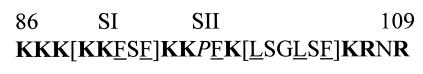Abstract
MARCKS-related protein (MRP) is a myristoylated protein kinase C substrate that binds calmodulin (CaM) with nanomolar affinity. To obtain structural information on this protein, we have engineered 10 tryptophan residues between positions 89 and 104 in the effector domain, a 24-residue-long amphipathic segment that mediates binding of MRP to CaM. We show that the effector domain is in a polar environment in free MRP, suggesting exposure to water, in agreement with a rod-shaped structure of the protein. The effector domain participates in the binding of MRP to CaM, as judged by the dramatic changes observed in the fluorescent properties of the mutants on complex formation. Intermolecular quenching of the fluorescence emission of the tryptophan residues in MRP by selenomethionine residues engineered in CaM reveals that the N-terminal side of the effector domain contacts the C-terminal domain of CaM, whereas the C-terminal side of the effector domain contacts the N-terminal domain of CaM. Finally, a comparison of the fluorescent properties of the myristoylated and unmyristoylated forms of a construct in which a tryptophan residue was introduced at position 4 close to the myristoylated N terminus of MRP suggests that the lipid moiety is also involved in the interaction of MRP with CaM.
The proteins of the myristoylated alanine-rich C kinase substrate (MARCKS) family are protein kinase C substrates that have been proposed to regulate the actin cytoskeleton (1). The family comprises two members: MARCKS itself is a ubiquitous 32-kDa protein, whereas MARCKS-related protein (MRP, also called MacMARCKS) is a 22-kDa protein expressed mainly in brain and reproductive tissues (2). MARCKS proteins share two conserved segments, namely the myristoylated N terminus and a central highly basic 24- to 25-residue-long segment, the effector domain, also called the “phosphorylation site domain” (1, 3). In vitro, MARCKS and MRP bind to calmodulin (CaM) with high affinity (Kd ≈ 5 nM) (4, 5). Although direct proof for an interaction between MARCKS proteins and CaM has so far not been obtained in cells, indirect evidence suggests that MARCKS proteins mediate crosstalk between the protein kinase C- and CaM-signal transduction pathways (for reviews, see refs. 6 and 7).
Two segments in MARCKS proteins are of interest with respect to their interactions with CaM.
(i) The Effector Domain.
The structure of the effector domain of MARCKS proteins has been the subject of several reports. Fig. 1 shows the amino acid sequence of the effector domain of MRP. This segment is highly basic (12 residues of 24 are lysines or arginines), but it also contains most of the large hydrophobic residues present in the intact protein (four phenylalanines and two leucines of seven). Although sequence analysis has previously led to the conclusion that this segment forms an α-helix (1), circular dichroic studies with peptides (8) and proteins (5, 9) suggest an extended structure for this segment. In the absence of information on the tertiary structure of MARCKS proteins, three different topologies can be proposed for the effector domain. The effector domain could be completely buried in a hydrophobic core (model A), which is unlikely because of the high content of charged amino acid residues. The effector domain could be on the surface of MARCKS proteins and partially exposed to water (model B). This model fits the proposed amphipathic structure of the effector domain (basic/hydrophobic residues). Finally, the effector domain could be completely exposed to water and either act as a hinge between the C- and N-terminal domains of MARCKS proteins (model C1) or form an exposed loop on the surface of the protein (model C2). The observation that MARCKS proteins are elongated molecules (5, 10) supports model C1. However, the presence of a proline at position 96 in the middle of the effector domain of MRP could potentially induce the formation of a kink or a turn (11), favoring model C2.
Figure 1.
Amino acid sequence of the effector domain of MRP. The amino acid residues K86-K109 are numbered starting from the N-terminal Gly residue of MRP, which becomes myristoylated. Basic amino acid residues are bold, and hydrophobic residues are underlined. P96, in the middle of the effector domain, is in italics. The two segments with mutated residues, SI (K89, K90, F91, S92 and F93) and SII (L99, S100, G101, L102 and F104), are shown in brackets.
(ii) The Myristoylated N Terminus.
Covalent modification of proteins with lipids, such as myristoylation in the case of MARCKS proteins, allows the interactions of these proteins with membranes (12). Myristoylation could also mediate protein–protein interactions. However, most proteins modified with lipids interact with other proteins on the membrane surface. It is therefore difficult to determine whether a loss in functionality after removal of the lipid moiety of a protein results primarily from a loss in protein–protein interaction or alternatively in protein–membrane interaction (13, 14). In this regard, MARCKS proteins can interact very efficiently with CaM in the absence of membranes (4, 5), which makes them appropriate proteins to investigate the role of myristoylation on protein–protein interaction.
MARCKS proteins are now available readily through Escherichia coli expression systems (4, 15). Furthermore, these proteins are highly soluble in aqueous solutions over a wide range of pH values. These conditions seem optimal for attempting a structural analysis of these proteins by x-ray crystallography or NMR spectroscopy. However, despite efforts from several investigators, the structure of MARCKS proteins has so far not been obtained. Their very irregular properties (unusual amino acid composition, lack of obvious secondary structures such as α-helices or β-stands, rod shape, myristoylation of the N terminus) (1, 3) are most certainly responsible for the lack of success in solving their structure. As to CaM, although the structures of the Ca2+ (16) and apo (17, 18) forms have been solved, high-resolution structures of ligands complexed to CaM have been obtained only with peptides corresponding to the binding site of myosin light chain kinase (19, 20), CaM-dependent protein kinase II (21), CaM-dependent kinase kinase (22), and the plasma membrane calcium pump (23); no data are available for an intact protein ligand.
