Abstract
The modified nucleoside 1-methyladenosine (m1A) is found at position 58 in the TΨC loop of many eukaryotic tRNAs. The absence of m1A from all tRNAs in Saccharomyces cerevisiae mutants lacking Gcd10p elicits severe defects in processing and stability of initiator methionine tRNA (tRNAiMet). Gcd10p is found in a complex with Gcd14p, which contains conserved motifs for binding S-adenosylmethionine (AdoMet). These facts, plus our demonstration that gcd14Δ cells lacked m1A, strongly suggested that Gcd10p/Gcd14p complex is the yeast tRNA(m1A)methyltransferase [(m1A)MTase]. Supporting this prediction, affinity-purified Gcd10p/Gcd14p complexes used AdoMet as a methyl donor to synthesize m1A in either total tRNA or purified tRNAiMet lacking only this modification. Kinetic analysis of the purified complex revealed KM values for AdoMet or tRNAiMet of 5.0 μM and 2.5 nM, respectively. Mutations in the predicted AdoMet-binding domain destroyed GCD14 function in vivo and (m1A)MTase activity in vitro. Purified Flag-tagged Gcd14p alone had no enzymatic activity and was severely impaired for tRNA-binding compared with the wild-type complex, suggesting that Gcd10p is required for tight binding of the tRNA substrate. Our results provide a demonstration of a two-component tRNA MTase and suggest that binding of AdoMet and tRNA substrates depends on different subunits of the complex.
The maturation of tRNA occurs through a series of posttranscriptional processing steps, including splicing, end-trimming, and base or ribose modifications. Modified nucleosides have been identified in all cellular RNAs, but the tRNAs by far contain the greatest number and variety of modifications. Of these, the most widely distributed and prevalent tRNA modification is base or ribose methylation. Several tRNA modification enzymes have been cloned and characterized from eubacteria and eukaryotes. Remarkably, only one tRNA modification, the subject of this report, has been found to be essential for cell growth under normal conditions (1). In other instances, the modifications are required for efficient decoding during protein synthesis (2). A 2′-O-phosphoribosyl modification of A64 in initiator tRNAMet (tRNAiMet) prevents inappropriate interaction of the initiator with translation elongation factor 1α (3).
The occurrence of 1-methyladenosine (m1A) at position 58 of the TΨC loop has been reported in tRNAs from all three kingdoms (2), suggesting an evolutionarily conserved role in tRNA structure or function. In the yeast Saccharomyces cerevisiae, m1A58 is widespread but not universal, occurring in 21 of 32 sequenced tRNAs (4). This modification seems to be crucial only for tRNAiMet because a null mutation in GCD10 that eliminates m1A in all yeast tRNAs leads specifically to reduced processing and stability of the initiator. This defect is lethal unless tRNAiMet is overproduced in the gcd10Δ mutant to compensate for reduced accumulation of mature initiator (5). It was proposed that m1A58 is singularly required for tRNAiMet production because it contributes to a unique substructure involving tertiary interactions between the TΨC and D loops (6).
Gcd10p resides in a nuclear complex with another essential protein, Gcd14p (5, 7), that also seems to be required primarily for production of mature tRNAiMet. Temperature-sensitive gcd14 mutations lead to accumulation of precursors and reductions in mature tRNAiMet, and exacerbate the effects of a temperature-sensitive gcd10 mutation on cell growth and tRNAiMet production. Furthermore, the lethal phenotype of a gcd14Δ allele was suppressed by overexpressing tRNAiMet (7). It was predicted that the defect in tRNAiMet processing observed in gcd14 mutants also derives from the absence of m1A in tRNA.
Most of the known tRNA methyltransferases (MTases) use S-adenosylmethionine (AdoMet) as the methyl donor, and most contain a domain comprised of four conserved motifs (I, post-I, II, and III) (8). The three-dimensional structures of several MTases reveal that motifs I and post-I directly contact AdoMet (9), and mutation of motif I impairs AdoMet binding (10). The presence of motifs I, post-I, and II in Gcd14p suggested that it can bind AdoMet and function directly as the yeast tRNA(m1A)MTase. Because all well-characterized tRNA MTases are single-subunit enzymes (1), the role of Gcd10p was more difficult to predict. The Gcd10p/Gcd14p complex could be a two-subunit tRNA(m1A)MTase, requiring Gcd10p for intrinsic enzymatic activity. Alternatively, Gcd10p might function in nuclear localization of the complex, or as a tRNA chaperone, and be required for m1A formation only in vivo.
In this report, we show that the purified Gcd10p/Gcd14p complex has tRNA(m1A)MTase activity. A mutation in motif I of the putative AdoMet binding domain in Gcd14p eliminated (m1A)MTase activity in vitro and abolished m1A formation in vivo. These results prove that Gcd10p and Gcd14p function directly in m1A formation in yeast tRNAs. Purified Gcd14p alone had no enzymatic activity and was defective for tRNA binding compared with the Gcd14p/Gcd10p complex. These findings, combined with the previous demonstration that Gcd10p alone has RNA binding activity (11), leads us to suggest that Gcd10p is required for tight binding of the tRNA substrate. Thus, the AdoMet and tRNA binding functions required for m1A formation may depend on distinct subunits of a two-component tRNA methyltransferase.
