Abstract
Bacterial photosynthesis relies on the interplay between light harvesting and electron transfer complexes, all of which are located within the intracytoplasmic membrane. These complexes capture and transfer solar energy, which is used to generate a proton gradient. In this study, we identify one of the factors that determines the organization of these complexes. We undertook a comparison of the organization of the light-harvesting complex 1 (LH1)/reaction center (RC) cores in the LH2− mutant of Rhodobacter sphaeroides in the presence or absence of the PufX protein. From polarized absorption spectra on oriented membranes, we conclude that PufX induces a specific orientation of the reaction center in the LH1 ring, as well as the formation of a long-range regular array of LH1-RC cores in the photosynthetic membrane. From our data, we have constructed a precise model of how the RC is positioned within the LH1 ring relative to the long (orientation) axis of the photosynthetic membrane.
When growing photosynthetically, the purple bacterium Rhodobacter (Rb.) sphaeroides uses the energy of light to drive the formation of ATP via a cyclic electron transfer. Light energy is captured by the light harvesting complex(es) and funneled toward the special pair of bacteriochlorophylls (BChls) (P) in the reaction center (RC). The energy is used for the release of an electron by the special pair, which is transferred via the accessory BChl and a bacteriopheophytin (BPheo) molecule to the initial ubiquinone acceptor QA and finally to the secondary ubiquinone QB. After two such photoreactions, doubly protonated ubiquinol (QH2) is formed that dissociates from the RC complex into the lipid phase of the membrane. The cytochrome bc1 complex utilizes ubiquinol and the oxidized cytochrome c2 as reductant and oxidant, respectively. The net result of this process is the formation of a proton gradient, resulting in the synthesis of ATP, which is used to power the metabolic reactions in the cell.
Light harvesting and subsequent cyclic electron transfer involve the interplay between at least four protein complexes, the peripheral light harvesting complex 2 (LH2), the LH1 complex, the reaction center (RC), and the cytochrome bc1 complex. There are structural models, to atomic resolution, for the LH2, RC, and cytochrome bc1 complexes (1–3). These data, together with structural information on LH1 and LH1-RC complexes from cryoelectron microscopy (4, 5), offer the possibility of building a model for the organization of the photosynthetic membrane. It is now clear that a protein, PufX, could hold the key to understanding the organization of these components within the membrane. The significance of PufX arises from the demonstration that it is necessary for photosynthetic growth of Rb. sphaeroides (6), and more specifically that it is required for ubiquinone/ubiquinol exchange between the RC QB site and the cytochrome bc1 complex (7, 8). When the LH1 complex is absent or is reduced in size, there is no such requirement for PufX (9, 10). Thus, there is an intriguing relationship between the RC, the LH1, and cytochrome bc1 complex, in which PufX plays a central role.
The pufX gene, which encodes a small integral membrane protein consisting of 82 amino acids, is found at the 3′ end of the puf operon (11), which also encodes the major components of the core complex, the LH1 α and β polypeptides, and the L and M subunits of the RC. The LH1 complex forms a ring apparently consisting of 16 α, β heterodimers (4), which then surrounds the RC, at least in the absence of PufX (9, 12, 10). Although there are no high resolution data for any LH1 complex, it is assumed that this complex contains a maximum of 32 BChls sandwiched between concentric rings of α and β subunit helices (4), much like arrangement found in the LH2 complex (3). In the presence of PufX, the number of BChls in the ring is reduced by approximately 2 (10). PufX is believed to form part of the LH1 ring (12); it associates with LH1 and the RC in vitro to form the core complex in Rb. sphaeroides (13, 14), and during photosystem assembly the insertion of PufX precedes the encirclement of the RC by LH1 (14).
