Abstract
Intact Escherichia coli ribosomes have been projected into the gas phase of a mass spectrometer by means of nanoflow electrospray techniques. Species with mass/charge ratios in excess of 20,000 were detected at the level of individual ions by using time-of-flight analysis. Once in the gas phase the stability of intact ribosomes was investigated and found to increase as a result of cross-linking ribosomal proteins to the rRNA. By lowering the Mg2+ concentration in solutions containing ribosomes the particles were found to dissociate into 30S and 50S subunits. The resolution of the charge states in the spectrum of the 30S subunit enabled its mass to be determined as 852,187 ± 3,918 Da, a value within 0.6% of that calculated from the individual proteins and the 16S RNA. Further dissociation into smaller macromolecular complexes and then individual proteins could be induced by subjecting the particles to increasingly energetic gas phase collisions. The ease with which proteins dissociated from the intact species was found to be related to their known interactions in the ribosome particle. The results show that emerging mass spectrometric techniques can be used to characterize a fully functional biological assembly as well as its isolated components.
In recent years mass spectrometry has become the method of choice for a number of important aspects of experimental structural biology. These include: the characterization of the stability and folding behavior of proteins under a wide range of conditions (1–3), the identification of subpicomole quantities of proteins from two-dimensional gels (4), the de novo sequencing of peptides and subsequent cloning of novel proteins (5), and the analysis of the components of single vesicles (6). These applications have driven mass spectrometry to new levels of detection from a range of complex biological matrices. In addition, high-resolution ion cyclotron resonance mass spectrometry has enabled the isolation of individual ions from polyethylene glycol (7) and DNA (8), with masses in excess of 108 Da. As well as being able to characterize highly charged polymers, it has been possible to detect signals from noncovalent complexes of small molecule ligands bound to proteins and of multiprotein complexes (9, 10). As the size of such assemblies increases, however, the number of charges acquired during the electrospray process increases less rapidly than the total mass. This phenomenon has been attributed to ionic interactions in the intermolecular interfaces and the appropriation of negatively charged counterions from the volatile buffers used in the analysis of such species (11). The net result of these effects is that large multimolecular complexes, such as the 2.3-MDa ribosome, have been outside the mass range of conventional mass spectrometers. In previous mass spectrometry experiments disruption of the ribosome enabled the identification of the contact sites between ribosomal proteins and RNA (12) and a novel protein component from the Saccharomyces cerevsiae ribosome (13). In addition we have shown that the component proteins and groups of up to five proteins in noncovalent complexes can be detected after exposure of solutions of ribosomes to the electrospray process (14). From the nature of the proteins observed in these experiments we inferred that they had dissociated from intact particles in the gas phase of the spectrometer. In this paper we show that this is indeed the case and demonstrate that it is possible to maintain the noncovalent associations in Escherichia coli ribosomes in the gas phase and record spectra by using time-of-flight (ToF) mass analysis.
Materials and Methods
Preparation of Ribosomes and Subunits.
Ribosomes were harvested from E. coli strain MRE600 by standard protocols (15), and buffer was exchanged in 10 mM ammonium acetate (Sigma) (pH 6.4) for the 30S and in 10 mM magnesium acetate (Sigma) (pH 6.4) for spectra of the 50S and intact 70S species. 50S subunits in the absence of the L7/L12/L10 complex were prepared by using a standard protocol (16). Cross-linked ribosomes were prepared by exposure to UV radiation (254 nm) at a distance of 5 cm from the UV lamp (Spectralight, Cambridge) for 10 min. The cross-linked sample was separated from small molecule contaminants by gel-filtration using a Sephacryl 5-300 column equilibrated with 10 mM ammonium acetate, pH 6.4.
Mass Spectrometry.