Materials and Methods
Yeast Strain and Plasmid Construction.
Transformation of yeast strains was carried out as described (24), as was the formulation of media for culturing yeast and bacteria (25, 26). Plasmid pJA148 (lcGCD14Flag) was constructed by PCR fusion (27) using oligonucleotides GCD14-A (5′-GAAACGAGGATGGCGTACCA) and GCD14-D (5′-GCTCTCGAAAAAAGCTCC), JA81 (5′-GCTAGCTTTGTCATCGTCGTCCTTGTAGTCTTTTTCCGTGGATCGAAGAATTTC), and JA82 (5′-GACTACAAGGACGACGATGACAAAGCTAGCTA ATTAAATGATTAACTACTTATA) as primers and p2599 (lcGCD14) (7) as template. The resulting PCR product digested with SphI and HpaI was subcloned into p2599 digested with the same enzymes. Plasmid pJA149 bearing lcgcd14–3Flag was constructed by using PCR fusion with oligos GCD14-D, JA83 (5′-TATGCCATGCCGTTA), JA84 (5′-CGAGGCAGCTACAGCCTCAGGATCA), and JA85 (5′-TGATCCTGAGGCTGTAGCTGCCTCG) as primers and pJA148 as template. The resulting PCR product digested with EcoRI and EcoNI was subcloned into EcoRI/EcoNI-digested pJA148. The PCR-amplified segments of pJA148 and pJA149 were confirmed by DNA sequence analysis. Plasmid pJA151 (hcGCD14Flag) was constructed by subcloning a ≈3.0-kb SalI/SacI fragment from pJA148 into SalI/SacI-digested YEpLac195 (28). Plasmid pLPYGCD10HisFlag was created by subcloning a fragment encoding the Flag epitope and six consecutive histidines, flanked by BglII (5′) and HindIII (3′) sites, into plasmid pLPY3 (5) linearized by digestion with BglII/HindIII. Yeast strain yJA203 is a Ura- derivative of F612 (MATα/gcd14ΔURA3/ura3–52/leu2–3/112/trp1–289/his3–111/can1–100/[p1775:hcIMT4]) that was isolated by growth on synthetic complete (SC) medium containing 1 μg/ml 5-fluoroorotic acid (29). Yeast strains yJA212 and yJA213 were constructed by introducing plasmid pJA148 (GCD14Flag) or pJA149 (gcd14–3Flag) into yJA203. Yeast strain yJA216 was constructed by introducing plasmid pJA151 into yJA146 (5) and selecting for a Ura+ transformant.
Protein Purification.
FlagGcd10p/Gcd14p or FlagGcd14p/Gcd10p complexes were affinity-purified from cell extracts prepared from 10 liters of yeast extract/peptone/dextrose or SC medium lacking uracil. All purification steps were carried out at 4°C. Yeast cells (≈50–100 g wet weight) were harvested, by centrifugation at 10,000 × g for 30 min, washed with water, and resuspended in 1× TBSC buffer [2 mM Tris, pH 7.5/14 mM NaCl/0.5 mM DTT/1× Complete protease inhibitor mixture (Boehringer Mannheim)] at 1.0 ml⋅g-1 of cells. Cells were disrupted by two passages through a French press. The whole-cell extract was clarified by centrifugation at 15,000 × g for 20 min and then at 274,000 × g for 30 min. The postribosomal supernatant, at 3–10 mg/ml, was batch-bound overnight to 0.1–0.3 ml of anti-Flag M2-agarose affinity gel (Sigma-Aldrich) in 50–200 ml of TBSC according to the manufacturer's recommendation. After washing four times in TBSC with constant mixing for 20 min each, the Flag-protein complexes were eluted from the resin twice with 0.3–1.0 ml of TBSC + 10% glycerol containing 100 μg/ml Flag peptide (Sigma-Aldrich). The eluted complexes were pooled and concentrated to a final volume of 0.15–0.3 ml by using a microcon-10 spin column (Amicon). The purified complexes were divided into aliquots and stored at −80°C. Further purification of the complex by Superose-6 gel filtration was done essentially as described (30) in SB-6 buffer (20 mM Tris, pH 7.5/50 mM NaCl/0.5 mM EDTA/1.0 mM DTT/10% glycerol).
MTase Assays and HPLC Analysis of m1A.
MTase activities were assayed in 100-μl reactions at 30°C containing 1× MTase buffer (100 mM Tris, pH 8.0/1.0 mM DTT/0.1 mM EDTA/10 mM MgCl2/20 mM NH4Cl), S-adenosyl-l-[methyl-14C]methionine (59.0 mCi/mmol) or S-adenosyl-l-[methyl-3H]methionine (500 mCi/mmol) (Amersham Pharmacia), and either total tRNA from yJA146 (gcd10Δ hcIMT4) or yJA158 (GCD10 hcIMT4) or purified tRNAiMet from yJA146 (gcd10Δ hcIMT4). The reactions were stopped by the addition of 500 μl of cold H2O and 3.0 ml of cold 5% trichloroacetic acid (TCA). The amount of radiolabeled methyl groups transferred to tRNA was determined by collecting the acid-insoluble material on 25-mm, 0.45-μm nitrocellulose filters (Millipore), washing with 10 ml of 5% TCA, drying the filters, and measuring the radioactivity by liquid scintillation using Econofluor-2 scintillation mixture (Packard Bioscience). HPLC analysis of total tRNA nucleosides was conducted as described (5).
tRNA Purification.