There is no direct structural evidence that PufX is part of the LH1-RC core complex; the cryoelectron microscopy data (4, 5) were obtained for LH1-RC complexes from Rhodospirillum (Rs.) rubrum and from an LH2− mutant of Rb. sphaeroides, respectively. In neither case was a protein, PufX, present, because it has not been found in Rs. rubrum (15), or because the gene encoding it was deleted in the Rb. sphaeroides mutant (9). The first step toward an analysis of a photosystem containing PufX can be seen in the work of Jungas et al. (16), which proposes that LH1-RC-PufX cores form a regular array in tubular membranes of an LH2− mutant of Rb. sphaeroides. Previously, the demonstration that PufX exerts its influence in facilitating the diffusion of reduced quinols out of the core complex led to the concept of a quinone pore in the LH1 ring, formed by PufX (7, 8, 12). It is implicit in this model of the core complex that the RC adopts a single orientation with respect to PufX to maximize the diffusion rate of the quinones into and out of the QB site. This, taken together with the existence of regular arrays of complexes seen in cryo-EM (16), would indicate a need to probe intact membranes using polarized light to identify any highly organized and oriented complexes, and to examine whether PufX influences this organization. We have used linear dichroism (LD) spectroscopy to examine membranes containing LH1-RC cores, with or without the PufX protein. Our data show a remarkable difference between the two types of membrane, which demonstrates that PufX promotes a well defined orientation of the RC with respect to the LH1 complex, and that this is accompanied by a long-range organization of the LH1-RC-PufX cores in the membrane.
Materials and Methods
Sample Preparation.
The DD13 double deletion strain (17), which does not synthesize LH2, LH1, or RC complexes, was complemented with the desired genes encoding RC, LH1, LH1-RC, or LH1-RC-PufX complexes as described earlier (14, 17, 18). Transconjugants were grown semiaerobically in the dark on M22 + medium supplemented with neomycin (20 μg⋅ml−1) and tetracycline (1 μg⋅ml−1). Mutant colonies were screened for by using a Guided Wave 260 spectrophotometer fitted with a home-built Petri dish holder. Selected mutants were then grown in liquid culture, and intracytoplasmic membranes prepared as described by Olsen et al. (19). The intracytoplasmic membranes were stored at −20°C until required for spectroscopy. The preparations were diluted with 10 mM Tris⋅HCI (pH 8.0) before use, and for low-temperature measurements glycerol was used at a concentration of 70% (vol/vol). For the LD measurements, the polyacrylamide gels contained 14.5% (wt/vol) acrylamide and 0.5% N,N′-methylbisacrylamide. After polymerization of the gels with 0.05% (wt/vol) ammonium persulfate and 0.03% TEMED (Sigma) in the dark, they were compressed in two perpendicular directions (x and y axis), and the gels expanded along the z axis. The samples were frozen in the dark for ≈15 min.
Spectroscopy.
Absorption, P+ minus P ΔA, LD, and P+ minus P ΔLD spectra were recorded on a home-built spectrapolarimeter, with a resolution of 1.5 nm. P+ minus P ΔLD spectra were obtained by laser excitation at 670 nm to avoid photoselection by the polarized light (CW dye-laser, DCM dye, Coherent CR599, bandwidth of ≈1 nm, pumped by an Ar+ laser, Coherent Innova 310). The power was attenuated to 15 mW⋅cm−2. The absorbance difference (LD) spectra were measured with two parallel lock-in amplifiers (one for the transmission signal and the other for the difference signal) and a monochromator, with 1-nm resolution. The ΔLD spectra were directly recorded by a third lock-in amplifier that was fed by the LD signal and locked at the 20-Hz laser modulation. For the ΔLD experiment, the LD signal was measured with a short, 3-ms integration time. The P+ minus P ΔA measurement was performed with two lock-in amplifiers in series with the laser at the same excitation wavelength as before, modulated with a chopper at a slow, 20-Hz frequency. In this case, the transmission signal was modulated with the use of two perpendicular placed polarizers with a PhotoElasticModulator (PEM 80, Hinds, Hillsboro, OR) that changes the polarization of the probing light from parallel to perpendicular with 20 kHz, placed in between.