Aliquots (1–2 μl) of solutions containing 2–5 μM of ribosomes or their subunits were placed in a nanoflow needle prepared in-house as described (17). Spectra were recorded on Micromass Q-ToF, LCT, and Platform mass spectrometers (Micromass, Manchester). The Q-ToF consists of a nanoflow electrospray interface (18), a quadrupole mass analyzer followed by a hexapole collision cell and a ToF mass analyzer. Ions were focused by a radio frequency (RF) lens before transmission to the quadrupole, which was used in RF-only mode as a wide bandpass filter. The ions then were conducted through the hexapole collision cell. Collisional cooling was used (19) such that the pressures in the quadrupole region were carefully balanced to allow optimal transmission of high-mass ions. A reduction of the source pumping speed in combination with an input of dry argon gas into the collision cell allowed the analyzer pressure to be increased from the normal operating pressure of 8.0 × 10−4 Pa to 9.0 × 10−3 Pa. Ions were pulsed into the ToF with an acceleration voltage of 8 kV. A pulse rate of 4 kHz was used for detection with a microchannel plate detector. Acquisition was achieved by using a time-to-digital converter operating at 1 GHz. The LCT mass spectrometer consists of a nanoflow source, a hexapole ion guide, and a ToF analyzer. The instrument was modified to allow the admission of nitrogen gas in the hexapole region of the spectrometer (pressure reading 4.7 × 10−6 mBar). The cross-linked ribosomes were subjected to increasing cone voltages up to 200 V and reduced pressure in the hexapole ion guide (pressure reading 2.6 × 10−6 torr). High-energy spectra recorded on the Micromass Platform single quadrupole mass spectrometer were acquired as described (14). For all mass spectrometers the tuning parameters were held constant, except where stated, with a needle voltage of 1.5 kV, a skimmer cone voltage of 150 V, and a skimmer offset of 4 V.
Results and Discussion
Mass Measurement of the Intact 30S Subunit.
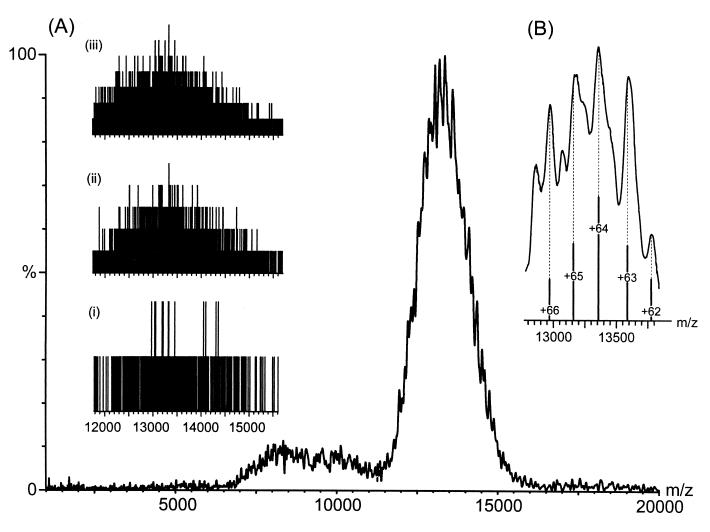
A solution containing the small subunit of the E. coli ribosome was placed in a nanoflow capillary and analyzed on a quadrupole ToF mass spectrometer. A carefully balanced regime of pressure gradients throughout the mass spectrometer was found to be effective in reducing the explosive nature of the initial desolvation step and minimizing the energy of collisional processes in the gas phase (20). Fig. 1 shows the raw data during acquisition of the small subunit of the E. coli ribosome collected in this manner. It reveals that few ions are detected in a single acquisition step but with signal averaging over successive acquisitions an overlapping series of peaks, centered at m/z 13,000, can be observed. A notable feature of this spectrum is the broad nature of the peaks (ca. 150 m/z units at 80% intensity). This value is much greater than predicted from simulation of the natural abundance isotopes and the resolving power of the instrument, which gives the width of each charge state as 0.3 m/z units at 80% intensity and the separation between neighboring charge states as greater than 200 m/z units. Simulations of the natural abundance isotope distributions were calculated from the elemental formulae by using two different software packages from masslynx (Micromass) and xmass (Bruker, Billerica, MA), which uses an algorithm described previously (34). One consequence of these broad peaks is that it is difficult to determine the mass and hence the charge of the peak. Iteration of the charge across the series of peaks was carried out to find the optimum fit of m/z values to the experimental data (21) from which the mass was determined as 852,187 ± 3,919 Da. The measured m/z values are 12,263.6, 12,398.5, 12,515.0, 12,689.4, 12,839.6, 13,046.7, 13,343.6, 13,577.6, 13,788.3, 13,954.3, and 14,197.8. The charge was iterated across the series of peaks from +54 to +78. From the values of the calculated molecular masses, based on these charge state assignments, the assignment that gave rise to the lowest value of the SD was taken to provide the best fit to the experimental data. The theoretical mass, calculated from the elemental formula¶ of the 21 proteins and 16S RNA molecule is 846,681 Da. The mass determined experimentally for the 30S subunit is therefore larger by 5,506 Da than that anticipated from the component macromolecules. This can be attributed to trapping of small molecules or counterions in the large intermolecular interfaces, a phenomenon observed in studies of smaller complexes (17) and anticipated from the long protein-free stretch of RNA visible from x-ray analysis of the small subunit of the ribosome (22). However, the measured mass is sufficiently close to the theoretical mass for the 30S subunit to reveal that all 21 proteins and the 16S RNA are present in the macromolecular complex.
