Abstract
Free extracellular DNA is abundant in many aquatic environments. While much of this DNA will be degraded by nucleases secreted by the surrounding microbial community, some is available as transforming material that can be taken up by naturally competent bacteria. One such species is Vibrio cholerae, an autochthonous member of estuarine, riverine, and marine habitats and the causative agent of cholera, whose competence program is induced after colonization of chitin surfaces. In this study, we investigate how Vibrio cholerae's two extracellular nucleases, Xds and Dns, influence its natural transformability. We show that in the absence of Dns, transformation frequencies are significantly higher than in its presence. During growth on a chitin surface, an increase in transformation efficiency was found to correspond in time with increasing cell density and the repression of dns expression by the quorum-sensing regulator HapR. In contrast, at low cell density, the absence of HapR relieves dns repression, leading to the degradation of free DNA and to the abrogation of the transformation phenotype. Thus, as cell density increases, Vibrio cholerae undergoes a switch from nuclease-mediated degradation of extracellular DNA to the uptake of DNA by bacteria induced to a state of competence by chitin. Taken together, these results suggest the following model: nuclease production by low-density populations of V. cholerae might foster rapid growth by providing a source of nucleotides for the repletion of nucleotide pools. In contrast, the termination of nuclease production by static, high-density populations allows the uptake of intact DNA and coincides with a phase of potential genome diversification.
The aquatic reservoirs in nature for Vibrio cholerae, the etiologic agent of cholera, are rivers, estuaries, and coastal waters, where it can be found free in the water column or associated with surfaces, including the chitinous exoskeletons of copepod molts (33, 19). Chitin, a polymer of N-acetylglucosamine, not only serves as a nutrient source but fosters horizontal gene transfer between genetic variants of V. cholerae by inducing natural competence for transformation (23). In laboratory microcosms, chitin-induced natural transformation has been shown to mediate the acquisition of multigene clusters of DNA coding for diverse functions, including the structure and antigenic characters of O serogroup determinants or the capacity to utilize specific carbohydrates as a nutrient source (3, 27). The kinds of genes which, in principle, can be acquired by this mechanism are apparently quite broad, requiring only the presence of conserved flanking segments by which incoming DNA is incorporated into the recipient chromosome by homologous recombination.
This mode of horizontal gene transfer is chitin induced and, thus, may be functionally restricted to niches and seasons where chitin is abundant in the environment, for example, during copepod blooms. It follows that genes acquired by these means are likely to be subject to natural selection by features of the habitat where these V. cholerae variants arise. With respect to the acquisition of novel O-antigen types or new catabolic functions, the former is arguably under powerful selection by phage predation, whereas the latter could promote the occupation of new niches in an environment where a particular nutrient is abundant (3, 27).
Competence entails the transfer of free extracellular DNA, present in the environment, into the cell. However, V. cholerae also secretes two extracellular nucleases, Xds and Dns (11), whose production would need to be terminated during the competence process to allow the uptake of intact DNA. xds, first cloned by Newland et al., encodes a single 100-kDa polypeptide (28) that can be detected in culture supernatants (32). dns was cloned and the protein was purified by Focareta and Manning, who showed that it is secreted across the outer membrane (13) as a mature 24-kDa processed protein (12). Dns was crystallized by Altermark et al. and designated V. cholerae EndA (2), and its structure was almost identical to the periplasmic endonuclease I (Vvn) of Vibrio vulnificus (17). Comparative studies of V. cholerae's Dns endonuclease (EndA) and its Vibrio salmonicida counterpart showed that the activity of each enzyme coincides with the optimal growth condition of the producing species. Thus, the Dns endonuclease of V. cholerae was found to be most active at 175 mM NaCl and pH 7.5 to 8.0 (1).
Quorum sensing governs a variety of cellular behaviors in V. cholerae. HapR is the major regulator of the V. cholerae quorum-sensing system. As cell density increases, HapR represses genes encoding virulence determinants (26, 39) and biofilm formation (14, 38), whereas it induces other genes required for natural competence (e.g., VC1917/comEA [23]; see below). The quorum-sensing signals are processed through a regulatory cascade (34). At low cell density, quorum-sensing receptor sensor kinases transfer phosphate via LuxU to LuxO (see Fig. 7). LuxO∼P stimulates the transcription of four small regulatory RNAs which, together with Hfq, destabilize hapR mRNA, leading to its degradation, a process that posttranscriptionally reduces the production of HapR (16). At high cell density, the stability of hapR mRNA is not affected due to the reversion of the phosphorylation cascade leading to dephosphorylated and, thus, inactive LuxO. Under these conditions, the abundance of HapR is controlled at the transcriptional level by cyclic AMP receptor protein-cyclic AMP complex (18), RpoS (29), and possibly other (transcription) factors.
FIG. 7.