Results and Discussion
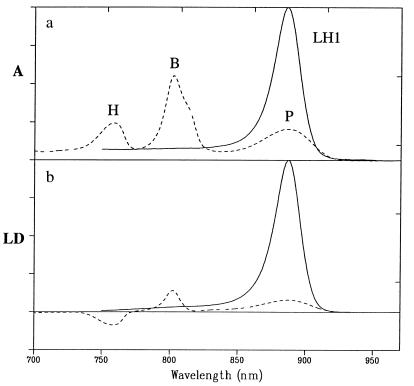
The main transitions of the RC pigments partially overlap with those of the LH1 complex. In Fig. 1a, we compare the 77 K absorption spectra of membranes containing only RCs with membranes containing only the LH1 complex. The LH1 complex has a single major absorption maximum at 887 nm and a long vibronic tail toward the blue. In the RC, the special pair (P) absorbs at 888 nm, the accessory BChls (B) at 802 nm, and the BPheos (H) at 758 nm. In the presence of PufX, the absorbance maximum of both LH1-only and LH1-RC cores are blueshifted by 1–2 nm with respect to their X-minus equivalents (see Fig. 2 and ref. 10). This effect can be modeled with exciton theory by assuming that in the presence of PufX LH1 is a curvilinear array rather than a complete ring of coupled pigments (20). When the membranes containing only RC or membranes containing only LH1 are oriented and probed with linearly polarized light, we observe the absorbance difference between horizontally and vertically polarized (LD) with respect to the axis of orientation (Fig. lb). This axis of orientation is the long axis of the particle, which in this case corresponds to membranes or membrane fragments that contain these complexes embedded in their native environment. For the RC-only membranes, we find a positive LD for the special pair P and the accessory BChls and a negative LD for the BPheos. This is what one would expect, assuming that the RC orients with its C2 symmetry axis perpendicular to the stretching axis of the gel. The P and B transition dipoles are perpendicular with respect to the normal of the membrane (which is coincident with the RC C2 symmetry axis) whereas H has a more parallel orientation (21). We can calculate the angles of the various transition dipoles with respect to the normal of the membrane plane (angle θ, see Appendix, section A) by calculating the ratio of the reduced LD (= LD/A) for B and H relative to P, assuming that the long-wavelength transition of P makes a 90° angle with this axis. This circumvents the problem of not knowing the orientation factor of the complex (see Table 1). The angles of B and H with respect to the membrane plane are calculated to be 24° and 55°, respectively. These values are in good agreement with the angles calculated from LD spectra and the structure by Breton (21), which were 20° and 50°.
Figure 1.
(a) The main Qy absorption of the LH1-only membranes (solid line) and the RC-only membranes (dashed line) at 77 K; the latter were frozen in the dark. P is the special pair, B the accessory BChls, and H the BPheos. The absorption of the RC complexes is enhanced for ease of comparison. (b) The LD spectra of these membranes at 77 K in the same spectral region.
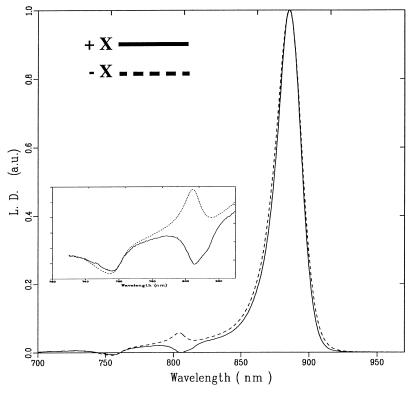
Figure 2.
LD spectra of cores at 77 K with (solid line) and without (dashed line) the PufX protein present. The core complexes were frozen in the dark. (Inset) Magnification of the LD spectra in the accessory BChl and BPheo region.
Table 1.
Relevant parameters from the LD measurements (see Appendix)
| RC only
|
X− cores
|
X+ cores
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P | B | H | LH1 | B | H | LH1 | B | H | P | |
| LDr* | 0.008 | 0.04 | −0.08 | 0.25 | 0.12 | −0.23 | 0.26 | −0.1 | −0.17 | — |
| LDr"† | — | 2 | −1 | — | 2 | −1 | — | −2.6 | −1.5 | — |
| θ‡ | 90° | 66° | 35° | 90° | 66° | 35° | 90° | 66° | 35° | — |
| φ‡ | — | — | — | — | — | — | — | 55° | 33° | 80°§ |
LDr values presented here as LD/A with A = Az (measured simultaneously with LD). Because we only use ratios for calculating angles, we do not need the isotropic absorption.
† LDr" is the measured LDr for RC-only membranes normalized to the LDr of P and for the PufX− and PufX+ membranes normalized to the LDr of LH1.
‡ θ and φ are calculated by comparing the measured values of LDr" with Eqs. 5 and 8, respectively. For RC-only membranes, the angle θ is assumed 90° for the longwavelength transition of P; for PufX+ and PufX− membranes, the angle θ is assumed 90° for the longwavelength transition of LH1.