Figure 1.
Nanoflow electrospray mass spectrum of the 30S subunit from E. coli ribosomes from m/z 2,000–20,000 with inserts of the raw data (A) and an expansion of the apex of the peak (>80% intensity) (B). The histograms recorded for the raw data on the Q-ToF mass spectrometer show a plot of m/z against ion counts and represent single ion hits on a microchannel plate detector. (i) A single 10-sec acquisition step in which single ions are visible above the level of background noise. (ii and iii) The summation of five and 15 acquisition steps, respectively. The most intense signal in iii is produced from seven ions per bin. The low yield of intact ions arises either from their decomposition during transmission or from a lack of focusing of high m/z species. The expansion of the apex of the peak shows five partially resolved charge states used in the calculation of the average mass.
The Stability of the Intact 70S Ribosome.
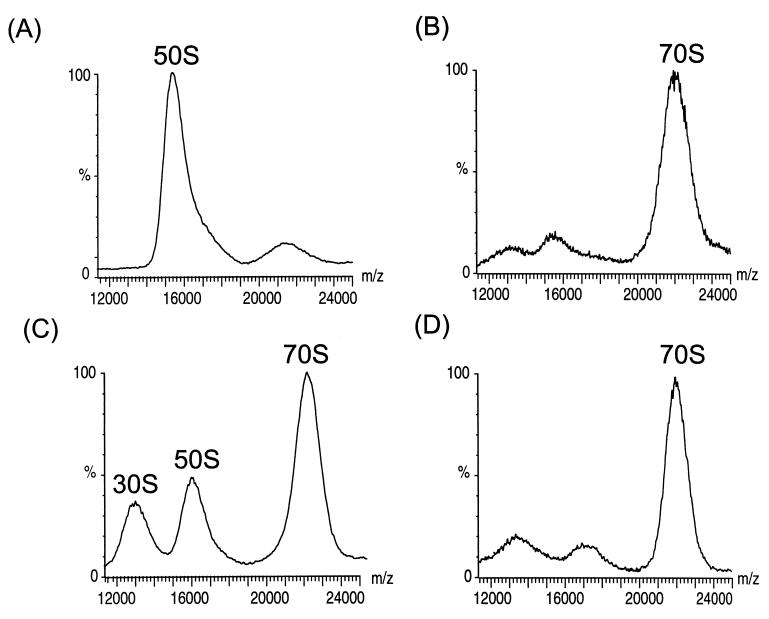
Fig. 2 A and B shows spectra obtained for the large (50S) subunits and intact (70S) ribosomes obtained in a similar manner to that described above. The assignment of the charge states is not possible for these species as their broad nature leads to considerable overlap. This broadening is increased by ion binding resulting from the addition of 5 mM Mg2+ to maintain the integrity of the 70S particle (23). Given the level of information available for the ribosome the masses of the intact subunits could be calculated¶. This information, together with the measured m/z values of 15.9 kDa/e and 22 kDa/e of the center of the peaks assigned to the 50S and 70S particles, allows the average charges to be calculated as +90 and +105, respectively. Because the isolated 30S has an average of 65 charges (Fig. 1) an average of 50 charges must be buried as a consequence of the formation of the 70S particle from the 50S and 30S subunits. This finding suggests that ionic interactions on the surfaces of the two subunits mediate their association into the intact particle. Further evidence for the importance of ionic interactions in the ribosome assembly comes from an experiment in which spectra were obtained after electrospray from the same solution used to observe the intact spectrum but after a 3-fold dilution to give a final Mg2+ concentration of 1.7 mM. Under these solution conditions, two lower m/z species are observed (Fig. 2C) with m/z ratios corresponding to the individual subunits. Previous studies of E. coli ribosomes have shown that as the concentration of Mg2+ in solution is reduced the two subunits dissociate (24).