To purify tRNAiMet, total tRNA isolated from strain yJA146 (gcd10Δ hcIMT4) as described (5) was fractionated on BD-cellulose by using a linear gradient from 450–650 mM NaCl in SB buffer (10 mM sodium acetate, pH 4.5/10 mM MgCl2/1 mM EDTA). Fractions containing tRNAiMet were identified by dot blot hybridization using a 32P-labeled oligonucleotide (JA11) complementary to tRNAiMet (5), ethanol-precipitated, and resuspended in water. The resulting sample was made 1.3 M (NH4)2SO4 and fractionated on Sepharose-4B by using a linear gradient from 1.3–0 M (NH4)2SO4 in 4B buffer (10 mM sodium acetate, pH 4.5/6 mM 2-mercaptoethanol/10 mM MgCl2/1 mM EDTA). The fractions containing tRNAiMet were pooled and dialyzed against an excess of DB buffer (1.0 mM Tris⋅HCl, pH 7.5/1.0 mM MgCl2), ethanol-precipitated, and resuspended in deionized distilled water. In the final step, the sample was resolved by RPC-5 chromatography using a linear gradient from 450–575 mM NaCl in SB buffer. Fractions containing tRNAiMet were pooled and ethanol-precipitated. To establish purity of the sample, an aliquot was separated on a 15%-PAGE-Urea gel (NOVEX, San Diego) and stained with ethidium bromide. More than 90% of the ethidium-stained tRNA on the gel migrated as a single band.
Filter Binding Assay.
FlagGcd14p/Gcd10p-tRNA complex formation was monitored by filter binding assay as described (12). Briefly, binding reactions (0.1 ml) were carried out in TB buffer (5 mM Tris⋅HCl, pH 7.5/5 mM MgCl2/1.0 mM DTT/25 mM NaCl), 50 μg/ml BSA, and 1 nM purified 32P-labeled tRNAiMet from yJA146 at ambient temperature for 20 min with purified protein complexes at the concentrations indicated in Fig. 5. Reactions were filtered through a three-membrane sandwich by using a slot blot apparatus and washed with 0.6 ml of TB buffer before being dried and analyzed by phosphorimaging on a Storm 860 (Molecular Dynamics) or by autoradiography.
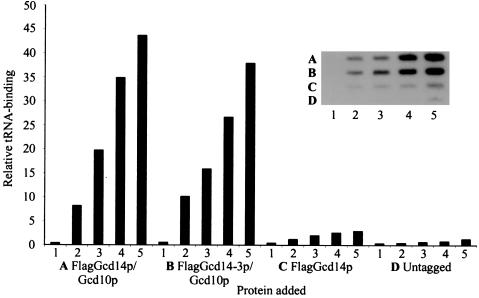
Figure 5.
Evidence that tRNA binding by Gcd14p/Gcd10p requires Gcd10p. The indicated purified FlagGcd14p/Gcd10p complexes (A and B) or FlagGcd14p alone (C) or comparable amounts of protein purified from untagged (GCD14, D) were combined in TB binding buffer with [32P]tRNAiMet purified from the gcd10Δ hcIMT4 strain. Reactions were filtered on a slot blot apparatus containing a three-membrane sandwich, as described in Materials and Methods. The cpm of 32P retained on the second membrane (nitrocellulose), which typically contains specific RNA-protein complexes, was quantified by phosphorimaging analysis using a Storm 860 apparatus and imagequant software. The graph shows the relative amount of bound [32P]tRNAiMet plotted against the protein concentration for each reaction, with columns 1–5 designating reactions containing protein concentrations of 0, 8, 16, 32, or 64 nM, respectively. (Inset) An autoradiogram of the nitrocellulose membrane used to generate the graphical data. The rows A–D and columns 1–5 designate the identities of the complex and protein concentrations, respectively, as described in the graph.
Results
Purified Gcd10p/Gcd14p Complexes Possess tRNA(m1A)MTase Activity.