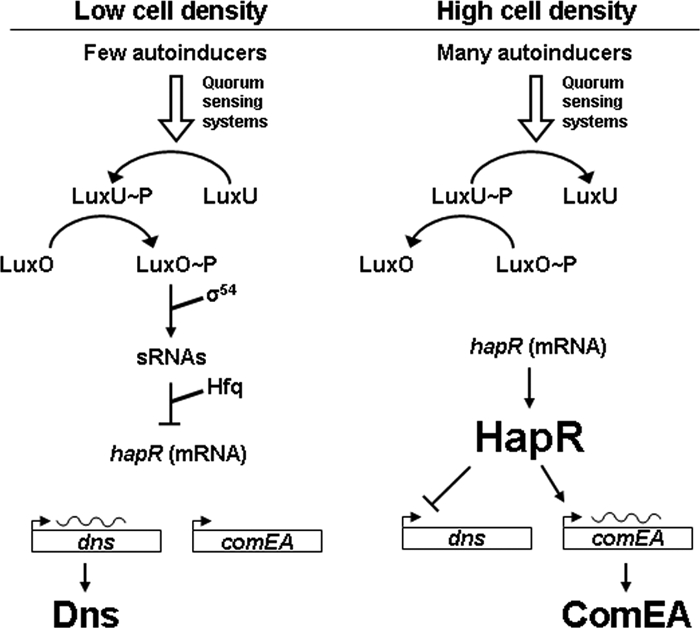
Model depicting the regulation of dns by quorum sensing (based on reference 34). At low cell density (left), few autoinducer molecules are present in the environment. This is sensed by the quorum-sensing systems (CqsS and LuxPQ; not shown), and phosphate is transferred via LuxU to LuxO. LuxO∼P is active in conjunction with RpoN (σ54) and inhibits the synthesis of the HapR regulator by inducing the expression of small regulatory RNAs (sRNAs) (posttranscriptional control). Under these conditions, the gene for the extracellular nuclease Dns is expressed. At high cell density (right), the phosphorylation cascade is reversed and LuxO is dephosphorylated. As a consequence, HapR accumulates, shutting down dns expression. HapR-mediated induction of comEA expression also occurs, provided that chitin is present (23).
Here, we explore how the production of Dns and Xds affects chitin-induced natural competence. We show that dns expression is repressed by increasing cell density via the quorum-sensing regulator HapR. This quorum-sensing-dependent regulatory scenario ensures substrate availability (DNA) at the onset of the competence process while allowing dns expression and DNA degradation to occur at low cell densities prior to induction of the competence phenotype.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The V. cholerae strains used in this study are listed in Table 1. Strains A1552Δdns, A1552Δxds, and derivatives of them were obtained using the gene disruption method described earlier (24), with the aid of the counterselectable plasmid pGP704-Sac28. The respective primer sequences for plasmid constructions are shown in Table 2. Escherichia coli DH5α was used as the host for cloning procedures, and SM10λpir and S17-1λpir were used to transfer plasmids from E. coli to V. cholerae by means of conjugation.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| A1552 | WT, O1 El Tor Inaba, Rifr | 37 |
| A1552Δdns | A1552ΔVC0470 | This study |
| A1552Δxds | A1552ΔVC2621 | This study |
| A1552ΔdnsΔxds | A1552ΔVC0470ΔVC2621 | This study |
| A1552ΔhapR (ATN140) | A1552ΔVC0583 | 23 |
| A1552ΔhapRΔdns | A1552ΔVC0583ΔVC0470 | This study |
| N16961 | N16961 (WT sequenced strain) | 15 |
| N16961Δdns | N16961ΔVC0470 | This study |
| FY_Vc_0003 (A1552ΔlacZ) | A1552ΔVC2338 (smooth) | 5 |
| A1552ΔluxO (ATN120) | A1552ΔVC1021 | 23 |
| A1552ΔluxOΔlacZ | A1552ΔVC1021ΔVC2338 | This study |
| A1552ΔhapRΔlacZ | A1552ΔVC0583ΔVC2338 | This study |
| Plasmids | ||
| pBR322 | Apr Tcr | 4 |
| pBR-Promo[dns]-lacZ | pBR322, −610 bp to ATG of VC0470 and lacZ | This study |
| pBR-Tet-VC0470-His | pBR322, Tet promoter, VC0470, and six-His tag sequence | This study |
| pBR-Promo-VC0470-His | pBR322, −610 bp and all of VC0470 and six-His tag sequence | This study |
Locus tags are given in accordance with reference 15.
TABLE 2.
Oligonucleotide primers used in this study
| Primer name | Sequence |
|---|---|
| VC0470 up for MCM67 | 5′-TTTTCTAGATGCCCAAGATGCAGATCGAGC-3′ |
| VC0470 up rev MCM79 | 5′-TGCCCGAACTGATGGGCAGA-3′ |
| VC0470 down for MCM92 | 5′-TCTGCCCATCAGTTCGGGCAAATCATCATAAAAGACGTAGAT-3′ |
| VC0470 down rev MCM105 | 5′-TTTCCATGGGAGCAGAAAATCAAACAATG-3′ |
| VC2621 up for MCM126 | 5′-TTTTCTAGAAAAGAAGCACAACTCGATCG-3′ |
| VC2621 up rev MCM138 | 5′-CGCCGTCGCTAGAAAGTG-3′ |
| VC2621 down for MCM150 | 5′-CACTTTCTAGCGACGGCGTGTTCTCATTTCCATGATGTAC-3′ |
| VC2621 down rev MCM162 | 5′-TTTCCATGGGTTAGATAGACCGATTCAGCA-3′ |
| VC0470-up-RBS | 5′-GGCAATTTATGAGTGTTAAACTGATAAAAAACC-3′ |
| VC0470-up-Promo#1 | 5′-GTTCGTTTTGATCTGCACTGCCAAGC-3′ |
| VC0470-His-down | 5′-TGCCCATCAATGATGATGATGATGATGGTTCGGGCATTGCTCACG-3′ |
| VC0470-Promo-lacZ#2 | 5′-CTCTACGGCGTACATAAAAGACGTAGATAAGTAGGTTTTTTATCAG-3′ |
| VC0470-Promo-lacZ#3 | 5′-TACGTCTTTTATGTACGCCGTAGAGCAAAGGCGTTATTGGCTTG-3′ |
| VC0470-Promo-lacZ#4 | 5′-TTATTGTGGGGATGACGCTTTCACACG-3′ |
| Dns-His-chrom-#2 | 5′-AGATTCTGCCCATCAATGATGATGATGATGATGGTTCGGGCATTGC-3′ |
| Dns-His-chrom-#3 | 5′-CATCATCATTGATGGGCAGAATCTCACCCTGCCCCCTTGTATTTTGC-3′ |
Media and growth conditions.