Figure 2.
Investigation of the stability of intact ribosomes in the presence of Mg2+ and after cross-liking of ribosomal proteins to rRNA. Nanoflow electrospray mass spectra of (A) 50S and (B) 70S ribosome particles in the presence of 5 mM Mg2+, (C) after a 3-fold dilution of the solution, and (D) after cross-linking by exposure to UV radiation. The overall quality of the spectrum is affected by the presence 5 mM Mg2+ in the sample buffer (B), an improvement in the signal-to-noise ratio is observed when the solution is diluted 3-fold (C) and in the absence of Mg2+ in the spectrum of the cross-linked particle (D). The mass spectra were recorded on the LCT mass spectrometer with collisional damping to maintain the integrity of the intact particles. The spectrum of the cross-linked ribosome (D) was recorded with a cone voltage of 200 V and with reduced pressure in the hexapole region.
The stability of the intact particle in the gas phase was investigated further by cross-linking of ribosomal proteins to rRNA by exposure to UV radiation. This procedure has been used previously and shown to link ribosomal proteins to the RNA molecules (25). From the recent x-ray analysis of the 50S subunit 12 proteins were located and shown to be evenly dispersed with protein α-helices and β-strands interacting with minor or major grooves of duplex RNA (26). A spectrum obtained after UV radiation is shown in Fig. 2D and indicates that the intact cross-linked ribosomes are stable even in the absence of Mg2+, which leads to an improved signal-to-noise ratio in the spectrum (compare to Fig. 2B) and is consistent with a higher stability of the intersubunit contacts as a consequence of cross-linking (compare to Fig. 2C). The stability of the cross-linked ribosomes was further investigated when subjected to increased acceleration and under reduced pressure within the spectrometer (see Materials and Methods). The results show that the particle remains intact under higher collision energies and lower vacuum than the intact particle from the unmodified sample. Moreover the width of the charge state distribution is reduced (1,300 m/z units instead of 2,400 m/z units at 50% intensity for the cross-linked and unmodified samples, respectively), presumably because of the absence of Mg2+ binding to charge states and the higher energy regime that could be used during the acquisition of the spectrum of the cross-linked ribosome. Furthermore, because the peaks in the spectra of the cross-linked and unmodified ribosome in the presence of Mg2+ occur at the same m/z value, this demonstrates that component proteins, even in the absence of cross-linking, do not dissociate from the 70S ribosome under the mass spectometric conditions used here.
Controlled Dissociation of the 50S Subunit.