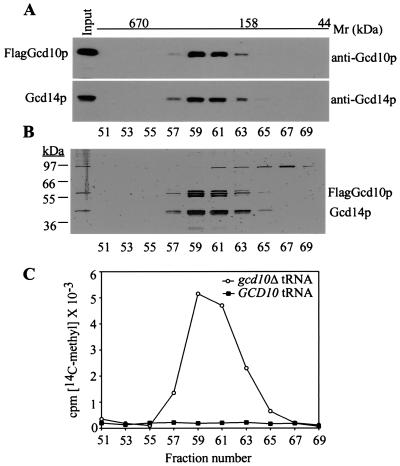
The absence of m1A in gcd10 mutants (5) and the presence of AdoMet binding motifs in Gcd14p (7) suggested that the Gcd10p/Gcd14p complex is the yeast tRNA(m1A)MTase. To test this hypothesis, we set out to purify the complex from a yeast strain expressing FLAG epitope-tagged Gcd10p. As there was no difference in growth rates between gcd10Δ strains bearing GCD10Flag or GCD10 on low-copy plasmids (data not shown), we concluded that GCD10Flag functions like wild-type GCD10 in vivo. We purified FlagGcd10p/Gcd14p complexes from whole-cell extracts with Flag affinity resin to approximately 70% homogeneity (data not shown) and resolved the preparation further on a gel filtration column. SDS/PAGE and immunoblot analysis of the fractions showed that FlagGcd10p and Gcd14p coeluted from the sizing column in a complex with an apparent molecular mass of ≈200 kDa (Fig. 1A). Silver staining of the gel showed that the peak fractions contained a series of three closely spaced bands close to the predicted Mr of FlagGcd10p (56 kDa) and a slightly heterogeneous band with approximate Mr of Gcd14p (44 kDa) (Fig. 1B). Closer inspection of the immunoblots revealed that all three bands with the approximate mobilities of FlagGcd10p were recognized by Gcd10p antibodies. Presumably the species of lowest mobility was full-length FlagGcd10p, and the two faster migrating species arose from the latter by proteolysis. Alternatively, posttranslational modifications might account for the different Gcd10p isoforms. We concluded that FlagGcd10p and Gcd14p were the only stoichiometric constituents of the purified complex. The apparent molecular weight of the complex is ca. 2-fold greater than that predicted for a FlagGcd10p/Gcd14p heterodimer, suggesting the possibility of a multimeric structure.
Figure 1.
The purified Gcd10p/Gcd14p complex has tRNA(m1A)MTase activity specific for gcd10Δ tRNA. Affinity-purified FlagGcd10p/Gcd14p complexes were resolved on a Superose-6 FPLC column precalibrated with commercial standards of 4–670 kDa (Bio-Rad). The elution positions and masses of the standards are shown on top above the line. Aliquots containing equal portions of every other column fraction in the range predetermined to contain the complexes were separated by using SDS/PAGE followed by immunoblot analysis with antibodies against Gcd10p or Gcd14p (A) or by silver staining (B). Input lane contains an aliquot of the material loaded on the column. (C) Aliquots (3%) of the indicated fractions were assayed for (m1A)MTase activity in reactions (0.05 ml) containing total tRNA (8.0 μM) isolated from yJA146 (gcd10Δ hcIMT4) or yJA158 (GCD10 hcIMT4) and S-adenosyl-l-[methyl-14C]methionine (10 μM). The acid-insoluble portion of each reaction was collected on nitrocellulose filters and dried, and the radioactivity was determined by liquid scintillation. The radioactivity incorporated is plotted against fraction number.
We asked whether the purified FlagGcd10p/Gcd14p complex could transfer a radiolabeled methyl group from Ado[methyl-14C]Met to total tRNA purified from a gcd10Δ strain, which lacks m1A (5). A peak of MTase activity eluted from the gel filtration column in exactly the same fractions that contained FlagGcd10p/Gcd14p complexes (Fig. 1C, gcd10Δ tRNA). Importantly, these fractions had no MTase activity with wild-type tRNA as substrate (Fig. 1C, GCD10 tRNA), as expected if FlagGcd10p/Gcd14p complexes catalyze only the m1A modification. Because gcd10Δ tRNAs differ from the wild-type only in lacking m1A (5), these results provide strong evidence that the FlagGcd10p/Gcd14p complex is the enzyme responsible for m1A modification of tRNAs.
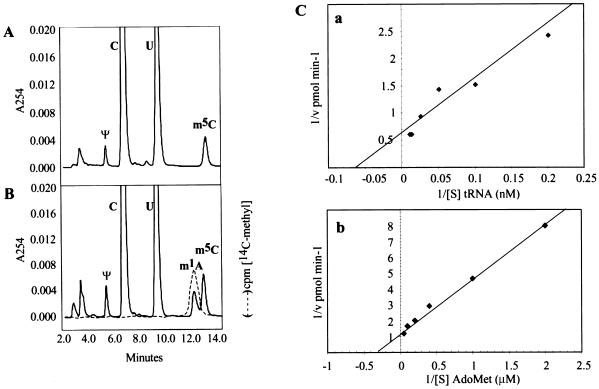
We next examined the ability of FlagGcd10p/Gcd14p complexes to synthesize m1A in tRNAiMet purified from a gcd10Δ mutant, which lacks this modification (5). The tRNAiMet (purified to at least 90% homogeneity) was incubated with FlagGcd10p/Gcd14p and Ado[methyl-14C]Met as above, and then hydrolyzed to nucleosides and resolved by HPLC to assay for the presence of radiolabeled m1A. As expected, the untreated tRNAiMet completely lacked m1A in the absorbance profile at 254 nm (A254) of its nucleosides (Fig. 2A), whereas the treated sample contained a peak of 14C radioactivity and A254 at the position expected for m1A (Fig. 2B). The identity of this peak was confirmed by coelution with commercially synthesized m1A (data not shown). We concluded that the FlagGcd10/Gcd14p complex has tRNA(m1A)MTase activity. By measuring initial reaction velocities over a range of AdoMet and initiator tRNAMet concentrations, we calculated Km values of 5 μM and 2.5 nM for AdoMet and tRNAiMet, respectively, for the purified FlagGcd10p/Gcd14p complex (Fig. 2C).
Figure 2.