For transformation experiments, V. cholerae precultures were grown aerobically at 30°C in LB medium. After being washed in defined artificial seawater (DASW) medium, cells were resuspended in the same medium and used to inoculate chitinous crab shell fragments as described previously (3, 23). LB medium was used for all β-galactosidase experiments. The antibiotics kanamycin, ampicillin, and tetracycline were added to the medium at concentrations of 75, 100, and 12.5 μg ml−1, respectively.
Chitin-induced natural transformation.
Natural transformation frequencies were determined using chitin-containing crab shell surfaces as described previously (3, 23).
Time course experiment with chitin surfaces.
V. cholerae strains A1552 (wild type [WT]), A1552ΔluxO (ATN120), and A1552Δdns were grown as day cultures and used to inoculate crab shell fragments as described previously (23). Two micrograms of donor genomic DNA (gDNA) (VCXB21) was added immediately after inoculation or 2, 4, or 6 h later. Cells were grown on the chitin surfaces for a total of 24 h before harvesting and plating and before determination of the numbers of CFU.
DNA recovery experiment.
Bacteria were inoculated onto crab shell surfaces as described above for the time course experiments. Two micrograms of donor gDNA (VCXB21) was simultaneously added and incubated for 2 hours. Subsequently, supernatants were withdrawn, sterile filtered, and used for gDNA isolation (DNeasy kit; Qiagen). DNA incubated on the chitin surface in the absence of any bacteria was used as an internal standard.
Nuclease activity experiment.
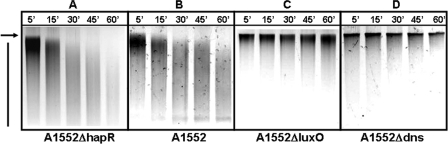
Bacteria were inoculated for 2 hours on chitin surfaces, whereupon supernatants were withdrawn, sterile filtered, and combined with 4 μg gDNA of strain VCXB21. Incubation took place between 5 and 60 min at 30°C (see Fig. 4). Subsequently, gDNA was recovered (DNeasy kit; Qiagen), and the quality of the DNA was scored on agarose gel.
FIG. 4.
The extracellular nuclease is active in vitro. Supernatants from 2-hour-old bacterial cultures on chitin surfaces were collected and tested for nuclease activity by adding 4 μg of gDNA. After incubation at 30°C for 5 to 60 min (as indicated at the top of the lanes), DNA was reisolated, and its quality was visualized on agarose gels. The bacterial cultures used were A1552ΔhapR (A), A1552 (WT) (B), A1552ΔluxO (C), and A1552Δdns (D). Undegraded (arrow) and degraded (bar) gDNA is indicated.
β-Galactosidase measurements.
β-Galactosidase activity of lacZ reporter fusions was measured by standard protocols, and results are given in Miller units (25).
Transformation in the absence of chitin (tfoX overexpression).
In order to induce chitin-independent transformation, tfoX, encoding one of the major positive regulators of competence, was overexpressed from plasmid pBAD-tfoX as described previously (23).
For time course experiments, strains were grown aerobically at 30°C in the presence of 0.2% arabinose. Duplicate samples were periodically obtained, supplemented with 2 μg donor gDNA (VCXB21), and incubated statically for 2 hours before resuming aerobic growth overnight. Kanr transformants were identified, and transformation frequencies were calculated as proportions of the total number of bacteria. Independent experiments were performed at least three times.
RESULTS
The extracellular nuclease Dns influences natural transformation.
V. cholerae produces two extracellular nucleases, Dns and Xds (12, 28). Both were shown to be secreted into the medium (12, 32) and therefore might have an effect on free DNA, the source for natural transformation. We constructed mutants with an in-frame deletion of dns (VC0470) or xds (VC2621). The growth of both mutants was similar to that of the WT parent in LB broth, and neither mutant showed an impaired biofilm phenotype in the microtiter plate assay (35) (data not shown). We further investigated the performances of these mutants in natural transformation experiments using the crab shell transformation assay system (23). As shown in Table 3, the transformation frequency of the dns mutant A1552Δdns was >2 orders of magnitude higher than that of the WT parent A1552. In contrast, a deletion of the xds gene increased the transformation frequency only ∼2.5-fold, and therefore, Xds was judged to be comparatively less important than Dns as a modulator of natural transformation. Consistent with the relative effects of each mutation, the transformation frequency for the double mutant (A1552ΔdnsΔxds) was found to be the product of each mutation (Table 3).
TABLE 3.
Transformation efficiencies of V. cholerae strains that are devoid of extracellular nucleases
| Strain | Genotype or description | Relative transformation frequency (±SD)a |
|---|---|---|
| A1552 | WT | 1b |
| A1552Δdns | ΔVC0470 | 339.4 (±179.4) |
| A1552Δxds | ΔVC2621 | 2.6 (±1.4) |
| A1552ΔdnsΔxds | ΔVC0470 ΔVC2621 | 910.2 (±399.8) |
Relative difference to WT transformation frequency; averages are of at least three independent experiments.
WT strain value set to 1; absolute transformation frequency equals 4.6 × 10−6.