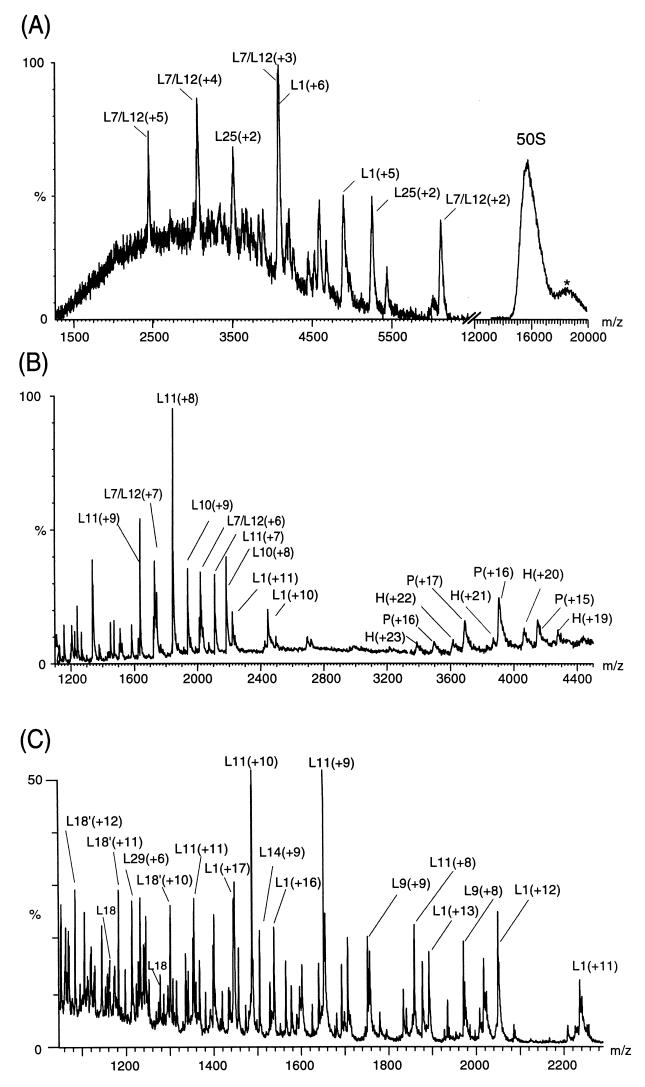
In a previous study of ribosomes using mass spectrometry we detected only the signals of proteins fully dissociated from the ribosome assembly (14). These proteins corresponded to those reported to bind most weakly to the particle in solution (16, 27). In particular, the stalk region of the 50S subunit is readily dissociated in solution as a noncovalent complex of its five component proteins. As well as the individual proteins from this region, the intact complex of five proteins could be detected in mass spectra resulting from both the 70S and 50S particles (14). This finding prompted us to explore in the present study whether controlled dissociation of the ribosome particles could be achieved by manipulating the energy of gas phase collisions. The results of this experiment are shown for the 50S subunit in Fig. 3. As the collision energy is increased there is a marked decrease in the intensity of the charge states assigned to the intact 50S subunit. This decrease is coupled with the appearance of signals at lower m/z values and with a broad distribution of charge states at higher m/z values than the intact particle (Fig. 3A). This finding is consistent with previous studies of smaller protein complexes where fragments are observed that are complementary in both mass and charge to the original species (28). Consequently the broad distribution of peaks at an average m/z value of 18,500 is assigned to a distribution of charge states from 50S subunits from which individual proteins have dissociated and with fewer positive charges than the intact 50S subunit. The most intense peaks at low m/z can be assigned on the basis of their m/z values to the individual proteins L1 and L7/L12. Two copies of L7/L12 are located at the tip of the stalk region and L1 forms a characteristic nodule seen in electron micrographs (29).
Figure 3.
Controlled dissociation of the 50S subunit (A) at low collision energy, (B) in the absence of collisional damping processes, and (C) under high energy conditions. The latter is from the 50S subunits from which the stalk complex had been removed before analysis. The peaks arising from individual proteins are labeled with their charge states in brackets. The spectra were obtained with a high cone voltage on the LCT (A), in the absence of argon gas in the collision cell and under low vacuum conditions in the quadrupole on the Q-ToF (B), and with no pressure control on the Platform (C) (14). The peak at m/z 18,500, labeled *, is attributed to the complementary fragment after dissociation of monomeric proteins from the intact 50S subunit whereas P and H are assigned to the pentamer, comprising of two copies of L7/L12 and L10, and the hexamer with the addition of protein L11. The peaks labeled L18′ in C correspond in mass to posttranslational modification of the protein L18. The protein L7 is N-acetyl L12 and although the species has been resolved previously (10, 14) no distinction is made in the spectra shown here because of the wide m/z range that has been displayed.