§ This value is estimated from the ΔLD signal and the absorption of P.
The LD for the LH1 complex is positive over the entire absorption band, which indicates that the particles are oriented such that the pigments within the ring have their Qy transitions parallel to the plane of the ring (which is in the membrane plane) (22). The LD spectra of membranes containing LH1-RC core complexes with or without PufX contains three regions of interest, around 870, 800, and 750 nm (Fig. 2). The LD in the 870 nm region for both PufX+ and PufX− core complexes is dominated by the positive signal of the LHl complex; the contribution of the special pair cannot be observed because the LH1 complex has 12 to 16 times more BChls absorbing around 870 nm than the RC. Nevertheless, a reduction of the LD/A value can be observed at the red edge of the 870-nm band in the presence of PufX, which may be indicative of a negative contribution of P. Although these spectra have been normalized to give the same maximum absorbance, the absolute magnitude of the LD spectra is almost equal for both PufX+ and PufX− core complexes, indicating that both types are embedded in similarly shaped membrane fragments. Around 750 nm, the LD signals almost coincide, and both PufX+ and PufX− complexes show the negative LD of the BPheos as observed for the RC-only complex. A striking difference between the two types of core complexes can be observed around 800 nm. Those complexes in membranes lacking PufX have a positive band similar to the LD signal of the accessory BChls in RC-only membranes whereas the membranes with PufX have a negative LD. Close inspection of this negative band (see Fig. 2 Inset) shows a shoulder on the red edge at 813 nm, which is the expected absorption maximum of the upper exciton band of the special pair. The contribution of this 813-nm transition is negative in both spectra, with and without PufX.
We can interpret the observed LD spectra by assuming that (i) membranes containing LH1-RC core complexes are oriented with their normal perpendicular to the stretching axis, (ii) that in these membranes PufX− cores have full rotational freedom around the normal to the membrane [meaning that the angle φ is free (see Fig. 5 and Appendix)], and (iii) that there is a fixed angle, φ, between the projection of the transition dipole moment in the membrane and the stretching axis for the PufX+ cores (see Appendix, section B).
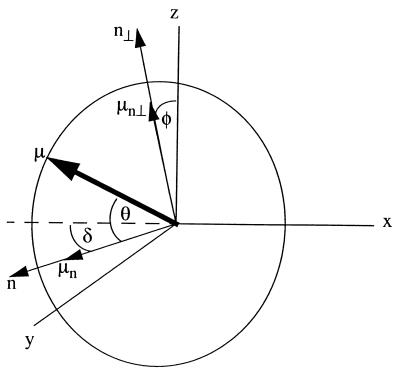
Figure 5.
The geometry of relevant vectors and angles for the LD experiment (measured is Az − Ax). The stretching axis coincides with the z axis. The long axis of the membrane fragment is supposed to coincide with the z axis. For a pigment in a complex embedded in an oriented membrane, we define a normal, n, and a vector perpendicular to this normal, n⊥, in the plane of the pigment transition dipole moment, μ, and n. We project the pigment transition dipole moment, μ, on n and n⊥. The orientation of μ is defined by three angles: (i) between the projection of μ to n⊥ and the z axis: φ; (ii) between μ and the normal, n: θ; and (iii) between the projection of μ to n and the x axis: δ.
The ratio of the reduced LD can be calculated for the PufX− cores in a similar fashion to that for the RC-only membranes. The ratio of the reduced LD of B (802 nm) and H (758 nm) is the same for both LH1-RC and RC-only membranes, and we conclude that, in both membranes, the RC is oriented in such a way that the transition dipole moment of P lies in the plane of the LH1 ring and that there exists no favored orientation of the RC in the ring. To explain the remarkable LD spectrum observed for the membranes containing PufX+ cores, we can safely assume that the RC structure has remained unchanged because no differences in the CD spectra for the accessory BChls were observed (data not shown). Furthermore, a large tilt of the RC relative to the plane of the LH1 ring can be ruled out because that would also affect the orientation of the BPheos. Therefore, we propose that, in the presence of PufX, a specific orientation of the RCs exists relative to the long axis of the particle. In other words, a fixed angle φ is assumed, which means that we cannot average over all orientations of this angle (Appendix; Fig. 5). As a consequence, the reduced LD now not only depends on θ, but also on φ. The same values for θ were used as for membranes containing PufX− cores or RC-only complexes. Again, the reduced LD of B and H relative to LH1 can be calculated assuming perfect orientation of the LH1 complex, yielding values for φ of 55° and 33°, respectively.