To confirm that the transformation phenotype of the dns mutant was due to the intended gene disruption, the hypertransformable mutant A1552Δdns was evaluated by complementation experiments. Two plasmids were constructed with pBR322 (4) as vectors: pBR-Tet-VC0470-His expressing dns constitutively from the vector-encoded tetracycline resistance promoter and pBR-Promo-VC0470-His harboring dns behind its indigenous promoter. These plasmids were transferred into V. cholerae A1552Δdns, and each complemented mutant was scored for its transformation frequency during growth on chitin surfaces (Fig. 1). Vector controls of WT A1552 and the dns knockout were performed to exclude the effects of either the vector per se or ampicillin, which was used as the plasmid-retaining antibiotic in these experiments (Fig. 1, lanes 2 and 4). Both of the dns-containing plasmids complemented the A1552Δdns mutant (Fig. 1, lanes 5 and 6), although differences in the effects of the two promoters driving dns expression were evident. No transformation was detected when dns was constitutively expressed (Fig. 1, lane 5). In contrast, although transformants were detected, the transformation frequency was reduced by about 4 orders of magnitude when dns expression from the multicopy plasmid was driven by its own promoter (Fig. 1, lane 6) in comparison to the vector control (Fig. 1, lane 4).
FIG. 1.
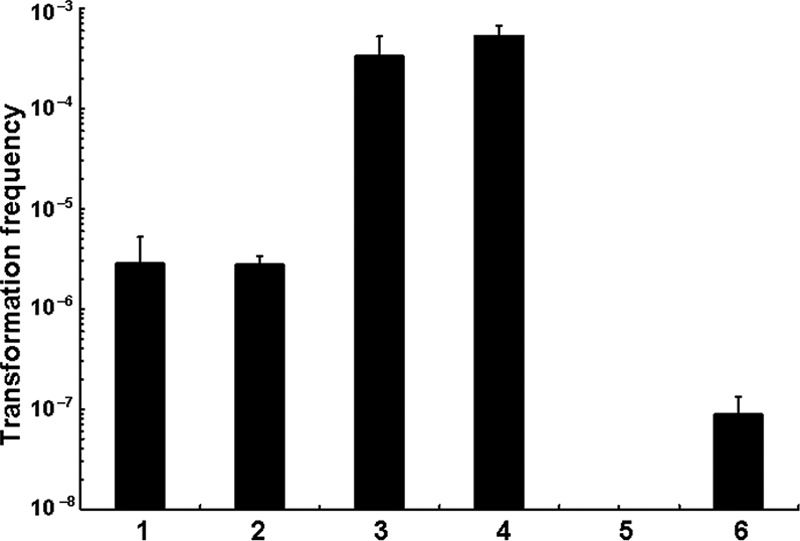
The dns-negative mutant is hypertransformable. The frequencies of chitin-induced transformation of a dns mutant and complemented strains were determined. Lane 1, A1552 (WT); lane 2, A1552/pBR322 (vector control); lane 3, A1552Δdns; lane 4, A1552Δdns/pBR322 (vector control); lane 5, A1552Δdns/pBR-Tet-VC0470-His (constitutive dns expression); lane 6, A1552Δdns/pBR-Promo-VC0470-His (indigenous promoter of dns). Results are from three or more independent experiments.
The availability of transforming DNA is regulated by quorum sensing.
Microarray expression data from our group and others indicated that dns expression is negatively regulated by HapR (23, 36) and thus led us to test whether dns expression is regulated in a cell density-dependent manner.
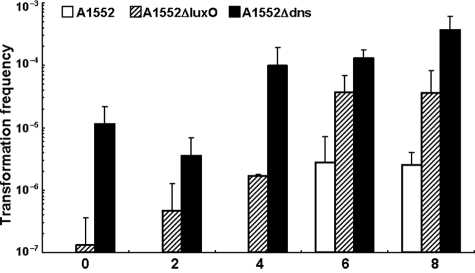
We reasoned that Dns may be selectively produced and secreted and thus able to degrade free DNA at low cell densities. If so, this could account in part for the low transformation frequencies observed early in the time course of low-density V. cholerae cultures growing on chitin-containing crab shells (23). In contrast, later in the time course, as cell densities increase, production of HapR occurs and would negatively regulate dns expression. As a consequence, the decreased production and secretion of Dns would permit the uptake of intact DNA by competent cells. To test this idea, we performed a time course experiment using chitin surfaces submerged in DASW medium and inoculated them either with the WT parent A1552 or with one of two mutants, A1552ΔluxO and A1552Δdns (Fig. 2). Donor gDNA, provided as a source of transforming DNA, was added between 0 and 8 h after inoculation, a time course that corresponds to a progressive increase in cell density. Figure 2 shows that the WT strain, which has an intact quorum-sensing system, is unable to become transformed by gDNA added under low-cell-density conditions (i.e., at 0, 2, and 4 h after inoculation). In contrast, transformants of the quorum-sensing defective strain A1552ΔluxO (Fig. 2) were detected even if gDNA was added at lower cell densities. Based on published expression profiling studies of a V. cholerae luxO mutant (39), we believe that the increased transformation frequency of the A1552ΔluxO mutant at low cell densities is due to the accumulation of the hapR transcript earlier in the time course than is the case for the WT parent. In turn, increased levels of HapR in the A1552ΔluxO mutant would negatively regulate dns expression, thus allowing transformation to occur earlier in the time course of the assay when cell density is still low. This prediction was tested by studying the dns mutant A1552Δdns (Fig. 2), which shows markedly less cell density-dependent transformability than the WT parent.
FIG. 2.
Cell density-dependent natural transformation. Transformation frequencies (y axis) were determined on chitin surfaces with donor gDNA (2 μg) added at 0, 2, 4, 6, and 8 h after inoculation, as indicated on the x axis. Results are from three independent experiments.
Extracellular DNA is degraded at low cell density.