In an additional series of experiments the effects of collisional damping were removed by manipulating the pressure gradients leading to an increased energy of the collision process. This gives rise to a spectrum (Fig. 3B) in which the five-protein stalk complex is maintained intact together with another species found to be the stalk complex associated with the protein L11. L11 has been found from cross-linking experiments to be located proximal to the stalk region and to interact strongly with the stalk complex in solution (30). Moreover, under the highest collision energy regime the 50S subunit from which the pentameric stalk complex had been removed revealed that the most intense signals were from L11 (Fig. 3C). The removal of the stalk is likely to have destabilized the interactions of L11 with the remainder of the subunit. A large number of individual proteins are present in this spectrum. An additional 15 proteins were identified from at least three consecutive charge states (Table 1). This set contains L18, which appears as two peaks for each charge state in the spectrum and suggests that a proportion of L18 is posttranslationally modified. Interestingly, the protein L1 was observed in all three spectra recorded under the different energy regimes and was found to have average charge states of +5, +10, and +17 under conditions of increasing collision energy. This finding reflects the production of a more highly charged monomeric protein and is consistent with the results observed for smaller protein complexes (28). It is noteworthy that the majority of the proteins that were assigned in all three spectra are among those with the lowest solution pI values (31). This observation suggests that their interactions with the negatively charged RNA might be weak and implies that these proteins not observed in the mass spectra remain attached to the RNA molecules, even under the highest energy regime. Many more peaks were observed in this high-energy spectrum; however, the extensive overlap of their charge states renders their assignment ambiguous. MS/MS strategies are in progress to assign fully these spectra. The results of these experiments using different collisional energies show that the proteins that dissociate most readily from the 50S subunit are located at the periphery of the particle and are not those involved in strong interactions with the two ribosomal RNA molecules (26, 32, 33).
Table 1.
Comparison of theoretical and experimental masses for the dissociation products observed in the spectra shown in Fig. 3
| Energy level | Protein | Experimental mass, Da | Theoretical mass, Da |
|---|---|---|---|
| Low | L1 | 24,588.2 ± 1.4 | 24,598.5 |
| L7 | 12,195.9 ± 8.8 | 12,206.1 | |
| Intermediate | L10 | 17,571.8 ± 7.3 | 17,580.4 |
| L11 | 14,863.3 ± 4.9 | 14,872.5 | |
| L12 | 12,154.1 ± 0.8 | 12,164.0 | |
| L7/L10/L12 | 66,349.3 ± 3.9 | 66,320.1 | |
| L7/L/10/L11/L12 | 81,214.5 ± 18.5 | 81,192.5 | |
| High | L5 | 20,159.8 ± 2.9 | 20,170.4 |
| L9 | 15,763.0 ± 1.2 | 15,769.1 | |
| L11 | 14,872.5 ± 9.5 | 14,872.5 | |
| L17 | 14,359.0 ± 4.0 | 14,364.6 | |
| L18 | 12,764.0 ± 1.4 | 12,768.6 | |
| L18′ | 12,995.3 ± 1.1 | ||
| L20 | 13,360.2 ± 0.3 | 13,365.7 | |
| L21 | 11,562.4 ± 6.4 | 11,564.3 | |
| L24 | 11,187.8 ± 6.5 | 11,185.0 | |
| L14 | 13,534.0 ± 0.7 | 13,540.9 | |
| L28 | 8,880.9 ± 14.1 | 8,875.3 | |
| L29 | 7,272.2 ± 3.4 | 7,273.5 | |
| L31 | 7,856.1 ± 3.5 | 7,871.1 | |
| L36 | 4,470.5 ± 4.3 | 4,364.4 |
Ribosomal proteins identified at low and intermediate collision energies are presented in the first seven rows. The remaining proteins were identified under high-energy conditions in the absence of the stalk complex. The series labeled L18′ corresponds in mass to addition of 233 mass units to L18. The differences in standard deviations of the experimental masses arise from the variation in intensity of the dissociated proteins and the extent of overlap in the spectra.
Conclusions
The results presented here show that a large biological assembly and its intact subunits can be projected into the gas phase from solution conditions of neutral pH and high Mg2+ concentration. Moreover, the results suggest that the particles have sufficient intrinsic stability to retain their integrity for detection, under high vacuum, in the absence of bulk water. This finding indicates the importance of intramolecular interactions in maintaining stability, and in addition it has important practical consequences. In particular, it enables the nature and stability of such complexes under specific conditions to be probed. For example, the results of the present study of the ribosome and its subunits illustrate the importance of Mg2+ in maintaining the integrity of the intact particle and demonstrate the increased stability afforded by cross-linking proteins to the RNA. Moreover, low-energy collisions of the subunits with neutral gas molecules enable weakly bound proteins and their complexes to be removed from the surface of the assembly whereas higher-energy collisions lead to successively greater disruption. The pattern of this dissociation illustrates the feasibility of initiating the mapping of subunit interactions through controlled dissociation. These results for the ribosome not only pave the way for more detailed studies of its properties under a variety of conditions, for example in the presence of tRNA and the nascent chain, but also indicate that mass spectrometry can be used to probe structural features of even the largest biological complexes.