Because neither MRP nor CaM contain tryptophan residues in their sequences, we have engineered tryptophan residues at selected sites in MRP and used them to probe the local conformation in MRP in the presence and absence of CaM. The conformation of the effector domain was investigated by creating a series of 10 mutants in which residues were systematically substituted by tryptophan. Similarly the conformation of the myristoylated N terminus of MRP was probed by introducing a tryptophan residue at position 4. Finally, we have also taken advantage of the ability of selenium engineered in CaM in the form of selenomethionine residues to quench the fluorescence of tryptophan engineered in MRP to investigate the MRP⋅CaM interface in more detail. We show that the use of intermolecular quenching of tryptophan fluorescence emission by selenomethionine allows a quite detailed molecular analysis of the interface between MRP and CaM.
Materials and Methods
Materials.
The plasmid pBB131NMT was a generous gift from Jeffrey Gordon (Washington University School of Medicine, St Louis, MO). The plasmid pET3dMRPHis has been described elsewhere (15). All oligonucleotides were synthesized by Microsynth (Balgach, Switzerland). Pfu polymerase was from Stratagene, the Qiaquick DNA purification kit from Qiagen, restriction enzymes and T4 DNA ligase from New England Biolabs, HisBind resin from Novagen, and phenylsepharose from Pharmacia Biotech. The CaM mutants and analogs were produced as described (24). Briefly, CaM* is a CaM construct with all four methionine residues in the C-terminal domain mutated to Leu, SeMet-CaM is a CaM construct with all nine methionine residues substituted by selenomethionine, and SeMet-CaM* is a CaM construct with all five methionine residues in the N-terminal domain substituted by selenomethionine and all four methionine residues in the C-terminal domain mutated to Leu. Dansylated CaM was obtained from Sigma, whereas wild-type CaM (bovine brain, high purity) was from Calbiochem. Single-use acrylamide fluorescence cuvettes (1-cm path length) were purchased from Sarstedt and acrylamide used for quenching experiments was from ICN. l-tryptophan for spectroscopic measurements was from Merck.
Mutagenesis.
Standard molecular biology techniques were used (25). All mutations in MRP were introduced by using the megaprimer method, which requires only one mutagenic primer per mutation (26, 27). We used two general nonmutagenic primers directed against the vector sequences surrounding the insert as described recently (28). Additionally, one mutagenic primer corresponding to the antisense sequence of the mrp gene but containing the sequence 5′-CCA-3′ coding for a tryptophan residue instead of the wild-type sequence was designed for each mutation (29). This mutagenic primer was used in a first PCR together with the general primer upstream of mrp and the plasmid pET3dMRPHis as the template to generate a megaprimer corresponding to the N-terminal part of MRP and already harboring the desired mutation. The PCR product was purified and used in a second PCR together with the second general primer downstream of mrp and the plasmid pET3dMRPHis as a template to generate full length mutated mrp.
The final PCR product was purified, double digested by NcoI/BamHI, purified again, and ligated into the NcoI/BamHI-digested pET3dMRPHis vector. The ligation mixture was used to electroporate E. coli JM109(DE3) cells. Colonies grown in the presence of 50 μg/ml ampicillin were characterized in respect to the presence of a plasmid containing an insert of appropriate size and their ability to overexpress recombinant protein after induction with isopropyl-d-thiogalactoside (IPTG). Plasmid DNA from selected colonies was sequenced (Microsynth AG) to confirm presence of the desired mutations.
Myristoylation, Expression, and Purification of MRP Mutants.
Plasmid DNA was electroporated into E. coli JM109(DE3) containing the plasmid pBB131NMT coding for N-myristoyl transferase. Colonies grown in the presence of 50 μg/ml ampicillin and 50 μg/ml kanamycin were tested for overexpression of recombinant protein after induction with IPTG. Large-scale cultures for protein production were grown from individual colonies and induced by IPTG as described (12, 15). Cells were harvested, lysed, and the recombinant protein isolated by chromatography on Ni2+ columns and phenylsepharose columns as described (15). The mutant proteins were extensively dialyzed against 10 mM Tris, pH 7.4, containing 1 mM EDTA, concentrated, flash frozen in liquid nitrogen, and stored at −20°C. The yield was determined by the Lowry assay (30) by using a correction factor of 1.6 (5). Note that coexpression of MRP with N-myristoyl transferase yields a protein that is correctly myristoylated as judged by the resistance of MRP to N-terminal sequencing by Edmann degradation and by the increase in mass of exact 210 Da measured by mass spectroscopy (unpublished data) (28).
All MRP mutants were found to be completely translated (the C-terminal histidine tag of MRP binds to the Ni2+ resin). The mutants were also quantitatively myristoylated by the N-myristoyl transferase expressed in E. coli as judged by the absence of unmyristoylated MRP on polyacrylamide gels (myristoylated MRP migrates slightly slower than unmyristoylated MRP during electrophoresis on 20% SDS polyacrylamide gels). Furthermore, possible contamination of the myristoylated MRP constructs with trace amounts of unmyristoylated proteins could be eliminated by chromatography on a phenylsepharose column (myristoylated MRP is retained on the column in the presence of 3 M NaCl, whereas unmyristoylated MRP is not) (15). The mutants were also recognized in Western blotting experiments by a polyclonal anti-MRP antibody. Purity was assessed by PAGE and found to be at least 95%. Although the additional 15-residue-long C-terminal histidine tag contains a thrombin cleavage site, the histidine tag was not removed because it does not quantitatively modify well-characterized properties of MRP such as phosphorylation by the catalytic subunit of protein kinase C (unpublished data), association with phospholipid membranes (15), and binding to CaM (this report).