The foregoing results are consistent with the notion that free, exogenously added gDNA is degraded by Dns secreted by the WT parent at low cell density, thus rendering this gDNA unavailable for transformation. As one test of this idea, we performed transformation experiments using DNA recovered from low-cell-density conditions. Chitin surfaces were inoculated with the WT parent A1552 or with the mutant A1552ΔluxO, A1552Δdns, or A1552ΔhapR, followed immediately by the addition of donor gDNA. After 2 hours of incubation (when cell density is low), the supernatant from each strain was removed, sterile filtered, and mixed with an “indicator strain” that had been pregrown to high density for 18 h. Then, the number of transformants was enumerated. In these experiments, we never obtained transformants using donor gDNA recovered from the supernatant of the WT parent or from the hapR mutant. In contrast, transformants were obtained in two of four experiments using donor gDNA recovered from the supernatant of A1552ΔluxO. More strikingly, gDNA recovered from the supernatant of the nuclease deletion strain A1552Δdns always yielded a positive transformation phenotype when tested by the competent indicator strain. Taken together, these results suggest that Dns is secreted by the WT parent and degrades transforming DNA under low-cell-density conditions of growth.
To directly test whether extracellular DNA is degraded under low-cell-density conditions in a dns- and quorum-sensing-dependent manner, we modified the DNA recovery experiment as described below. The WT strain A1552 was inoculated onto chitin surfaces with 2 μg of donor gDNA. After 2 h of incubation, the gDNA was reisolated from the supernatant, and the size range of the recovered DNA was analyzed by agarose gel electrophoresis (Fig. 3). Within the first 2 hours of growth (a low-cell-density condition), donor gDNA was found to have been partially degraded (Fig. 3, lanes 2) by the WT strain, thus explaining in part why transformation is undetectable under this condition. To determine whether this DNA degradation phenotype is dns dependent, the same experimental system was used to study the DNA-degrading capacity of A1552Δdns, which does not produce the extracellular nuclease Dns but harbors an intact xds gene. No DNA degradation was apparent (Fig. 3, lane 3). If dns is negatively regulated by HapR, we reasoned that disruption of hapR should lead to sustained production of Dns and to increased DNA degradation. This prediction was confirmed by the demonstration that A1552ΔhapR completely degraded high-molecular-weight DNA (Fig. 3, lane 5). The DNA-degrading phenotype of the luxO mutant (Fig. 3, lane 4) varied slightly between experimental replicates: it either partially degraded DNA in a manner comparable to the WT parent (Fig. 3, lane 2), or it did not significantly degrade DNA in a manner comparable to the dns mutant (Fig. 3, lane 3, and data not shown). The cause of variation in the luxO mutant's DNA-degrading phenotype was not further investigated.
FIG. 3.
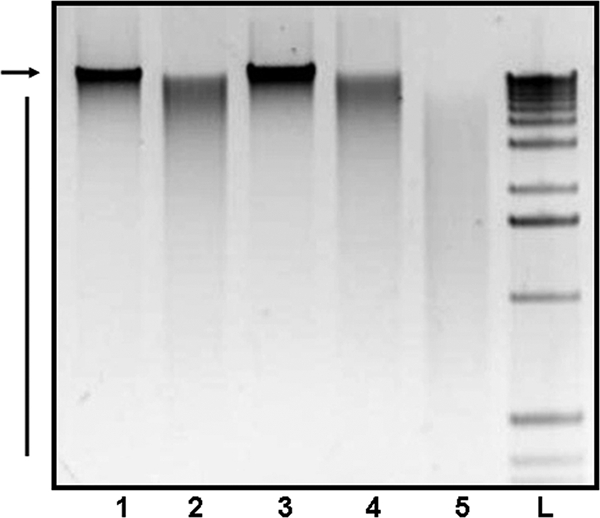
The nuclease Dns degrades extracellular DNA. Crab shell fragments immersed in DASW medium were inoculated without bacteria (control; lane 1) or with V. cholerae A1552 (WT; lane 2), A1552Δdns (lane 3), A1552ΔluxO (lane 4), and A1552ΔhapR (lane 5), respectively. Donor gDNA (2 μg) was added immediately at the time of inoculation; DNA was recovered from the supernatant after a 2-hour incubation period and visualized on an agarose gel. Undegraded donor gDNA (arrow) and degradation products (bar) are shown. Lane L, 1-kb DNA ladder (Invitrogen).
Extracellular nuclease activity is abundant in the supernatants of low-cell-density cultures on a chitin surface.
In the experiments depicted in Fig. 3, DNA-degrading activity was measured by introducing gDNA into a V. cholerae culture growing on a chitin surface. These experiments clearly showed that such cultures have DNA-degrading activity and that this activity varies as a function of mutations in dns and in genes in the quorum-sensing system (hapR and luxO). However, the experimental design we used for those experiments was not able to determine whether the DNA-degrading activity is cell surface- or chitin surface associated or instead is released into the culture supernatant. Nor did it determine whether the presence of exogenous DNA was required to induce the production and secretion of the DNA-degrading activity. To address these questions, we performed an experiment similar to the one shown in Fig. 3 but without the addition of donor gDNA to the culture. Instead, the supernatant of chitin-grown cultures at low cell density was tested for nuclease activity (Fig. 4). The degradation of high-molecular-weight DNA was evident in the culture supernatant from the WT parent strain (Fig. 4B). In contrast, apparently intact high-molecular-weight gDNA was identified in the supernatant of the dns mutant A1552Δdns. Taken together, these results indicate that Dns, but not Xds, is very likely responsible for the DNA-degrading activity seen in Fig. 4B, that it is secreted rather than surface associated (as previously shown by Focareta and Manning [12]), and that its production and secretion neither require nor are induced by exogenous gDNA. The highest degree of DNA degradation was again apparent for the supernatant of the hapR deletion mutant (Fig. 4A), whereas the DNA-degrading activity was almost abolished in the A1552ΔluxO mutant (Fig. 4C).