FlagGcd10p/Gcd14p complexes specifically catalyze synthesis of m1A in purified tRNAiMet. Initiator tRNAMet purified from yJA146 (gcd10Δ hcIMT4) was incubated with purified FlagGcd10p/Gcd14p complexes and [methyl-14C]AdoMet as described in Fig. 1, hydrolyzed to nucleosides, and resolved by HPLC chromatography as described (5). The fractions were monitored for UV absorbance at 254 nm (A254; solid line) and assayed for cpm [14C-methyl] by flow scintillation analysis as described (31) (dotted line). (A) Mock-treated tRNA. (B) tRNA treated with FlagGcd10p/Gcd14p complexes. (C) Double reciprocal plots of tRNA(m1A)MTase activity of purified FlagGcd10p/Gcd14p complexes as a function of tRNAiMet concentration (1.2–60 nM) (a) or Ado[14C]Met concentration (0.5–20 μM) (b). Assays contained 10 μM Ado[14C]Met (a) or 10 μM tRNAiMet (purified from the gcd10Δ hcIMT4 strain) (b) and approximately 0.3 μg of enzyme in each reaction (100 μl). Lines were fitted by using linear regression analysis.
The AdoMet Binding Motif Is Required for Gcd14p Function in Vivo and (m1A)MTase Activity in Vitro.
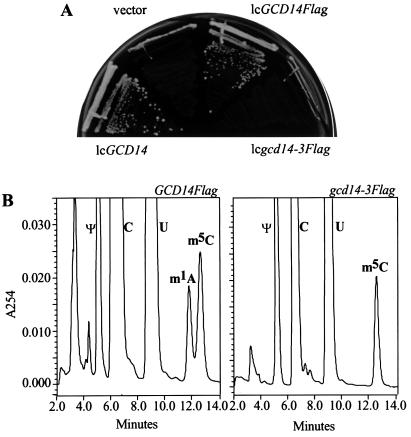
Gcd14p contains motifs I, post-I, and II found in AdoMet-MTases, suggesting that it binds the AdoMet in the Gcd10p/Gcd14p complex. To test this prediction, mutations were introduced into GCD14 that replaced two highly conserved glycine residues in motif I (Gly-118 and Gly-120) with alanines (gcd14–3Flag). One of the corresponding glycines in the mRNA-cap MTase of yeast (Abd1p) was shown to be essential for cap MTase activity and cell viability (10). The results in Fig. 3A demonstrate that gcd14–3Flag on a low-copy plasmid (lcgcd14–3Flag) failed to complement the temperature-sensitive phenotype of a gcd14–2 mutant. Note also that the Flag-tagged wild-type allele (GCD14Flag), and native GCD14 complemented gcd14–2 equally well (Fig. 3A). Immunoblot analysis of FlagGcd14p and FlagGcd14–3p using anti-Flag antibodies showed no difference in the expression levels of these two proteins in whole-cell extracts (data not shown). Thus, it appeared that gcd14–3Flag was defective for GCD14 function in vivo.
Figure 3.
Mutations in the AdoMet-binding motif I of Gcd14p eliminate m1A synthesis in vivo. (A) Transformants of yeast strain F516 (gcd14–2) containing low-copy URA3 plasmids bearing wild-type GCD14 (p2599), GCD14Flag (pJA148), gcd14–3Flag (pJA149), or vector alone, were streaked on SC medium lacking uracil and incubated at 36°C for 2 days. (B) HPLC chromatographs of total tRNA isolated from yeast strain yJA203 (gcd14Δ hcIMT4) harboring pJA148 (GCD14Flag) or pJA149 (gcd14–3Flag). The A254 was plotted against the time of elution. The identities of known nucleosides or their modified counterparts are indicated above or within the corresponding peaks (Ψ, pseudouridine; C, cytidine; U, uridine; m1A; m5C, 5-methylcytidine).
To confirm this last conclusion, GCD14Flag and gcd14–3Flag plasmids were introduced into a strain lacking chromosomal GCD14 (yJA203), which depends for viability on overexpression of initiator tRNAMet from the IMT4 gene on a high-copy plasmid (hcIMT4) (7). The presence of lcgcd14–3Flag in this strain (yJA213) did not permit loss of the hcIMT4 plasmid at 26°C, whereas lcGCD14Flag in strain yJA212 led to more than 80% plasmid loss under the same conditions (data not shown). Thus, gcd14–3Flag cannot overcome the lethality of deleting GCD14. HPLC analysis of nucleosides in total tRNA isolated from the strains bearing hcIMT4 and either GCD14-Flag or gcd14–3Flag showed that m1A was readily detectable in the former but absent in the latter strain (Fig. 3B). Because the point mutations in motif I of FlagGcd14p completely eliminated m1A from tRNAs in vivo, we conclude that gcd14–3Flag is a null allele.