Steady-State Fluorescence Spectroscopy.
We used an SLM–Aminco (Urbana, IL) spectrofluorometer for anisotropy measurements. All other fluorescence measurements were performed with a Jasco FP-777 spectrofluorometer. Usual settings included an excitation spectral bandwidth of 5 nm, an emission spectral bandwidth of 3 nm, a scan speed of 50 nm/min, and a “high” photomultiplier gain. The incubation buffer was composed of 10 mM Tris, pH 7.4/1 mM EDTA/100 mM NaCl/1.2 mM CaCl2/1 mM DTT. The samples were stirred continuously during titrations, and the temperature was maintained at 20°C. Stirring was interrupted to record the spectra. The MRP samples (1.25 μM) were preincubated at room temperature for at least 15 min in 1 ml of incubation buffer in single-use acrylamide cuvettes. The spectra recorded with an excitation wavelength of 280 nm were corrected for background and for dilution, if applicable. Maxima were determined by fitting a Gaussian function to the data after smoothing. This method gave the same results as a peak search using the numeric first derivative (not shown). To determine anisotropy values, measurements were performed as above at an excitation wavelength of 300 nm. Values obtained from all four polarisator settings were used to calculate the anisotropy, r, as given by Lakowicz (31). Quenching by acrylamide was achieved by addition of a 8-M acrylamide stock solution to the fluorophore solution, and emission was recorded at 350 nm. To reduce absorption by the quencher, the excitation wavelength was set to 295 nm. The data were corrected for background, dilution, and the inner filter effect (32, 33). The ratio of the fluorescence emission intensity of the samples in the absence vs. presence of acrylamide, F0/F, was plotted as a function of the acrylamide concentration, and the dynamic Stern–Volmer quenching constant, KSV, was obtained as the slope of a linear fit of the data at quencher concentrations up to 100 mM. Determination of the affinity of MRP for dansylated CaM was performed essentially as described (5), except that dissociation constants were determined by the algorithm of Creighton (34), assuming the formation of a 1:1 complex between dansylated CaM and MRP (5). Data were analyzed by using the programs sigmaplot (Jandel, San Rafael, CA), easyplot (Spiral Software, Chinle, AZ), and coreldraw (Corel, Ottawa, Canada).
Measurements with Met-CaM and SeMet-CaM.
Quenching of the fluorescence of tryptophan residues in MRP by CaM constructs containing selenomethionine residues was measured essentially as described previously (24). The MRP mutants (0.75 μM) were incubated at 20°C for 15 min in a buffer containing 10 mM Tris, pH 7.4, 0.2 mM CaCl2, and 1 mM DTT. A fluorescence emission spectrum was then recorded. CaM (CaM, CaM*, SeMet-CaM, or SeMet-CaM*) was added to MRP to obtain final concentrations of 2.1 μM and 0.7 μM for CaM and MRP, respectively. After a 15-min incubation, a fluorescence emission spectrum of the solution was recorded at 20°C. The emission spectra of the free MRP constructs were corrected for the contribution of the background by subtracting the spectrum of buffer. Similarly, the spectra of the MRP⋅CaM complexes were corrected for the contribution of free CaM. All spectra were further corrected for shifts along the y axis (fluorescence emission intensity) by setting the fluorescence emission intensity to zero at 450 nm. The ability of the selenomethionine residues in CaM to quench the fluorescence of tryptophan residues in MRP was quantified by calculating the quenching factor, i.e., the ratio of the maximal fluorescence emission intensity of the MRP constructs complexed to SeMet-CaM or SeMet-CaM* to the intensity of the same constructs complexed to CaM or CaM*, respectively. Finally, to correct for small differences in the concentrations of MRP measured in the presence of each of the four CaM constructs, all spectra were normalized so that the fluorescence emission intensity of the free MRP constructs at λmax is 1.
Results and Discussion
Tryptophan as a Selective Fluorescent Probe to Monitor Locally the Environment in MRP.
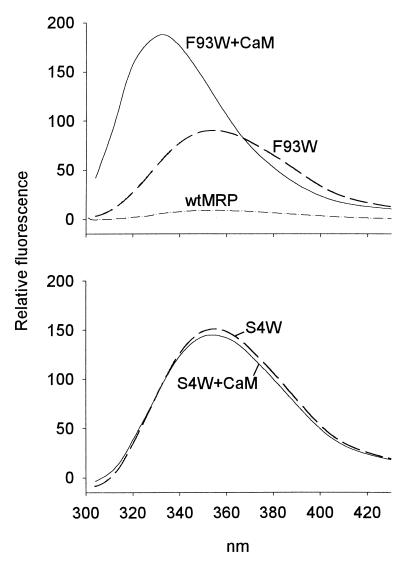
The successful engineering of tryptophan residues in MRP is illustrated with the unmyristoylated form of F93W, a mutant with a conservative substitution (Fig. 2 Upper). F93W (dashed line) exhibits a typical tryptophan emission spectrum with a λmax at 351 nm. In contrast, wild-type MRP (solid line), which does not contain tryptophan residues, shows only a background signal.
Figure 2.