Expression of dns is regulated at the transcriptional level by HapR.
From the experiments described above, we concluded the following: exogenously added gDNA is extensively degraded by a hapR deletion V. cholerae mutant, and this mutant produces abundant extracellular nuclease activity that can be demonstrated in culture supernatants. We were curious to know whether this HapR-dependent regulation of dns occurs at the transcriptional level. For this purpose, we constructed a transcriptional reporter fusion of the dns promoter region and lacZ and thus could correlate the level of dns transcription with β-galactosidase activity measured at mid-exponential phase (Fig. 5). Significantly higher β-galactosidase activity was detected when this dns transcriptional reporter was introduced into the hapR mutant (Fig. 5, lane 1) than in the WT parent (Fig. 5, lane 2), clearly indicating that the HapR repression of dns is relieved in this mutant and that HapR negatively regulates dns expression at the transcriptional level. The lowest level of β-galactosidase activity was detected when the dns transcriptional reporter was harbored in the A1552ΔluxO mutant (Fig. 5, lane 3), a result that is consistent with the luxO-dependent destabilization of hapR mRNA. As a consequence, the luxO mutant produces more HapR and therefore more negatively regulates dns expression. However, in the absence of HapR, no repression occurs (Fig. 5, lane 1).
FIG. 5.
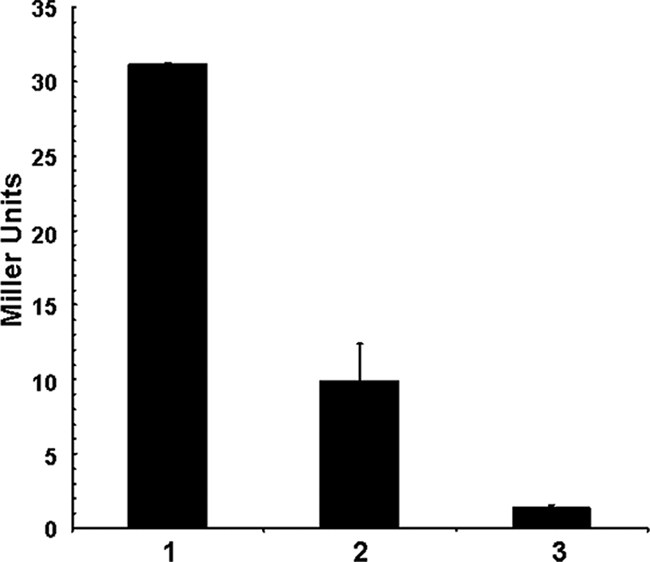
Quorum-sensing-dependent expression of dns. A transcriptional fusion between the promoter of dns (VC0470) and lacZ was constructed (plasmid pBR-Promo[dns]-lacZ) and transferred into strains A1552ΔhapRΔlacZ (lane 1), A1552ΔlacZ (FY_Vc_0003; lane 2), and A1552ΔluxOΔlacZ (lane 3). The bacterial strains were grown in LB medium until they reached an OD600 of ∼0.8. dns expression is reflected by β-galactosidase activity (given in Miller units).
The dns deletion can partly restore the transformability of an hapR-negative mutant and of a WT strain with a defective hapR gene.
We recently showed that hapR-negative WT V. cholerae strains with an inactive or defective variant of hapR are not naturally transformable (Table 4) (23). From the results presented above, we hypothesized that this effect is due largely to the degradation of the added gDNA by failure of these hapR variants to repress dns expression, thus resulting in the sustained production of Dns and the degradation of free DNA. However, because HapR regulates a variety of other important functions, it was not clear whether the disruption of hapR abolished competence solely because of continued expression of dns or whether it positively regulates additional genes that are critical for the competence phenotype like comEA (renamed from VC1917) (23), which is predicted to encode a periplasmic DNA binding protein. To address this question, we constructed a double knockout mutant of hapR and dns and tested its transformation phenotype. As shown in Table 4, the double mutant A1552ΔhapRΔdns regained transformability with a frequency in the range of that of the WT strain A1552. However, the transformation frequency of this double mutant is around 2 orders of magnitude lower than A1552Δdns (Table 4), which retains a functional hapR gene. The greater transformation frequency of the A1552Δdns mutant than that of the A1552ΔhapRΔdns mutant is probably caused by a lack of HapR-dependent comEA expression in the double mutant. Support for this explanation comes from experiments showing that a V. cholerae comEA deletion mutant is markedly impaired in its transformation phenotype, i.e., either below the detection limit of the assay or completely transformation negative (23). Moreover, in Neisseria gonorrhoeae, mutation of an orthologous comEA was associated with a 4 × 104-fold reduction in transformation frequency (6). Thus, even though ComEA appears to increase the efficiencies of DNA uptake and concomitantly transformation, it might not be completely essential. The same holds true for the first sequenced V. cholerae strain, N16961 (15), which was also naturally transformable once dns was deleted from the large chromosome (Table 4). This strain is known to possess a frameshift mutation within the hapR gene (39), and we showed earlier that it can also be complemented for natural transformation by the functional hapR gene of the WT strain used in this study (A1552) (23). Thus, although HapR's positive regulation of comEA contributes to the transformation phenotype, these data show that the principal reason that transformability is lost in hapR mutants is the failure of HapR-dependent repression of dns as a function of increasing cell density.
TABLE 4.