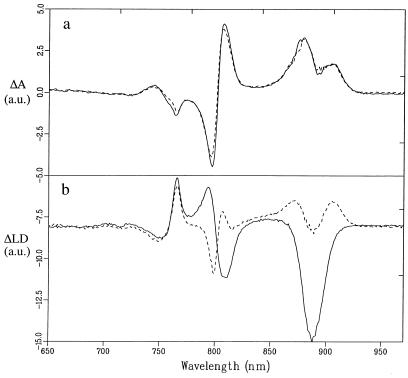
When P is assumed to be in the plane of the LH1 ring (i.e., θP = 90°), the expected value of φ for P (from the RC structure and based on the values for B and H as given above) would be ≈80°, a geometry that leads to a value for the reduced LD of −0.2. This would imply that, in each RC, the long wavelength transition dipole of P is almost perfectly oriented perpendicular to the stretching axis. To observe this predicted orientation of the long wavelength P transition within a membrane with PufX+ cores, we designed a double absorption difference experiment that measured the difference in LD due to the light-induced bleaching of P. The LD signal was modulated with a 20-Hz laser flash so that the difference in LD (ΔLD) could be measured directly. Fig. 3 shows the resulting dark minus light absorption and LD spectra. The sign of the latter spectrum was checked by measuring the LD with continuous laser excitation and subtracting the spectrum obtained in the light from the dark LD spectrum. The dark-minus-light absorption difference spectra for the PufX+ and PufX− cores (Fig. 3a) are almost identical and show the well known bleachings and bandshift signals due to the formation of P + QB−. In contrast, the ΔLD spectra are dramatically different, but all of the features are fully consistent with the LD spectra for PufX+ and PufX− membranes shown in Fig. 2. In the two spectra, the peaks for H are similar; in contrast, the signals have opposite sign for B. Furthermore, a negative signal is seen at 888 nm for the PufX+ cores whereas in the same region a split positive band is observed for the PufX− cores. In the latter case, the spectrum is interpreted as attributable to the positive P signal on which an LH1 bandshift signal, caused by the electric field associated with charge separation, is superimposed. For the PufX+ cores, the P-spectrum has been inverted, which explains the difference. These features are also consistent with our predictions for the LD of P mentioned earlier. Although some cancellation effects are expected because of the bandshift of B, one can conclude that P has stronger negative LD than B for the PufX+ cores. We roughly estimate the reduced LD for P to be about twice the magnitude of B, which is consistent with the predicted value of −0. 2.
Figure 3.
77 K dark minus light absorption (Upper) and dark minus light LD (Lower) of cores with X (solid line) and without (dashed line) X. Both spectra are normalized on OD.
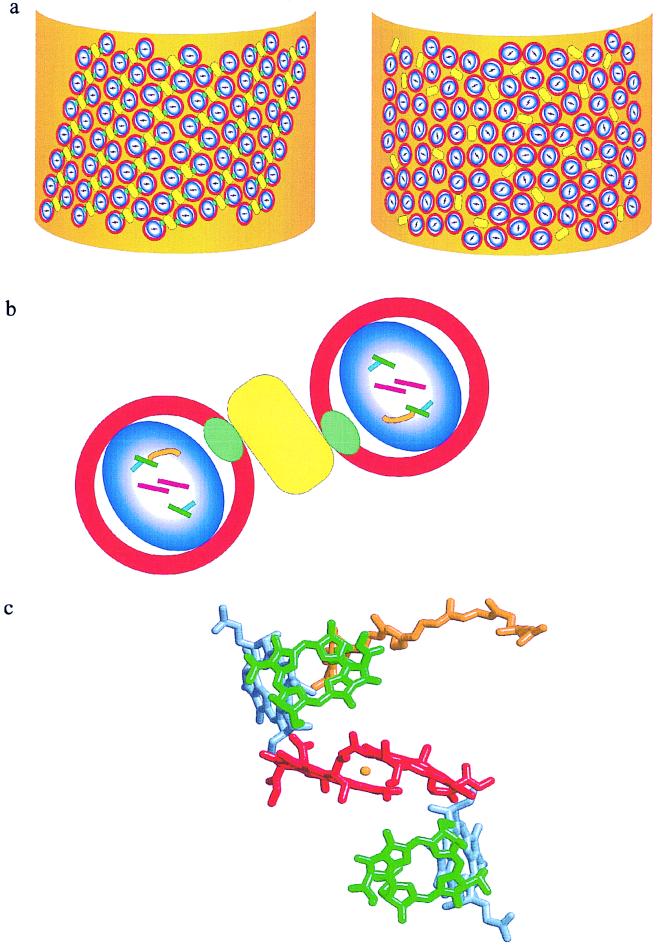
The LD experiments carried out in this study have enabled us to calculate the arrangement of the core complexes in the PufX-containing membrane, and to demonstrate that PufX exerts a decisive effect on the orientation of the RC. A model depicting the organization of core complexes within their native membrane is presented in Fig. 4. The membranes of this LH2-minus strain are assumed to constitute a pronounced long axis, as previously observed for such strains (23–25). When PufX is absent, the LH1-RC cores have a random orientation with respect to each other (Fig. 4a Right) whereas in the presence of PufX the LH1-RC cores form regular arrays within the