Acknowledgments
This paper is a contribution from the Oxford Centre for Molecular Sciences, which is supported by the Biotechnology and Biological Sciences Research Council, Engineering and Physical Sciences Research Council, and Medical Research Council. A.A.R. is grateful for support from Zeneca Pharmaceuticals and a Biotechnology and Biological Sciences Research Council studentship. D.R.B. was supported by a Hitchings-Elion Fellowship from the Burroughs Wellcome Fund. The research of C.M.D. is supported in part by a program grant from the Wellcome Trust and an International Research Scholars award from the Howard Hughes Medical Institute. C.V.R. acknowledges support from the Royal Society.
Abbreviation
- ToF
time of flight
Footnotes
The masses of the ribosomal particles were calculated from the RNA modification database (35) and from the known modifications of E. coli proteins found in the Swiss-Prot database. The small subunit is composed of 21 proteins (total mass 348,291.8 Da) and 1,542 bases in the 16S RNA (predicted mass 498,389 Da), giving a total of 846,680.8 Da. For the 50S subunit the 5S and 23S RNA give rise to masses of 38,731.4 Da and 938,935.7 Da, respectively, and the 33 proteins to a mass of 473,702.6 Da. This gives a total of 1,451,370 Da. Summation of the two subunits gives 2,298,050.4 Da for the 70S particle. The elemental formulae of the two subunits were calculated as C 30040, H 41783, N 10706, O 15206, P 1542, S 77 for the 30S subunit and as C 49801, H 67325, N 18181, O 27061, P 3024, S 112 for the 50S subunit.
References
- 1.Miranker A, Robinson C V, Radford S E, Aplin R T, Dobson C M. Science. 1993;262:896–900. doi: 10.1126/science.8235611. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Z, Smith D L. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson R S, Walsh K A. Protein Sci. 1994;3:2411–2418. doi: 10.1002/pro.5560031224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuster B, Mann M. Curr Opin Struct Biol. 1998;8:393–400. doi: 10.1016/s0959-440x(98)80075-4. [DOI] [PubMed] [Google Scholar]
- 5.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 6.Rubakhin S S, Garden R W, Fuller R R, Sweedler J V. Nat Biotechol. 2000;18:172–175. doi: 10.1038/72622. [DOI] [PubMed] [Google Scholar]
- 7.Smith R D, Cheng X, Bruce J E, Hofstadler S A, Anderson G A. Nature (London) 1994;369:137–139. [Google Scholar]
- 8.Chen R D, Cheng X H, Mitchell D W, Hofstadler S A, Wu Q Y, Rockwood A L, Sherman M G, Smith R D. Anal Chem. 1995;67:1159–1163. [Google Scholar]
- 9.Smith R D, Bruce J E, Wu Q, Lei Q P. Chem Soc Rev. 1997;26:191–202. [Google Scholar]
- 10.Rostom A A, Robinson C V. Curr Opin Struct Biol. 1999;9:135–141. doi: 10.1016/s0959-440x(99)80018-9. [DOI] [PubMed] [Google Scholar]
- 11.Rostom A A, Sunde M, Richardson S J, Schreiber G, Jarvis S, Bateman R, Dobson C M, Robinson C V. Proteins Struct. Funct. Genet. 1998. , Suppl. 2, 3–11. [DOI] [PubMed] [Google Scholar]
- 12.Thiede B, Urlaub H, Neubauer H, Wittmann-Liebold B. Methods Mol Biol. 1999;118:63–72. doi: 10.1385/1-59259-676-2:63. [DOI] [PubMed] [Google Scholar]