Fluorescence emission spectra of unmyristoylated MRP constructs (1 μM) containing a single tryptophan residue at position 93 (Upper) or 4 (Lower). The spectra were recorded in the absence (dashed line) or presence (solid line) of 1.5 μM CaM. As a control, wild-type MRP, which does not contain a tryptophan residue, is shown (dashed-doted line, Upper).
Because CaM binds to MRP with nanomolar affinity (5), and because the effector domain is a major determinant of this interaction (4), the fluorescence properties of a tryptophan engineered in the effector domain of MRP should be sensitive to the formation of a complex between MRP and CaM. Indeed, Fig. 2 Upper also shows that the fluorescence emission spectrum of unmyristoylated F93W is blue shifted from 351 to 330 nm on binding to CaM, indicating the formation of a less polar environment in the vicinity of this residue. This shift is accompanied by an almost 2-fold increase in the fluorescence emission intensity. The specificity of this assay is demonstrated in Fig. 2 Lower: the fluorescence properties of an unmyristoylated MRP construct in which a tryptophan residue was introduced at position 4 close to the N terminus (S4W) are not modified after association of MRP with CaM. This lack of sensitivity is not because of a disruptive effect of the mutation on the formation of the complex, because unmyristoylated S4W as well as myristoylated S4W binds to CaM (5) with the same affinity (low nanomolar range) as wild-type MRP (data not shown).
Probing the Structure of the Effector Domain of MRP: The Tryptophan Scan.
To investigate the structure of the effector domain of MRP, a series of myristoylated MRP constructs, each containing a single tryptophan residue, were expressed in E. coli, purified to homogeneity, and their fluorescent properties were investigated. The mutations were divided in two segments (SI and SII), each segment sequentially mutated at five different positions, on either side of Pro-96 in the middle of the effector domain (see Fig. 1). Note that we have chosen to mutate Phe-104 rather than Ser-103, to provide an additional conservative mutation.
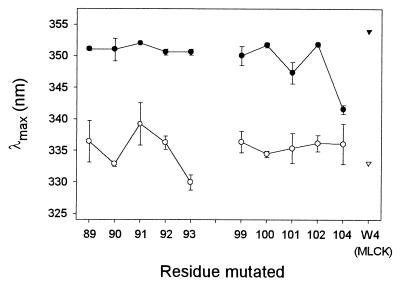
The 10 residues investigated in the effector domain show emission spectra with very similar λmax (range: 347–352 nm; mean: 350.8 nm) with the exception of F104W, whose λmax is at 342 nm (Fig. 3 ●). For comparison, the λmax of an aqueous solution of the free amino acid tryptophan was measured at 355 nm (data not shown). These results therefore show that all of the residues in SI and SII are in a polar environment, suggesting exposure to the aqueous phase. This conclusion is in agreement with models C1 or C2 presented in the Introduction.
Figure 3.
Fluorescence emission maxima, λmax, of myristoylated MRP constructs (1 μM) containing a single tryptophan residue at various positions in the effector domain. The spectra were recorded in the absence (●) or presence (○) of 1.5 μM CaM. The results are averaged from six to nine experiments in the absence and five experiments in the presence of CaM. Errors bars along the y axis are shown and are not apparent if smaller than the size of the symbols. As controls, the λmax of a peptide corresponding to binding site of myosin light chain kinase, which contains a single tryptophan residue at position 4, is shown for the free peptide (▾) as well as for the peptide bound to CaM (▿) (taken from ref. 24).
Taking advantage of the fluorescent properties of the tryptophan residues engineered in MRP, we have investigated how CaM interacts with the effector domain in the intact MRP protein. To exclude artifacts resulting from disruptive effects of the mutations on the formation of the complex, we have first measured the affinity of our mutants for CaM by using dansylated CaM as a fluorescent probe (5). The dissociation constants (Kd) for the mutants are increased only marginally (range: 5–27 nM; mean: 14 nM; individual data not shown) compared with wild-type MRP (4 nM). Thus, as already reported for a peptide corresponding to the binding site of myosin light chain kinase (36), replacing wild-type residues by tryptophan in the CaM-binding site does not significantly alter the strength of the complex formation.
For each of the residues investigated in SI and SII, binding of MRP to CaM results in a blue shift in the emission spectrum as well as an increase in the fluorescence emission intensity. On average, the λmax of the 10 mutants is blue shifted by 15 nm (range in the presence of CaM: 330–339 nm; mean: 335.3 nm), showing that complex formation reduces the polarity in the proximity of the effector domain (Fig. 3). Interestingly, the blue shift induced on binding of F104W to CaM (5.5 nm) is significantly smaller than obtained for the other mutants (12–21 nm). This effect can be attributed clearly to the emission spectrum of F104W in the absence of CaM: this spectrum is already blue shifted compared with the spectra of the other mutants, suggesting a somewhat “restricted” environment for residue 104 in free MRP. For comparison, the λmax of the single tryptophan in myosin light chain kinase and CaM kinase I peptides is shifted from 353 nm to 330 nm on binding to CaM (see also Fig. 3 Right ▾, ▿) (24).
Information on the mobility of the side chains of the effector domain was obtained by measuring the fluorescence emission anisotropy (r) for each of the mutants in the absence and presence of CaM. In the absence of CaM, all r values are close to zero (range: −0.001 − +0.016; mean: 0.007; individual data not shown). The average fluorescence anisotropy of the mutants is increased 7-fold on complex formation (range: 0.036–0.073; mean: 0.049; individual data not shown), showing that CaM partially immobilizes the residues in SI and SII.
