nucleoside is nonproductive for the Michael mechanism (Figure 1, panel A) since C6 is distant from the active site Asp. Furthermore, it appears that the U is untouched by Y76F TruB (although Phannachet et al. do not explicitly address the issue and the top of the U and C peaks in HPLC traces are cut off, which prevents precise analysis of the published data in this regard). General bond energy considerations indicate that the formation of the C–C glycosidic bond should be heavily favored over the initial C–N bond, and heats of formation calculated by the method of Joback and Read (50) as implemented by ChemDraw Pro® (version 7.0.1, CambridgeSoft, Cambridge, MA) indicate that the last intermediate depicted in Figure 1 is favored over U by 15 kcal/mol. U should, therefore, be converted to the last intermediate depicted in Figure 1 if, as proposed by Phannacet et al.. the final deprotonation is the only step that Y76F TruB cannot carry out. Since the intermediate is more polar than U, it seems safe to assume that it would elute distinctly earlier than U from a C18 HPLC column, but the data of Phannachet et al. show no such new peak (40). The pH profile for TruB (39), to which Phannachet et al. did not refer, shows a descending limb with pKa of 9, suggesting that deprotonation of Tyr-76 deactivates wild-type TruB. We therefore propose that all of the data are best accommodated if Tyr-76 donates a proton or a hydrogen bond to the uracil(ate) leaving group to facilitate glycosidic bond cleavage. Unable to carry out this early step of the mechanism, Y76F TruB leaves U unchanged. On the other hand, the glycosidic bond of f5U can undergo cleavage without the assistance provided by Tyr-76 because the inductive effect of fluorine makes the anion of 5-fluorouracil a better leaving group than that of uracil. Clearly, the elucidation of the function of Tyr-76 requires additional experiments. Given the uncertainty over its role, we have omitted Tyr-76 from the depictions of the alternate mechanisms to focus on the issues addressed by the experiments presented in this report.
TSL (with U) is as good a substrate for TruB as full-length tRNA (48).
S1 nuclease is the same enzyme as nuclease P1, which we used previously (45).
References
- 1.Grosjean H, Benne R. Modification and Editing of RNA. Washington, DC: ASM Press; 1998. [Google Scholar]
- 2.Koonin EV. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 1996;24:2411–2415. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gustafsson C, Reid R, Greene PJ, Santi DV. Identification of new RNA modifying enzymes by iterative genome search using known modifying enzymes as probes. Nucleic Acids Res. 1996;24:3756–3762. doi: 10.1093/nar/24.19.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaya Y, Ofengand J. A novel unanticipated type of pseudouridine synthase with homologs in bacteria archaea, and eukarya. RNA. 2003;9:711–721. doi: 10.1261/rna.5230603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma XJ, Zhao XL, Yu YT. Pseudouridylation of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 2003;22:1889–1897. doi: 10.1093/emboj/cdg191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kammen HO, Marvel CC, Hardy L, Penhoet EE. Purification, structure, and properties of Escherichia coli tRNA pseudouridine synthase I. J. Biol. Chem. 1988;263:2255–2263. [PubMed] [Google Scholar]
- 7.Nurse K, Wrzesinski J, Bakin A, Lane BG, Ofengand J. Purification, cloning, and properties of the tRNA Ψ55 synthase from Escherichia coli. RNA. 1995;1:102–112. [PMC free article] [PubMed] [Google Scholar]
- 8.Wrzesinski J, Nurse K, Bakin A, Lane BG, Ofengand J. A dual-specificity pseudouridine synthase: An Escherichia coli synthase purified and cloned on the basis of its specificity for Ψ746 in 23S RNA is also specific for Ψ32 in tRNAPhe. RNA. 1995;1:437–448. [PMC free article] [PubMed] [Google Scholar]
- 9.Wrzesinski J, Bakin A, Nurse K, Lane BG, Ofengand J. Purification, cloning, and properties of the 16S RNA pseudouridine-516 synthase from Escherichia coli. Biochemistry. 1995;34:8904–8913. doi: 10.1021/bi00027a043. [DOI] [PubMed] [Google Scholar]