membrane (Fig. 4a Left). The angles of the projection of the transition dipole moments of the RC pigments as calculated above for the pufX+ cores allow us to state how the RC is oriented within the LH1 ring (Fig. 4B). The angle between the projection of the transition dipole moment on n⊥ and the z axis of the RC pigments and the stretching axis (i.e., the long axis of the membrane fragment) is estimated: for P, ≈80°; for B, 55°; and for H, 33°. Our calculations do not predict the sign of these angles. When φH is taken to be −33° (see Fig. 5 and Appendix), the calculated angles coincide with the orientation of the RC as depicted in Fig. 4c. Combining this orientation with the hypothesis that pufX forms a pore in the LH1 ring for the quinones and the structure reported by Jungas et al. (16), we arrive at a full description of the arrangement of the core complexes within the membrane. This model fully explains the reversal of sign around 800 and 888 nm seen in the LD and ΔLD spectra, respectively, between cores with and without PufX. No large differences in LD were expected or observed for the BPheos at 750 nm or for the LH l pigments at 870 nm, and this is reflected in the model. Thus, the LD spectrum of pufX containing membranes can be explained by a regular array of cores aligned in a membrane fragment with a well defined long axis, as illustrated in Fig. 4a. In contrast, in the absence of pufX, the cores are randomly oriented in the membrane. Finally, we note that an explanation of the observed LD data requires a macroscopic organization of the LH1-RC-PufX core complexes relative to the long axis of the membrane fragment as any organizational disorder would dramatically attenuate the LD signals at 800 and 880 nm.
Figure 4.
A model of the organization of RC-LH1 core complexes in a membrane fragment of an LH2-minus mutant of Rb. sphaeroides. The core complexes in the left-hand representation contain PufX whereas those on the right do not. (a) The intracytoplasmic membrane is depicted as a membrane fragment in which the circular RC-LH1-PufX cores (blue and white oval enclosed by red circle) and cytochrome bc1 complex (ES) (yellow rectangle) are embedded. The long-range order of these complexes with respect to the long axis (y) of the membrane fragment is apparent when the PufX protein (green oval) is present in the core complex, whereas the cores that contain only RC-LH1 can adopt any orientation. The Qy transitions of the special pair of bacteriochlorophylls (P) are represented by the bars within the blue and white oval. (b) An enlarged view of a pair of core complexes flanking one or two cytochrome bc1 complexes, as suggested by Jungas et al. (16). The RC pigments P are in red, B in green, and H in blue, and the position of QB is marked by the orange curve. The bars represent the Qy transitions of these pigments, approximated by drawing a line between the NB and ND atoms within the macrocycles. (c) The pigments P, B, and H plus QB and the nonheme iron from the crystal structure of Rb. sphaeroides (Protein Data Bank ID code 1PCR) viewed along the C2 axis of symmetry. The color coding is as used in b with the nonheme iron represented by the orange sphere.