To verify that the FlagGcd14–3p/Gcd10p complex was defective for m1A synthesis in vitro, we used Flag affinity resin to purify the wild-type FlagGcd14p/Gcd10p and mutant FlagGcd14–3p/Gcd10p complexes from strains yJA212 and yJA213 as described above. Coomassie staining and immunoblot analysis of SDS/PAGE separations of the resulting purified complexes suggested that Gcd10p and FlagGcd14p or FlagGcd14–3p were the major polypeptides present in these preparations (Fig. 4 A, lanes 1 and 2, and B, lanes 1–6). In parallel, we purified FlagGcd14p alone from a gcd10Δ+hcIMT4 strain bearing GCD14Flag on a high-copy plasmid to examine the requirement for Gcd10p in the (m1A)MTase activity of the complex. The resulting preparation of FlagGcd14p lacked Gcd10p, as expected, but contained a large amount of an unknown protein of 90 kDa (Fig. 4 A, lane 3, and B, lanes 7–9). Attempts to remove this protein from FlagGcd14p by gel filtration were unsuccessful (data not shown), leading us to suspect that it may be a protein chaperone that forms a stable complex with FlagGcd14p in the absence of Gcd10p.
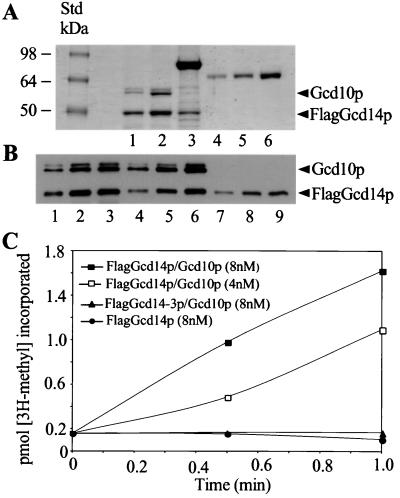
Figure 4.
In vitro tRNA(m1A)MTase activity requires wild-type FlagGcd14p and Gcd10p. (A) FlagGcd14p/Gcd10p (lane 1), FlagGcd14–3p/Gcd10p (lane 2), or FlagGcd14p alone (lane 3) were affinity-purified from yeast cell extracts, separated by SDS/PAGE, and stained with Coomassie brilliant blue (Bio-Rad). Included on the gel are known amounts of BSA (lanes 4–6), and SeeBlue molecular weight standards (NOVEX). The positions of FlagGcd14p or Gcd10p are indicated. (B) Immunoblot analysis of increasing amounts of FlagGcd14p/Gcd10p (lanes 1–3), FlagGcd14–3p/Gcd10p (lanes 4–6), or FlagGcd14p alone (lanes 7–9) by using antibodies that recognize either FlagGcd14p or Gcd10p. (C) Samples of the purified proteins shown in A at the indicated concentrations were assayed for tRNA(m1A)MTase activity by using Ado[methyl-3H]Met (30 μM) and tRNAiMet (150 nM) purified from yJA146 (gcd10Δ hcIMT4) as substrate. The acid-insoluble cpm produced in each reaction was determined at the indicated times, converted into pmol of [methyl-3H]tRNAiMet, and plotted against time.
Samples of the three preparations containing equimolar amounts of FlagGcd14p were tested for (m1A)MTase activity by using the assay described above. The FlagGcd14p/Gcd10p complex had a high specific activity (≈18 pmol of [methyl-3H]-tRNAiMet synthesized μg-1⋅min-1), whereas the FlagGcd14–3p/Gcd10p mutant complex and FlagGcd14p alone had no measurable activity, even at twice the enzyme concentration at which the FlagGcd14p/Gcd10p complex showed substantial (m1A)MTase activity (Fig. 4C). The activity of wild-type FlagGcd14p/Gcd10p was not impaired when mixed with twice the molar amount of FlagGcd14p alone (data not shown), indicating that the 90-kDa contaminant in the FlagGcd14p preparation did not interfere with (m1A)MTase function. The results in Fig. 4 demonstrate that Gcd10p and motif I of Gcd14p both are required for m1A synthesis in vitro. They also show unequivocally that the activity of our purified Gcd14p/Gcd10p preparation is not conferred by a copurifying (m1A)MTase.
Evidence that Gcd10p Is Required for tRNA Binding.
The requirement for motif I of Gcd14p for (m1A)MTase activity implies that Gcd14p is responsible for transferring the methyl group from AdoMet to tRNA. What essential role does Gcd10p serve in the production of m1A? As Gcd10p contains an RNA recognition motif and exhibits mRNA binding activity in vitro (11), we explored whether Gcd10p is required for stable binding of the tRNA substrate. The purified FlagGcd14p/Gcd10p, FlagGcd14–3p/Gcd10p, and FlagGcd14p preparations were assayed for binding radiolabeled tRNAiMet by using a filter binding assay (12). The wild-type and FlagGcd14–3p/Gcd10p mutant complexes bound tRNAiMet in a dose-dependent manner at levels far above the background level observed by using an equal amount of protein obtained from a mock purification using a strain containing untagged Gcd14p (Fig. 5, compare A and B with D). Thus, the mutations in motif I of FlagGcd14–3p had little or no effect on tRNA binding by the complex. In contrast, FlagGcd14p alone showed very little binding of tRNAiMet above background levels, supporting the notion that Gcd10p is required for high-affinity binding of tRNA substrates by the Gcd14p/Gcd10p complex.