- 10.Foster PG, Huang L, Santi DV, Stroud RM. The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I. Nature Struct. Biol. 2000;7:23–27. doi: 10.1038/71219. [DOI] [PubMed] [Google Scholar]
- 11.Hoang C, Ferré-D'Amaré AR. Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107:929–939. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- 12.Pan H, Agarwalla S, Moustakas DT, Finer-Moore J, Stroud RM. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12648–12653. doi: 10.1073/pnas.2135585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phannachet K, Huang RH. Conformational change of pseudouridine 55 synthase upon its association with RNA substrate. Nucleic Acids Res. 2004;32:1422–1429. doi: 10.1093/nar/gkh287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Campo M, Ofengand J, Malhotra A. Crystal structure of the catalytic domain of RluD, the only rRNA pseudouridine synthase required for normal growth of Escherichia coli. RNA. 2004;10:231–239. doi: 10.1261/rna.5187404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sivaraman J, Iannuzzi P, Cygler M, Matte A. Crystal structure of the RIuD pseudouridine synthase catalytic module, an enzyme that modifies 23S rRNA and is essential for normal cell growth of Escherichia coli. J. Mol. Biol. 2004;335:87–101. doi: 10.1016/j.jmb.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Mizutani K, Machida Y, Unzai S, Park SY, Tame JRH. Crystal structures of the catalytic domains of pseudouridine synthases RluC and RluD from Escherichia coli. Biochemistry. 2004;43:4454–4463. doi: 10.1021/bi036079c. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhuri BN, Chan S, Perry J, Yeates TO. Crystal structure of the apo forms of Ψ55 tRNA pseudouridine synthase from Mycobacterium tuberculosis. J. Biol. Chem. 2004;279:24585–24591. doi: 10.1074/jbc.M401045200. [DOI] [PubMed] [Google Scholar]
- 18.Kaya Y, Del Campo M, Ofengand J, Malhotra A. Crystal structure of TruD, a novel pseudouridine synthase with a new protein fold. J. Biol. Chem. 2004;279:18107–18110. doi: 10.1074/jbc.C400072200. [DOI] [PubMed] [Google Scholar]
- 19.Ericsson UB, Nordlund P, Hallberg BM. X-ray structure of tRNA pseudouridine synthase TruD reveals an inserted domain with a novel fold. FEBS Lett. 2004;565:59–64. doi: 10.1016/j.febslet.2004.03.085. [DOI] [PubMed] [Google Scholar]
- 20.Hoang C, Ferré-D'Amaré AR. Crystal structure of the highly divergent pseudouridine synthase TruD reveals a circular permutation of a conserved fold. RNA. 2004;10:1026–1033. doi: 10.1261/rna.7240504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoang C, Hamilton CS, Mueller EG, Ferré-D'Amaré AR. Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain. Prot. Sci. 2005;14:2201–2206. doi: 10.1110/ps.051493605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller EG. Chips off the old block. Nature Struct. Biol. 2002;9:320–322. doi: 10.1038/nsb0502-320. [DOI] [PubMed] [Google Scholar]
- 23.Huang LX, Pookanjanatavip M, Gu XG, Santi DV. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry. 1998;37:344–351. doi: 10.1021/bi971874+. [DOI] [PubMed] [Google Scholar]
- 24.Ramamurthy V, Swann SL, Paulson JL, Spedaliere CJ, Mueller EG. Critical aspartic acid residues in pseudouridine syntheses. J. Biol. Chem. 1999;274:22225–22230. doi: 10.1074/jbc.274.32.22225. [DOI] [PubMed] [Google Scholar]
- 25.Raychaudhuri S, Conrad J, Hall BG, Ofengand J. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA. 1998;4:1407–1417. doi: 10.1017/s1355838298981146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conrad J, Niu LH, Rudd K, Lane BG, Ofengand J. 16S ribosomal RNA pseudouridine synthase RsuA of Escherichia coli: Deletion, mutation of the conserved Asp102 residue, and sequence comparison among all other pseudouridine synthases. RNA. 1999;5:751–763. doi: 10.1017/s1355838299990167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillips B, Billin AN, Cadwell C, Buchholz R, Erickson C, Merriam JR, Carbon J, Poole SJ. The Nop60B gene of Drosophila encodes an essential nucleolar protein that functions in yeast. Mol. Gen. Genet. 1998;260:20–29. doi: 10.1007/s004380050866. [DOI] [PubMed] [Google Scholar]