Quenching of the fluorescence emission by acrylamide was also measured to assess the solvent accessibility of the residues mutated in the effector domain. Their fluorescence can be readily quenched in the absence of CaM with similar KSV values (range: 6.5–8.7 M−1; mean: 7.2 M−1; individual data not shown) with the exception of F104W, which has a KSV of 4 M−1. Almost three times more acrylamide is required on average to quench the fluorescence emission of the MRP mutants after their binding to CaM, as judged by the 3-fold decreased average KSV (range: 1.5–3.4 M−1; mean: 2.4 M−1; individual data not shown), demonstrating a decreased access of SI and SII to water in the complex.
Taken together, our data strongly suggest that all residues investigated in the effector domain are at the interface between MRP and CaM. These side chains are, however, still significantly less “restricted” by the environment compared with side chains present in the hydrophobic core of proteins [for comparison, the embedded Trp-187 in calcium-free annexin V has a λmax of 326 nm and a KSV of 0.4 M−1 at neutral pH (35)]. Thus we conclude that SI and SII are not completely shielded from the aqueous phase in the MRP⋅CaM interface.
Secondary Structure of the Effector Domain in the MRP⋅CaM Complex.
The technique of the tryptophan scan has been used to probe the secondary structure of the myosin light chain kinase peptide bound to CaM: the periodicity in the fluorescent properties of the series of mutated peptides was indicative of an α-helical conformation (36). Can this technique be applied to an analysis of the secondary structure of the effector domain of MRP? The data presented in Fig. 3 raise two comments regarding the secondary structure of SI and SII in the MRP⋅CaM complex: (i) Although the λmax values for the residues mutated in SI vary significantly in the MRP⋅CaM complex as a function of the position along the primary sequence of MRP, the length of SI (five residues) is not sufficient to allow us to identify a secondary structure. (ii) In contrast to the data obtained with the myosin light chain kinase peptide, the λmax for the residues mutated in SII are very similar in the MRP⋅CaM complex, suggesting a lack of secondary structure. Thus, in agreement with previous findings (5, 8, 9), our data indicate that SII is not part of an α-helix in the MRP⋅CaM complex.
Determining the Orientation of the Effector Domain of MRP in the CaM-Binding Site.
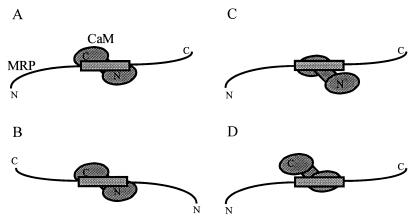
The N- and C-terminal domains of CaM each contain a cluster of methionine residues that mediate the binding of CaM to its ligands (19–23). Several topologies have been reported with respect to the association of peptide ligands with the N- and C-terminal domains of CaM (see also Fig. 4): With myosin light chain kinase (19, 20, 24) and CaM kinase I (24), the N-terminal side of the peptides associates with the C-terminal domain of CaM, whereas the C-terminal side of the peptides associates with the N-terminal domain of CaM (topology A). This orientation is not observed for other CaM ligands: a peptide encompassing the binding site of CaM-dependent kinase kinase associates with CaM in the opposite orientation (22) (topology B), whereas a peptide corresponding to the binding site of the plasma membrane calcium pump binds solely to the C-terminal domain of CaM (23) (topology C). No report has been published that describes a ligand binding solely to the N-terminal domain of CaM (topology D).
Figure 4.
Schematic representation of possible orientations of the effector domain of MRP in the MRP⋅CaM complex. (A) The C-terminal domain of CaM binds to the N-terminal side of the effector domain of MRP. (B) Opposite orientation. (C) The effector domain interacts solely with the C-terminal domain of CaM. (D) The effector domain interacts solely with the N-terminal domain of CaM. Note that MRP is shown in an extended conformation (model C1 in the Introduction).
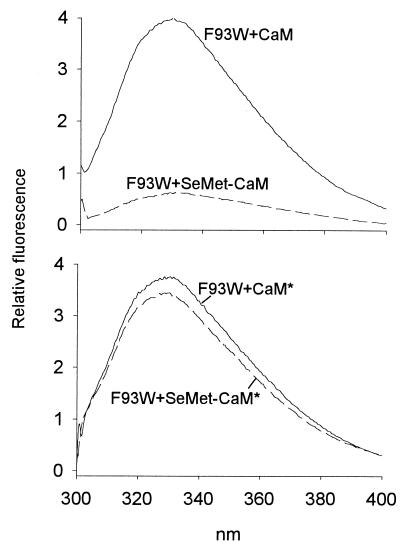
Which one of these topologies could be valid for the effector domain in the MRP⋅CaM complex (Fig. 4)? Because selenomethionine can be biosynthetically incorporated into recombinant CaM (37), and because selenium can act as an efficient quencher of tryptophan fluorescence (31), we have previously taken advantage of these properties to determine the orientation of the complex between the myosin light chain kinase peptide and CaM by fluorescence spectroscopy (24). In agreement with NMR and x-ray studies (19, 20), we have shown that the myosin light chain kinase peptide binds to CaM with topology A (see Fig. 4A). We now apply this technique to the intact MRP protein. Fig. 5 illustrates this approach for myristoylated F93W. In the presence of SeMet–CaM (see Materials and Methods for a description of the CaM constructs), the fluorescence emission intensity of F93W is reduced dramatically, evidently as a result of the quenching of Trp-93 fluorescence by the selenomethionine residues in CaM. This effect is lost completely when SeMet-CaM* is used instead of SeMet-CaM. Quenching of the fluorescence of F93W by SeMet-CaM must therefore be because of the selenomethionine residues present in the C-terminal domain of CaM. We therefore conclude that residue 93 in the effector domain of MRP is associated with the C-terminal methionine cluster of CaM.