Conclusions
Our data demonstrate the ability of spectroscopy, not only to determine the arrangement of pigment molecules, but also to use these molecules as reporters for the macromolecular organization of the pigment-protein complexes in the photosynthetic membrane. Our data lead to the conclusion that, although PufX is quantitatively a minor component of the bacterial photosynthetic unit, it can exert a profound influence on the organization of the bacterial photosynthetic apparatus by promoting the formation of regular arrays of LH1-RC core complexes in the intracytoplasmic membrane. Recently, electron-micrographs have been reported (16), demonstrating the presence of quasi-crystalline arrays in the photosynthetic membrane of LH2- strains of Rb. sphaeroides. The elementary unit was proposed to be a dimer of LH1-RC-PufX cores separated by a region of electron density that is tentatively ascribed to the cytochrome bc1 complex. We have probed such membranes with polarized light, have identified the presence of organized arrays of reaction centers, and have demonstrated that this organization depends on the presence of PufX. If PufX does act as a portal between the RC and the cytochrome bc1 complexes, it is reasonable to suggest that the cytochrome bc1 complex is also present in organized arrays within this membrane. An elucidation of the organization of bacterial photosynthetic membranes will enhance our understanding of how multicomponent protein supercomplexes function in vivo. As membrane shape is determined by its constituents and their organization, our findings indicate that eventually the pufX protein plays a major role in the full appearance of the photosynthetic membrane.
Acknowledgments
C.N.H., J.D.O., and W.H.J.W. thank the Biotechnology and Biological Sciences Research Council (United Kingdom) and the Human Frontier Science Program for financial support. This project is supported by the Netherlands Organisation for Scientific Research (NWO) through the Council of Earth and Life Science (ALW).
Abbreviations
- BChl
bacteriochlorophyll
- BPheo
bacteriopheophytin
- RC
reaction center
- LD
linear dichroism, LH1, light-harvesting complex 1
Appendix
The geometry of relevant vectors in a LD experiment is shown in Fig. 5. We assume perfect orientation of the membranes so the long axis of the membranes lies along the stretching axis z. n is the normal to the membrane in which the pigment-proteins are embedded, and μ represents the transition dipole moment of the pigment under consideration. We derive an expression for the reduced linear dichroism considering two cases: with full rotational freedom around n (part A) and with a fixed orientation with respect to n (part B).
A.
The geometry as in Fig. 5 leads to the expressions of the x and z component of the transition dipole moment as in Eqs. 1 and 2:
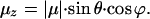 |
1 |
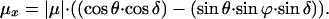 |
2 |
Averaging over all angles φ and δ leads to the following expressions of the z and x components of the absorption:
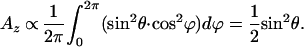 |
3 |
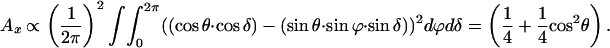 |
4 |
The reduced LD is defined as
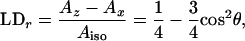 |
5 |
where Aiso is the isotropic absorption. Note that if θ is 90° the reduced LD is 0.25. The values in Table 1 for θ are obtained by calculating the ratio of LDr of P with B or H from the spectra and comparison with Eq. 5, assuming the LDr of P is 0.25.
B.
The expressions for the z and x component of the absorption when φ is fixed (and not averaged out) are
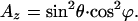 |
6 |
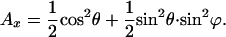 |
7 |
The reduced LD is
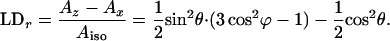 |
8 |
Like for θ (Eq. 5), we obtain the values for φ by comparing the reduced LD of B and H with LH1, assuming perfect orientation for LH1, meaning LDr = 0.25.