- 28.Lafontaine DLJ, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nature Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 30.Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giordano E, Peluso I, Senger S, Furia M. minifly, a Drosophila gene required for ribosome biogenesis. J. Cell Biol. 1999;144:1123–1133. doi: 10.1083/jcb.144.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doong SL, Dolnick BJ. 5-Fluorouracil substitution alters pre-mRNA splicing in vitro. J. Biol. Chem. 1988;263:4467–4473. [PubMed] [Google Scholar]
- 33.Sierakowska H, Shukla RR, Dominski Z, Kole R. Inhibition of pre-messenger RNA splicing by 5-fluorouridine, 5-chlorouridine, and 5-bromouridine. J. Biol. Chem. 1989;264:19185–19191. [PubMed] [Google Scholar]
- 34.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santi DV, McHenry CS, Sommer H. Mechanism of interaction of thymidylate synthetase with 5-fluorodeoxyuridylate. Biochemistry. 1974;13:471–481. doi: 10.1021/bi00700a012. [DOI] [PubMed] [Google Scholar]
- 36.Zhao XM, Horne DA. The role of cysteine residues in the rearrangement of uridine to pseudouridine catalyzed by pseudouridine synthase I. J. Biol. Chem. 1997;272:1950–1955. doi: 10.1074/jbc.272.3.1950. [DOI] [PubMed] [Google Scholar]
- 37.Ramamurthy V, Swann SL, Spedaliere CJ, Mueller EG. Role of cysteine residues in pseudouridine synthases of different families. Biochemistry. 1999;38:13106–13111. doi: 10.1021/bi9913911. [DOI] [PubMed] [Google Scholar]
- 38.Gu XR, Liu YQ, Santi DV. The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14270–14275. doi: 10.1073/pnas.96.25.14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamilton CS, Spedaliere CJ, Ginter JM, Johnston MV, Mueller EG. The roles of the essential Asp-48 and highly conserved His-43 elucidated by the pH dependence of the pseudouridine synthase TruB. Arch. Biochem. Biophys. 2005;433:322–334. doi: 10.1016/j.abb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 40.Phannachet K, Elias Y, Huang RH. Dissecting the roles of a strictly conserved tyrosine in substrate recognition and catalysis by pseudouridine 55 synthase. Biochemistry. 2005;44:15488–15494. doi: 10.1021/bi050961w. [DOI] [PubMed] [Google Scholar]
- 41.Spedaliere CJ, Mueller EG. Not all pseudouridine synthases are potently inhibited by RNA containing 5-fluorouridine. RNA. 2004;10:192–199. doi: 10.1261/rna.5100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spedaliere CJ, Ginter JM, Johnston MV, Mueller EG. The pseudouridine synthases: Revisiting a mechanism that seemed settled. J. Am. Chem. Soc. 2004;126:12758–12759. doi: 10.1021/ja046375s. [DOI] [PubMed] [Google Scholar]
- 43.Gehrke CW, Kuo KC, McCune RA, Gerhardt KO, Agris PF. Quantitative enzymatic hydrolysis of tRNAs: reversed-phase high-performance liquid chromatography of tRNA nucleosides. J. Chromatogr. 1982;230:297–308. [PubMed] [Google Scholar]
- 44.Buck M, Connick M, Ames BN. Complete analysis of tRNA modified nucleosides by high-performance liquid chromatography: the 29 modified nucleosides of Salmonella typhimurium and Escherichia coli tRNA. Anal. Biochem. 1983;129:1–13. doi: 10.1016/0003-2697(83)90044-1. [DOI] [PubMed] [Google Scholar]
- 45.Mueller EG, Buck CJ, Palenchar PM, Barnhart LE, Paulson JL. Identification of a gene involved in the generation of 4-thiouridine in tRNA. Nucleic Acids Res. 1998;26:2606–2610. doi: 10.1093/nar/26.11.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horne DA, Rood K, Levine E, Ofengand J. RNA substrate recognition by E. coli pseudouridine synthase (RluA) with dual substrate specificity. Abstr. Pap. Am. Chem. Soc. 1997;213:308. [Google Scholar]
- 47.Tworowska I, Nikonowicz E. General method of pseudouridylation of RNA: sequence specificity of the pseudouridine synthase RluA. Abstr. Pap. Am. Chem. Soc. 2004;228:U202-U202. [Google Scholar]
- 48.Gu XR, Yu M, Ivanetich KM, Santi DV. Molecular recognition of tRNA by tRNA pseudouridine 55 synthase. Biochemistry. 1998;37:339–343. doi: 10.1021/bi971590p. [DOI] [PubMed] [Google Scholar]
- 49.Krause E, Wenschuh H, Jungblut PR. The dominance of arginine-containing peptides in MALDI-derived tryptic mass fingerprints of proteins. Anal. Chem. 1999;71:4160–4165. doi: 10.1021/ac990298f. [DOI] [PubMed] [Google Scholar]
- 50.Joback KG, Reid RC. Estimation of pure component properties from group contributions. Chem. Eng. Commun. 1987;57:233–243. [Google Scholar]