- 13.Link A J, Eng J, Schieltz D M, Carmack E, Mize G J, Morris D R, Garvik B M, Yates J R., 3rd Nat Biotechol. 1999;7:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 14.Benjamin D R, Robinson C V, Hendrick J P, Hartl F U, Dobson C M. Proc Natl Acad Sci USA. 1998;95:7391–7395. doi: 10.1073/pnas.95.13.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bommer U A, Burkhardt N, Junemann R, Spann C M T, Triana-Alonso F J, Nierhaus K H. In: Subcellular Fractionation: A Practical Approach. Graham J, Rickwoods D, editors. Oxford: Oxford Univ. Press; 1996. pp. 271–301. [Google Scholar]
- 16.Hamel E, Koka M, Nakamoto T. J Biol Chem. 1972;247:805–814. [PubMed] [Google Scholar]
- 17.Nettleton E J, Sunde M, Lai V, Kelly J W, Dobson C M, Robinson C V. J Mol Biol. 1998;281:553–564. doi: 10.1006/jmbi.1998.1937. [DOI] [PubMed] [Google Scholar]
- 18.Wilm M, Mann M. Int J Mass Spectrom Ion Processes. 1994;136:167–180. [Google Scholar]
- 19.Krutchinsky I V, Spicer V L, Ens W, Standing K G. J Am Soc Mass Spectrom. 1998;9:569–579. [Google Scholar]
- 20.Rostom A A, Robinson C V. J Am Chem Soc. 1999;121:4718–4719. [Google Scholar]
- 21.Tito, M. A., Tars, K., Valegard, K., Hadju, J. & Robsinson, C. V. (2000) J. Am. Chem. Soc., in press.
- 22.Clemmons W, May J, Wimberly B, McCutcheon J, Capel M, Ramakrishnan V. Nature (London) 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- 23.Gesteland R. J Mol Biol. 1966;16:67–84. doi: 10.1016/s0022-2836(66)80263-2. [DOI] [PubMed] [Google Scholar]
- 24.Hapke B, Noll H. J Mol Biol. 1976;105:97–109. doi: 10.1016/0022-2836(76)90196-0. [DOI] [PubMed] [Google Scholar]
- 25.Urlaub H, Kruft V, Bischof O, Muller E C, Liebold-Wittmann B. EMBO J. 1995;18:4578–4588. doi: 10.1002/j.1460-2075.1995.tb00137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ban N, Nissen P, Hansen J, Capel M, Moore P, Steitz T. Nature (London) 1999;400:841–847. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- 27.Petterson I, Hardy S J S, Liljas A. FEBS Lett. 1976;64:135–138. doi: 10.1016/0014-5793(76)80267-0. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz B L, Bruce J E, Anderson G A, Hofstadler S A, Rockwood A L, Smith R D, Chilkoti A, Stayton P S. J Am Soc Mass Spectrom. 1995;6:459–465. doi: 10.1016/1044-0305(95)00191-F. [DOI] [PubMed] [Google Scholar]
- 29.Malhotra A, Penczek P, Agrawal R K, Gabashvili I S, Grassucci R A, Junemann R, Burkhardt N, Nierhaus K H, Frank J. J Mol Biol. 1998;280:103–116. doi: 10.1006/jmbi.1998.1859. [DOI] [PubMed] [Google Scholar]
- 30.Walleczek J, Schuler D, Stoffler-Meilicke M, Brimacombe R, Stoffler G. EMBO J. 1988;7:3571–3576. doi: 10.1002/j.1460-2075.1988.tb03234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaltschmidt E. Anal Biochem. 1971;43:25–31. doi: 10.1016/0003-2697(71)90103-5. [DOI] [PubMed] [Google Scholar]
- 32.Nierhaus K. In: Ribosomes and Protein Synthesis: A Practical Approach. Spedding G, editor. Oxford: Oxford Univ. Press; 1990. pp. 161–188. [Google Scholar]
- 33.Moore P B. Annu Rev Biophys Biomol Struct. 1998;27:35–58. doi: 10.1146/annurev.biophys.27.1.35. [DOI] [PubMed] [Google Scholar]
- 34.Rockwood A L, VanOrden S L. Anal Chem. 1996;68:2027–2030. doi: 10.1021/ac951158i. [DOI] [PubMed] [Google Scholar]
- 35.Rozenski J, Crain P, McCloskey J. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]