Discussion
Previously we found that Gcd10p is required for m1A formation in all tRNAs in vivo (5) and that it resides in a stable nuclear complex with Gcd14p, a protein with strong sequence similarities to AdoMet-dependent MTases (7). Here we showed that mutants lacking Gcd14p also contain no detectable m1A, leading us to predict that the Gcd10p/Gcd14p complex represents the tRNA(m1A)MTase in yeast. We have provided strong support for this hypothesis by showing that affinity-purified Gcd10p/Gcd14p complex can transfer a 14C-methyl group from AdoMet to mutant tRNAs lacking only m1A at position 58, and we showed that the [14C]methyl group was incorporated into m1A. Because the purified complex could not generate m1A in wild-type tRNA, it appears to be specific for adenine residues at position 58. The (m1A)MTase activity coeluted with Gcd10p/Gcd14p complex from a sizing column; moreover, a mutation in motif I of the putative AdoMet-binding domain of Gcd14p produced a null allele in vivo (gcd14–3) and abolished (m1A)MTase activity of the purified complex in vitro. These last findings demonstrate conclusively that (m1A)MTase activity is conferred by the Gcd10p/Gcd14p complex and not by a copurifying contaminant. Because the gcd14–3 mutation did not reduce Gcd14p expression, alter its ability to form a stable complex with Gcd10p, or impair the tRNA binding activity of the complex, the simplest explanation is that the mutant complex was inactive because it failed to bind AdoMet.
Considering that all known tRNA MTases have only a single polypeptide component harboring AdoMet binding motifs (1), it is important to determine whether Gcd10p is required for the intrinsic (m1A)MTase activity of the Gcd10p/Gcd14p complex. It was conceivable that Gcd10p would be essential for efficient methylation of tRNA in the intact cell nucleus but dispensable for the reaction using purified components in vitro. For example, Gcd10p might be required to localize Gcd14p to the nuclear membrane where other tRNA processing and modification enzymes have been found (13, 14). We showed that purified Gcd14p alone had no (m1A)MTase activity and was greatly impaired for the ability to form a stable complex with tRNAiMet compared with the wild-type Gcd10p/Gcd14p complex. These observations suggest that Gcd10p is required for tight binding of tRNA substrates by the Gcd10p/Gcd14p complex. In this respect, it would resemble Arc1p, a protein that binds tRNA and functions as a cofactor for methionyl-tRNA and glutamyl-tRNA synthetases (15, 16). One caveat to this argument is that FlagGcd14p copurified with an abundant unknown contaminant. Although we showed that the contaminant did not inhibit (m1A)MTase activity of the FlagGcd14p/Gcd10p complex, it might interfere with the ability of FlagGcd14p to bind tRNA in the absence of Gcd10p. It remains possible that Gcd10p has additional functions in vivo, such as nuclear localization of the Gcd10p/Gcd14p complex or in channeling the m1A-containing tRNAs to other processing/modification enzymes (17).
All yeast tRNAs where the sequence has been determined contain adenine at position 58, but only 21 of 32 sequenced tRNAs contain m1A at this position (4). Thus, it will be important to determine how the Gcd10p/Gcd14p complex discriminates against the subset of tRNAs that lack m1A. One possibility is that Gcd10p specifically binds only those nascent tRNA transcripts destined for m1A methylation and presents them to Gcd14p for modification. However, considering that isolated Gcd10p binds globin mRNA (11), it may not be capable of distinguishing between different tRNAs. An alternative possibility is that Gcd10p contributes only nonspecific RNA binding affinity to the complex and that Gcd14p imposes tRNA-substrate specificity at the step of methyl group transfer. This would be akin to certain aminoacyl-tRNA synthetases that distinguish cognate from noncognate tRNAs primarily at the catalytic step rather than at the tRNA binding step (18).
Database searches reveal the presence of Gcd10p and Gcd14p orthologs in several multicellular eukaryotes, including humans (J.A. and A.G.H., unpublished observations), suggesting that Gcd10p- and Gcd14p-related proteins comprise the tRNA(m1A)MTase in diverse eukaryotes. Purification of tRNA(m1A)MTases from bovine liver (19) and extreme thermophilic archaebacteria (20, 21) indicated that the activity resided within single polypeptides of 95 kDa and 78 or 60 kDa, respectively; however, the genes encoding these proteins have not been described. It is possible that the bovine enzyme represents a higher molecular weight Gcd14p homolog that does not require a Gcd10p-related subunit for activity. Alternatively, those workers may have purified adenosine methylase activities whose natural substrate is not tRNA, but another cellular RNA. For example, there are reports that the 16S rRNA from species of Streptomyces contain 1-methyladenosine (22, 23). The bovine enzyme may represent the (m1A)MTase that is responsible for the rare m1A modification of cytoplasmic phenylalanine tRNA at position 14 or of several mitochondrial tRNAs at position 9 in mammals (4).
Interestingly, Gcd10p and Gcd14p orthologs were identified as components of a complex purified from trypanosomes that catalyzes the unique 2′-O-methylation of 4 nucleotides in the cap structure of the spliced leader RNA (17). Accordingly, Gcd10p- and Gcd14p-related proteins may participate in multiple RNA methylation reactions in different organisms. Although m1A is the only tRNA modification catalyzed by Gcd10p/Gcd14p in S. cerevisiae, and this appears to be its essential function, it may be responsible for additional, nonessential methylation events in other classes of RNA in yeast.