Figure 5.
Quenching of the fluorescence emission of myristoylated F93W by selenomethionine residues incorporated into CaM. Fluorescence spectra of 0.7 μM F93W were recorded separately in the presence of 2.1 μM CaM (Upper, solid line), SeMet-CaM (Upper, dashed line), CaM* (Lower, solid line), or SeMet-CaM* (Lower, dashed line). To correct for small variations in the concentrations of F93W measured in the presence of each of the four CaM constructs, all spectra were normalized so that the fluorescence emission intensity at λmax of F93W in the absence of the CaM constructs is 1.
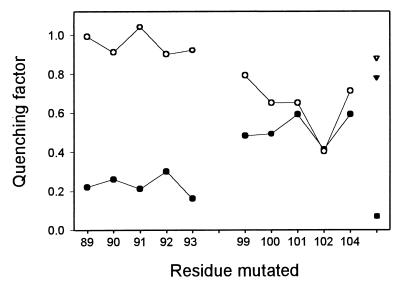
The experiment presented with F93W was extended to our entire set of myristoylated mutants. Fig. 6 shows that the fluorescence of all residues in SI is quenched dramatically by SeMet-CaM (range: 0.16–0.30; mean: 0.23) but not by SeMet-CaM* (range: 0.90–1.04; mean: 0.95), demonstrating that the side chains in SI are close to the C-terminal methionine cluster of CaM. The fluorescence of the mutated residues in SII is also quenched by SeMet-CaM (range: 0.41–0.59; mean: 0.51), however significantly less compared with the residues in SI. Importantly, replacing SeMet-CaM by SeMet-CaM* only weakly reduces the average quenching in SII (range: 0.40–0.79; mean: 0.64). Of particular interest is the observation that the fluorescence of L102W is not further decreased when SeMet-CaM is replaced by SeMet-CaM*. These observations strongly suggest that SII interacts with the N-terminal domain of CaM.
Figure 6.
Quenching of the fluorescence emission of myristoylated constructs of MRP, containing a single tryptophan residue at various positions in the effector domain, by constructs of CaM, containing either methionine or selenomethionine clusters. For each mutant, 0.7 μM MRP was separately incubated with 2.1 μM CaM, SeMet-CaM, CaM*, and SeMet-CaM*, and the fluorescence spectra were recorded. The ability of the selenomethionine residues to quench the fluorescence of tryptophan residues in MRP was quantified by calculating the quenching factor, i.e., the ratio of the maximal fluorescence emission intensity of the MRP constructs complexed to SeMet-CaM or SeMet-CaM* to the intensity of the same constructs complexed to CaM (●) or CaM* (○), respectively (see Materials and Methods). A quenching factor of 1 indicates that the tryptophan residue of interest is remote from selenomethionine residues, whereas lower numbers indicate proximity. For comparison, the quenching factors of myristoylated S4W complexed to CaM (▾) or CaM* (▿) as well as of the myosin light chain kinase peptide complexed to CaM (■) (taken from ref. 24) are shown.
On the basis of these results, we propose that the effector domain of MRP is associated with CaM in an orientation that is similar to myosin light chain kinase (19, 20, 24) and CaM kinase I (24) (topology A in Fig. 4).
The primary sequence of the effector domain of MRP and MARCKS is highly conserved with one notable exception: Pro-96 in the middle of the effector domain of MRP (see Fig. 1) is replaced by a serine residue in MARCKS. However, this difference apparently has no significant effect on the affinity of these proteins for CaM or on their secondary structure, either as free proteins or bound to CaM (5, 9). Using the technique described in this section, it would be informative to determine whether MARCKS binds to CaM with the same orientation as MRP. If different, determining the contribution of Pro-96 to the orientation of the effector domain in the MRP⋅CaM complex would be highly interesting.
Modulation of the Conformation of MRP by Myristoylation.
To assess the impact of the myristoyl moiety on the conformation of MRP complexed to CaM, we have produced previously both myristoylated and unmyristoylated MRP in E. coli (5, 12). The consensus sequence for myristoylation of proteins by N-myristoyl transferase is Gly(1)-X(2)-X(3)-X(4)-S/T(5) (38). Thus, a tryptophan residue could be potentially incorporated at position 2–4 without preventing myristoylation. Indeed, the behavior of S4W during purification by phenylsepharose chromatography as well as during electrophoresis on polyacrylamide gels clearly indicates that this mutant can be efficiently myristoylated (see Materials and Methods).