Transformation efficiencies of V. cholerae dns deletion strains
| Strain | Genotype or description | Relative transformation frequency (±SD)a |
|---|---|---|
| Crab shell associated | ||
| A1552 | WT | 1b |
| A1552Δdns | ΔVC0470 | 339.4 (±179.4) |
| A1552ΔhapR | ΔVC0583 | BDc |
| A1552ΔhapRΔdns | ΔVC0583 ΔVC0470 | 2.5 (±1.5) |
| N16961 | First sequenced WT strain (15) | BDc |
| N16961Δdns | ΔVC0470 | 1.5 (±1.2) |
| tfoX overexpressing | ||
| A1552/pBAD-tfoX | Parental strain; tfoX overexpression | 1d |
| A1552Δdns/pBAD-tfoX | ΔVC0470; tfoX overexpression | 166 (±99) |
| A1552ΔhapR/pBAD-tfoX | ΔVC0583; tfoX overexpression | BDe |
| A1552ΔhapRΔdns/pBAD-tfoX | ΔVC0583 ΔVC0470; tfoX overexpression | 1.9 (±2) |
Relative difference to WT transformation frequency; averages are of three independent experiments. BD, below detection limit.
WT strain value set to 1; absolute transformation frequency equals 4.6 × 10−6.
Transformation is below the detection limit of 6.7 × 10−8.
Parental strain A1552/pBAD-tfoX value set to 1; absolute transformation frequency equals 1.5 × 10−7.
Transformation is below the detection limit of 4.0 × 10−9.
The role of Dns in natural transformation is biofilm independent.
Because these experiments were performed on chitin surfaces, the transformation phenotypes of the mutants reported above could, in principle, be due directly to the role of the product of the disrupted gene or, alternatively, could be secondary to an effect of the mutation on some aspect of biofilm development or structure, as previously reported for hapR deletion strains (39). To address this issue, we used the tfoX overexpressing transformation method described earlier (23) to study the effect of these mutations on transformation frequency in stirred, homogeneous liquid cultures. In the experiments described below, we used arabinose added to stirred cultures to induce tfoX, the gene encoding the major regulator of competence. Using this experimental system, competence could be induced in the absence of chitin or in a chitin-containing substratum (23). We overexpressed tfoX in the WT parent and in each of the following mutants: A1552Δdns, A1552ΔhapR, and A1552ΔhapRΔdns. As shown in Table 4, the transformation frequency of the nuclease deletion mutant again is 2 orders of magnitude higher than that of the parental strain. No transformants were detected for the hapR-negative mutant, and transformation could be partly restored in this strain by deleting dns in addition to hapR (Table 4).
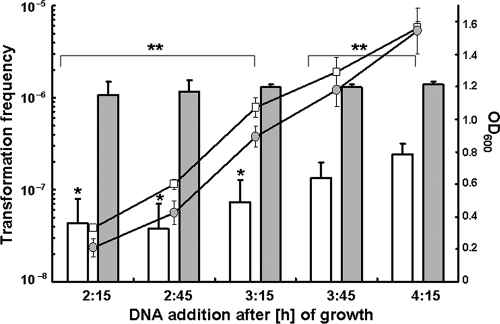
We further used this tfoX overexpression experimental system for a time course experiment in which donor gDNA is provided at different cell densities (Fig. 6). In contrast to the chitin surface biofilm system described in Fig. 2, this setup not only allows transformation to occur independently of chitin availability and biofilm formation, but in addition, cell density can be directly determined by measuring the optical density at 600 nm (OD600) of the liquid culture. We found that the transformation frequency of the parental strain A1552/pBAD-tfoX is significantly lower if the donor gDNA is added at low cell density (three earliest time points) than at high cell density (two latest time points) (Fig. 6). In contrast, the transformation efficiency of the dns deletion mutant is the same at low (OD600 of 0.2), intermediate (OD600 of 0.9), and high (OD600 of 1.6) cell densities (Fig. 6).
FIG. 6.
Cell density-dependent degradation of extracellular DNA is a result of the nuclease Dns. Bacterial strains A1552/pBAD-tfoX (parental strain; open bars) and A1552Δdns/pBAD-tfoX (nuclease deletion mutant; filled bars) were rendered competent by artificially inducing tfoX. Samples were taken at the time points indicated on the x axis and checked for their OD600 (secondary y axis) and their frequency of transformation (primary y axis). The results are from three independent experiments. Student's t test results: *, statistically significant difference relative to latest time point at 4:15 (P < 0.05); **, statistically significant difference between early time points (2:15, 2:45, 3:15) and late time points (3:45, 4:15) (P < 0.001).
DISCUSSION
V. cholerae becomes naturally competent upon exposure to chitin surfaces (23) and thus able to internalize free DNA in the environment. Recognizing that the extracellular nucleases produced by Vibrio have the capacity to degrade free DNA and thus render it unsuitable for transformation, we examined the relationship between natural competence and nuclease production and how these are regulated as a function of time and cell density. We showed that V. cholerae strains devoid of dns exhibit a significant increase in transformation frequency (>2 orders of magnitude). Thus, Dns hinders natural transformation in a cell density-dependent manner, while Xds had no such effect. This indicates that the downregulation of dns is crucial for the uptake of undegraded free DNA by competent V. cholerae cells. Through the study of mutants in the quorum-sensing system, we were able to demonstrate that dns is negatively regulated in a quorum-sensing-dependent manner by HapR acting as a repressor of dns transcription (Fig. 7). Because HapR is produced only at high cell densities (as the abundance of hapR mRNA is posttranscriptionally decreased at low cell densities), V. cholerae switches from the production of Dns early in the time course when cell densities are low to the termination of Dns production later in the time course when cell densities are high. Because this switch is both cell density dependent and HapR mediated, it was informative to examine the regulation and role in natural transformation of another gene, comEA (VC1917), which appears to be controlled by the same conditions and regulator. ComEA, a putative periplasmic DNA binding protein that is required for transformation in V. cholerae (23), is also regulated in a cell density-dependent manner by HapR but in the opposite direction. That is, in contrast to dns, comEA is positively regulated by HapR when cell density increases. The counterregulation of dns and comEA by the same condition (high cell density) and regulator (HapR) ensures that the availability of nondegraded extracellular DNA is coordinated with the presence of the periplasmic DNA binding protein ComEA (Fig. 7).