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090083797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090083797
References
- 1.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. Nature (London) 1985;318:618–624. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- 2.McDermott G, Prince S M, Freer A A, Hawthornthwaite-Lawless A M, Papiz M Z, Cogdell R J, Isaacs N W. Nature (London) 1995;374:517–521. doi: 10.1016/s0969-2126(96)00050-0. [DOI] [PubMed] [Google Scholar]
- 3.Xia D, Yu C A, Kim H, Xia J Z, Kachurin A M, Zliang L, Yu L, Deisenhofer J. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PubMed] [Google Scholar]
- 4.Karrasch S, Bullough P A, Ghosh R. EMBO J. 1995;14:631–638. doi: 10.1002/j.1460-2075.1995.tb07041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walz T, Jamieson S J, Bowers C M, Bullough P A, Hunter C N. J Mol Biol. 1998;282:833–845. doi: 10.1006/jmbi.1998.2050. [DOI] [PubMed] [Google Scholar]
- 6.Farchaus J W, Grünberg H, Oesterhelt D. J Bacteriol. 1990;172:977–985. doi: 10.1128/jb.172.2.977-985.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lilburn T G, Haith C E, Prince R C, Beatty J T. Biochim Biophys Acta. 1992;110:160–170. doi: 10.1016/0005-2728(92)90077-f. [DOI] [PubMed] [Google Scholar]
- 8.Barz W P, Verméglio A, Francia E, Venturoli G, Melandri B A, Oesterhelt D. Biochemistry. 1995;34:15248–15258. doi: 10.1021/bi00046a033. [DOI] [PubMed] [Google Scholar]
- 9.McGlynn P, Hunter C N, Jones M R. FEBS Lett. 1994;349:349–353. doi: 10.1016/0014-5793(94)00701-2. [DOI] [PubMed] [Google Scholar]
- 10.McGlynn P, Westerhuis W H J, Jones M R, Hunter C N. J Biol Chem. 1996;271:3285–3292. doi: 10.1074/jbc.271.6.3285. [DOI] [PubMed] [Google Scholar]
- 11.Lee J K, Dehoff B S, Donohue T J, Gumport R I, Kaplan S. J Biol Chem. 1989;264:19354–19365. [PubMed] [Google Scholar]
- 12.Cogdell R J, Fyfe P K, Barrett S J, Prince S, Freer A A, Isaacs N W, Hunter C N. Photosynth Res. 1996;48:55–63. doi: 10.1007/BF00040996. [DOI] [PubMed] [Google Scholar]
- 13.Francia F, Wang J, Venturoli G, Melandri B A, Barz W P, Oesterhelt D. Biochemistry. 1999;38:6834–6845. doi: 10.1021/bi982891h. [DOI] [PubMed] [Google Scholar]
- 14.Pugh R J, McGlynn P, Jones M R, Hunter C N. Biochim Biophys Acta. 1998;1366:302–316. doi: 10.1016/s0005-2728(98)00131-5. [DOI] [PubMed] [Google Scholar]
- 15.Walz T, Ghosh R. J Mol Biol. 1997;265:107–111. doi: 10.1006/jmbi.1996.0714. [DOI] [PubMed] [Google Scholar]
- 16.Jungas C, Ranck J, Rigaud J, Jolliot P, Verméglio A. EMBO J. 1999;18:534–542. doi: 10.1093/emboj/18.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones M R, Fowler G J S, Gibson L C D, Grief G G, Olsen L, Crielaard W, Hunter C N. Mol Microbiol. 1992;6:1173–1184. doi: 10.1111/j.1365-2958.1992.tb01556.x. [DOI] [PubMed] [Google Scholar]
- 18.Jones M R, Visschers R W, van Grondelle R, Hunter C N. Biochemistry. 1992;31:4458–4465. doi: 10.1021/bi00133a011. [DOI] [PubMed] [Google Scholar]
- 19.Olsen J D, Sockalingum G D, Robert B, Hunter C N. Proc Natl Acad Sci USA. 1994;91:7124–7128. doi: 10.1073/pnas.91.15.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westerhuis W H J, Hunter C N, van Grondelle R, Niederman R A. J Phys Chem B. 2000;103:7733–7742. [Google Scholar]
- 21.Breton J. Biochim Biophys Acta. 1985;810:235–245. [Google Scholar]
- 22.Kramer H J M, Grondelle R van, Hunter C N, Westerhuis W H J, Amesz J. Biophys Acta. 1984;765:156–165. [Google Scholar]
- 23.Hunter C N, Pennoyer J D, Sturgis J N, Farrelly D, Niederman R A. Biochemistry. 1988;27:3459–3467. [Google Scholar]
- 24.Kiley P J, Varga A, Kaplan S. J Bacteriol. 1988;170:1103–1115. doi: 10.1128/jb.170.3.1103-1115.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson L C D, McGlynn P, Chaudhri M, Hunter C N. Mol Microbiol. 1992;6:3171–3186. doi: 10.1111/j.1365-2958.1992.tb01773.x. [DOI] [PubMed] [Google Scholar]