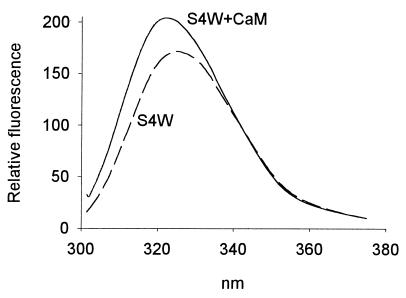
Very interestingly, binding of myristoylated S4W to CaM modifies the fluorescence emission spectrum of Trp-4: the λmax is blue shifted by 7 nm from 352 nm to 345 nm, and the intensity is increased significantly (Fig. 7). In marked contrast, binding of unmyristoylated S4W to CaM reduces the λmax by only 1 nm (356 nm in the absence of CaM; 355 nm in the presence of CaM) and hardly affects the fluorescence intensity (see Fig. 2 Lower). Note that the λmax values presented above represent the average of two experiments with CaM (Fig. 7 shows the spectra of one of these experiments); these results were confirmed in three additional experiments in which SeMet-CaM, CaM*, and SeMet-CaM* were used instead of CaM. Importantly, myristoylation does not affect the fluorescence spectrum of either free F93W or F93W complexed to CaM (data not shown), strongly suggesting that the tryptophan residue introduced at position 4 in MRP serves as a specific probe to locally monitor the environment of the N terminus of MRP. Thus we conclude that the myristoylated N terminus of MRP interacts with CaM in the complex, whereas the unmyristoylated N terminus does not. The lack of significant quenching of the fluorescence of either unmyristoylated or myristoylated S4W by SeMet-CaM shows that this interaction does not involve the N- or C-terminal methionine clusters of CaM (see data, Fig. 6 Right).
Figure 7.
Fluorescence emission spectra of the myristoylated form of S4W (1 μM) in the absence (dashed lines) and presence (solid lines) of 1.5 μM CaM. The corresponding spectra for unmyristoylated S4W are shown in Fig. 2 Lower.
In line with our finding, it has been reported recently that myristoylation is involved in the interaction of NAP-22, a neuron-specific protein kinase C substrate, with CaM (39). However, whereas the myristoylated and unmyristoylated forms of MRP bind to CaM with similar dissociation constants (5), myristoylation of NAP-22 is required for the formation of the complex.
Outlook.
Because of their very irregular structural properties (see Introduction), we propose that MARCKS proteins belong to the family of so-called “natively unfolded” proteins (40). These proteins have no significant amount of regular secondary structure, are rod shaped, lack a hydrophobic core, and remain soluble under extreme conditions such as high temperature and low pH. Furthermore, they exist as a mixture of rapidly equilibrating extended conformers, many of them being able to work as mediators of protein–protein and protein–membrane interactions. In agreement with this concept, we have shown previously by circular dichroic spectroscopy that MRP has a random coil conformation with a maximal α-helical content of 15%. Also, the secondary structure of MRP is not changed on binding to CaM (5). Because of the dynamic properties mentioned above, the structure of “natively unfolded” proteins is unlikely to be solved by x-ray or NMR spectroscopy, and other techniques must therefore be developed. The current state of biotechnologies (random mutagenesis, heterologous expression and single-step purification of proteins, sensitive fluorescence spectroscopy, high-throughput screening) theoretically allows a matrix analysis in which a large number of intermolecular interactions involving tryptophan and selenomethionine residues could be investigated. This could ultimately lead to a detailed map of interfaces in protein complexes. This report on the CaM⋅MRP complex provides an early step in that direction.
Acknowledgments
A.U. was the recipient of a predoctoral fellowship from the Austrian Department of Science, Vienna. T.Y. was supported by a studentship from the Alberta Heritage Foundation for Medical Research (AHFMR) and H.J.V. is the recipient of an AHFMR scientist award. The authors thank Mr. A. Weljie, Calgary, and Dr. A. I. P. M. de Kroon, Utrecht, for stimulating discussions. This work was funded by the Swiss National Foundation for Scientific Research (Grant 3100–042045.94 to Prof. G. Schwarz, Biozentrum, Basel) and by the Medical Research Council of Canada (Grant MT 15237 to H.J.V.).
Abbreviations
- CaM
calmodulin
- CaM*
CaM with all four methionine residues in the C-terminal domain mutated to Leu
- MARCKS
myristoylated alanine-rich C kinase substrate
- MRP
MARCKS-related protein
- SeMet-CaM
CaM with all nine methionine residues substituted by selenomethionine
- SeMet-CaM*
CaM with all five methionine residues in the N-terminal domain substituted by selenomethionine and all four methionine residues in the C-terminal domain mutated to Leu
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090500397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090500397
References
- 1.Aderem A. Cell. 1992;27:713–716. doi: 10.1016/0092-8674(92)90546-o. [DOI] [PubMed] [Google Scholar]
- 2.Lobach D F, Rochelle J M, Watson M L, Seldin M F, Blackshear P J. Genomics. 1993;17:194–204. doi: 10.1006/geno.1993.1301. [DOI] [PubMed] [Google Scholar]
- 3.Blackshear P J. J Biol Chem. 1993;268:1501–1504. [PubMed] [Google Scholar]