Acknowledgments
We thank G. R. Björk and Kerstin Jacobsson for HPLC analysis of modified nucleotides. We are indebted to Dolph Hatfield and Bradley Carlson for their guidance in purifying tRNAs. G. R. Björk and K. Jacobsson are supported by grants (BU-2930) and (proj 680) from the Swedish Natural Science Research Council and Swedish Cancer Society.
Abbreviations
- m1A
1-methyladenosine
- AdoMet
S-adenosylmethionine
- MTase
methyltransferase
- tRNAiMet
initiator methionine tRNA
- SC
synthetic complete
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090102597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090102597
References
- 1.Björk G R. In: tRNA: Structure Biosynthesis and Function. Söll D, RajBhandary U L, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 165–206. [Google Scholar]
- 2.Björk G R, Ericson J U, Gustafsson C E, Hagervall T G, Jönsson Y H, Wikström P M. Annu Rev Biochem. 1987;56:263–287. doi: 10.1146/annurev.bi.56.070187.001403. [DOI] [PubMed] [Google Scholar]
- 3.Åström S U, Byström A S. Cell. 1994;79:535–546. doi: 10.1016/0092-8674(94)90262-3. [DOI] [PubMed] [Google Scholar]
- 4.Sprinzl M, Horn C, Brown M, Loudovitch A, Steinberg S. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson J, Phan L, Cuesta R, Carlson B A, Pak M, Asano K, Björk G R, Tamame M, Hinnebusch A G. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basavappa R, Sigler P B. EMBO J. 1991;10:3105–3111. doi: 10.1002/j.1460-2075.1991.tb07864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvo O, Cuesta R, Anderson J, Gutierrez N, Garcia-Barrio M T, Hinnebusch A G, Tamame M. Mol Cell Biol. 1999;19:4167–4181. doi: 10.1128/mcb.19.6.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kagan R M, Clarke S. Arch Biochem Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- 9.Niewmierzycka A, Clarke S. J Biol Chem. 1999;274:814–824. doi: 10.1074/jbc.274.2.814. [DOI] [PubMed] [Google Scholar]
- 10.Mao X, Schwer B, Shuman S. Mol Cell Biol. 1996;16:475–480. doi: 10.1128/mcb.16.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Barrio M T, Naranda T, Cuesta R, Hinnebusch A G, Hershey J W B, Tamame M. Genes Dev. 1995;9:1781–1796. doi: 10.1101/gad.9.14.1781. [DOI] [PubMed] [Google Scholar]
- 12.Bovee M L, Yan W, Sproat B S, Francklyn C S. Biochemistry. 1999;41:13725–13735. doi: 10.1021/bi991182g. [DOI] [PubMed] [Google Scholar]
- 13.Westaway S K, Abelson J. In: tRNA: Structure, Biosynthesis, and Function. Soll D, RajBhandary U L, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 79–92. [Google Scholar]
- 14.Rose A M, Belford H G, Shen W C, Greer C L, Hopper A K, Martin N C. Biochemie. 1995;77:45–53. doi: 10.1016/0300-9084(96)88103-x. [DOI] [PubMed] [Google Scholar]
- 15.Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt E C. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 16.Simos G, Sauer A, Fasiolo F, Hurt E C. Mol Cell. 1998;1:235–242. doi: 10.1016/s1097-2765(00)80024-6. [DOI] [PubMed] [Google Scholar]
- 17.Wolin S L, Matera A G. Genes Dev. 1999;13:1–10. doi: 10.1101/gad.13.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Schimmel P R, Soll D. Annu Rev Biochem. 1979;48:601–648. doi: 10.1146/annurev.bi.48.070179.003125. [DOI] [PubMed] [Google Scholar]
- 19.Glick J M, Leboy P S. J Biol Chem. 1977;252:4790–4795. [PubMed] [Google Scholar]
- 20.Morozov I A, Gambaryan A S, Lvova T N, Nedospasov A A, Venkstern T V. Eur J Biochem. 1982;129:429–436. doi: 10.1111/j.1432-1033.1982.tb07068.x. [DOI] [PubMed] [Google Scholar]
- 21.Yamazaki N, Hori H, Ozawa K, Nakanishi S, Ueda T, Kumagai I, Watanabe K, Nishikawa K. Biosci Biotechnol Biochem. 1994;58:1128–1133. doi: 10.1271/bbb.58.1128. [DOI] [PubMed] [Google Scholar]
- 22.Beauclerk A A, Cundliffe E. J Mol Biol. 1987;193:661–671. doi: 10.1016/0022-2836(87)90349-4. [DOI] [PubMed] [Google Scholar]
- 23.Ballesta J P, Cundliffe E. J Bacteriol. 1991;173:7213–7218. doi: 10.1128/jb.173.22.7213-7218.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito H, Fukada Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherman F, Fink G R, Lawrence C W. Methods of Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1974. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 27.Yon J, Fried M. Nucleic Acids Res. 1989;17:4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gietz R D, Sugino A. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 29.Boeke J D, LaCroute F, Fink G R. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 30.Phan L, Zhang X, Asano K, Anderson J, Vornlocher H P, Greenberg J R, Qin J, Hinnebusch A G. Mol Cell Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian Q, Curran J F, Björk G R. J Bacteriol. 1998;180:1808–1813. doi: 10.1128/jb.180.7.1808-1813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]