- 4.Verghese G M, Johnson J D, Vasulka C, Haupt D M, Stumpo D J, Blackshear P J. J Biol Chem. 1994;269:9361–9367. [PubMed] [Google Scholar]
- 5.Schleiff E, Schmitz A, McIlhinney R A J, Manenti S, Vergères G. J Biol Chem. 1996;271:26794–26802. doi: 10.1074/jbc.271.43.26794. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz A, Schleiff E, Vergères G. In: Molecular Mechanisms of Signaling and Membrane Transport. Wirtz K W A, editor. Berlin: Springer; 1997. pp. 127–150. [Google Scholar]
- 7.Chakravarthy B, Morley P, Whitfield J. Trends Neurosci. 1999;22:12–16. doi: 10.1016/s0166-2236(98)01288-0. [DOI] [PubMed] [Google Scholar]
- 8.Porumb T, Crivici A, Blackshear P J, Ikura M. Eur Biophys J. 1997;25:239–247. doi: 10.1007/s002490050036. [DOI] [PubMed] [Google Scholar]
- 9.Matsubara M, Yamauchi E, Hayashi N, Taniguchi H. FEBS Lett. 1998;421:203–207. doi: 10.1016/s0014-5793(97)01557-3. [DOI] [PubMed] [Google Scholar]
- 10.Hartwig J H, Thelen M, Rosen A, Janmey P A, Nairn A C, Aderem A. Nature (London) 1992;356:618–622. doi: 10.1038/356618a0. [DOI] [PubMed] [Google Scholar]
- 11.Richardson J S, Richardson D C. In: Prediction of Protein Structure and the Principles of Protein Conformation. Fasman G D, editor. New York: Plenum; 1989. pp. 1–98. [Google Scholar]
- 12.Vergères G, Manenti S, Weber T, Stürzinger C. J Biol Chem. 1995;270:19879–19887. doi: 10.1074/jbc.270.34.19879. [DOI] [PubMed] [Google Scholar]
- 13.Kamps M P, Buss J E, Sefton B M. Proc Natl Acad Sci USA. 1985;82:4625–4628. doi: 10.1073/pnas.82.14.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallego C, Gupta S K, Eisfelder B J, Johnson G L. Proc Natl Acad Sci USA. 1992;89:9695–9699. doi: 10.1073/pnas.89.20.9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitz A A P, Schleiff E, Loidl-Stahlhofen A, Röhring C, Vergères G. Anal Biochem. 1999;268:343–353. doi: 10.1006/abio.1998.3080. [DOI] [PubMed] [Google Scholar]
- 16.Babu Y S, Sack J S, Grennhough T J, Bugg C E, Means A R, Cook W J. Nature (London) 1985;315:37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- 17.Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee C B, Bax A. Nat Struct Biol. 1995;2:768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 18.Zhang M, Tanaka T, Ikura M. Nat Struct Biol. 1995;2:758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- 19.Ikura M, Clore G M, Gronenborn A M, Zhu G, Klee C B, Bax A. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 20.Meador W E, Means A R, Quiocho F A. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 21.Meador W E, Means A R, Quiocho F A. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 22.Osawa M, Tokumitsu H, Swindells M B, Kurihara H, Orita M, Shibanuma T, Furuya T, Ikura M. Nature Struct Biol. 1999;6:819–824. doi: 10.1038/12271. [DOI] [PubMed] [Google Scholar]
- 23.Elshorst B, Hennig M, Forsterling H, Diener A, Maurer M, Schulte P, Schwalbe H, Griesinger C, Krebs J, Schmid H, et al. Biochemistry. 1999;38:12320–12332. doi: 10.1021/bi9908235. [DOI] [PubMed] [Google Scholar]
- 24.Yuan T, Weljie A M, Vogel H J. Biochemistry. 1998;37:3187–3195. doi: 10.1021/bi9716579. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 26.Sarkar G, Sommer S S. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 27.Barik S. Methods Mol Biol. 1997;67:173–182. doi: 10.1385/0-89603-483-6:173. [DOI] [PubMed] [Google Scholar]
- 28.Corradin S, Ransijn A, Corradin G, Roggero M A, Schmitz A A P, Schneider P, Mauël J, Vergères G. J Biol Chem. 1999;274:25411–25418. doi: 10.1074/jbc.274.36.25411. [DOI] [PubMed] [Google Scholar]
- 29.Umekage T, Kato K. FEBS Lett. 1991;286:147–151. doi: 10.1016/0014-5793(91)80961-2. [DOI] [PubMed] [Google Scholar]
- 30.Lowry O H, Rosebrough N J, Farr A L, Randall R J. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 31.Lakowicz J. Principles of Fluorescence Spectroscopy. New York: Plenum; 1983. [Google Scholar]
- 32.Killian J A, Keller R C A, Struyvé M, de Kroon A I P M, Tommassen J, de Kruijff B. Biochemistry. 1990;29:8131–8137. doi: 10.1021/bi00487a021. [DOI] [PubMed] [Google Scholar]
- 33.de Kroon A I P M, Soekarjo M W, de Gier J, de Kruijff B. Biochemistry. 1990;29:8229–8240. doi: 10.1021/bi00488a006. [DOI] [PubMed] [Google Scholar]
- 34.Creighton T E. Proteins, Structures and Molecular Properties. New York: Freeman; 1993. [Google Scholar]
- 35.Sopkova J, Vincent M, Takahashi M, Lewitt-Bentley A, Gallay J. Biochemistry. 1998;37:11962–11970. doi: 10.1021/bi980773o. [DOI] [PubMed] [Google Scholar]
- 36.O'Neil K T, Wolfe H R, Erickson-Viitanen S, DeGrado W F. Science. 1987;236:1454–1456. doi: 10.1126/science.3589665. [DOI] [PubMed] [Google Scholar]
- 37.Zhang M, Vogel H J. J Mol Biol. 1994;239:545–554. doi: 10.1006/jmbi.1994.1393. [DOI] [PubMed] [Google Scholar]
- 38.Johnson D R, Bhatnagar R S, Knoll L J, Gordon J I. Annu Rev Biochem. 1994;63:869–914. doi: 10.1146/annurev.bi.63.070194.004253. [DOI] [PubMed] [Google Scholar]
- 39.Takasaki A, Hayashi N, Matsubara M, Yamauchi E, Taniguchi H. J Biol Chem. 1999;274:11848–11853. doi: 10.1074/jbc.274.17.11848. [DOI] [PubMed] [Google Scholar]
- 40.Weinreb P H, Zhen W, Poon A W, Conway K A, Lansburry P T., Jr Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]