The results of this study may begin to disclose how V. cholerae growth, V. cholerae DNA utilization as a nutrient, and natural transformation are integrated in aquatic habitats. In marine ecosystems, high-molecular-weight extracellular DNA is present in considerable quantities, and turnover times range from 6 to 28 h (7, 8, 21, 30, 31). DNA-degrading microorganisms can be readily isolated and reach concentrations of up to 105 DNA-hydrolyzing bacteria per ml of seawater (22).
V. cholerae, autochthonous in aquatic habitats of this kind, degrades free DNA by producing two extracellular nucleases, Dns and Xds (11, 12, 28). Beyond their capacity to degrade DNA, the roles of these nucleases in the ecology and pathogenesis of V. cholerae are largely unexplored. Focareta and Manning investigated the possibility that Dns and Xds might facilitate efficient colonization by degrading DNA-rich, viscous mucus in the small intestine (11). However, the virulence of the respective nuclease deletion mutants was not reduced in comparison to that of the WT in an infant-mouse cholera model (11). Taken together, these observations suggest that neither dns nor xds is likely to have a role in the pathogenesis of cholera. This conclusion is supported by the presence of both dns and xds in two sequenced environmental isolates of V. cholerae, RC385 and 2740-80 (>98% identical; obtained from shotgun sequences provided by the Vibrio Genome Project [www.tigr.org]). Further study of these genome sequences shows that both environmental isolates lack the gene for cholera toxin, an essential virulence determinant of pathogenic clones of V. cholerae. The absence of this virulence factor in environmental isolates that retain the dns and xds genes indicates that the extracellular nucleases they encode likely have a role in the fitness of V. cholerae in the aquatic habitats from which these environmental strains were isolated.
One such nonpathogenic role could be nutrient acquisition, since degraded DNA can be used as a source of carbon, nitrogen, phosphorous, and nucleotides. This idea is supported by the findings of Maeda and Taga, who used an isolated marine Vibrio strain to show the following results. (i) The addition of DNA to cultures in seawater supplemented with amino acids stimulated growth. (ii) DNA was hydrolyzed, releasing guanine and thymine into the medium, whereas cytosine was assimilated by the bacteria. (iii) Inorganic phosphate was first released into the medium and later taken up by the bacteria (22). Thus, some of the released phosphate could enter the phosphate cycle in marine ecosystems (9) and the remainder could be assimilated, together with other DNA-derived nutrients, for growth.
We did not directly evaluate the role of DNA as a nutrient source. However, in our experimental system, which used a chitin surface to grow V. cholerae in nutrient-free seawater, entry into log-phase growth occurred and coincided with Dns production. From this observation, it seems likely that during bursts of rapid replication, such as those that occur in chitin-associated biofilms, it will be necessary to replenish the nucleotide pool either by de novo synthesis, which is an energetically costly multistep biosynthetic pathway, or by scavenging nucleotides from external sources. Thus, the combination of chitin or other nutrient sources in aquatic habitats, high environmental concentrations of DNA, and nuclease secretion during periods of rapid growth may account, in part, for the capacity of vibrios to grow rapidly when conditions are favorable (10).
Our observation that dns is downregulated prior to the onset of transformation indicates that both nuclease production and natural competence can coexist in the same strain, provided that they are expressed at different times. This idea is reinforced by Lorenz et al., who, in studies of the naturally competent bacterium Bacillus subtilis, showed that nuclease secretion precedes induction of DNA secretion and natural competence in that species (20). On balance, the results of our studies and those of Lorenz et al. (20) would seem to favor a nutrient acquisition role for the extracellular nucleases in naturally competent bacteria. The apparent paradox that some competent bacteria also secrete DNA-degrading enzymes is resolved by the recognition that while the same strain may have both capacities, they are deployed at different times. That this is orchestrated in V. cholerae by the same quorum-sensing regulatory circuit is remarkable, elegant in its simplicity, and a reflection of the evolutionary processes that shaped this system.
Acknowledgments
We thank Cheng-Yen Wu and Michael Miller for the construction of the nuclease deletion strains, and we are grateful to Karin Meibom for initial discussions.
This work was supported by grants to G.K.S. from the Ellison Foundation, the National Institutes of Health (AI053706), an Environmental Venture Project grant from the Stanford Institute for the Environment, and a research fellowship from the German Research Foundation (BL 786/1-1) and the Stanford University School of Medicine Dean's Fellowship Award to M.B.
Footnotes
Published ahead of print on 29 August 2008.
REFERENCES
- 1.Altermark, B., L. Niiranen, N. P. Willassen, A. O. Smalas, and E. Moe. 2007. Comparative studies of endonuclease I from cold-adapted Vibrio salmonicida and mesophilic Vibrio cholerae. FEBS J. 274252-263. [DOI] [PubMed] [Google Scholar]
- 2.Altermark, B., A. O. Smalas, N. P. Willassen, and R. Helland. 2006. The structure of Vibrio cholerae extracellular endonuclease I reveals the presence of a buried chloride ion. Acta Crystallogr. D 621387-1391. [DOI] [PubMed] [Google Scholar]
- 3.Blokesch, M., and G. K. Schoolnik. 2007. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 3E81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, H. W. Boyer, J. Crosa, and S. Falkow. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 295-113. [PubMed] [Google Scholar]
- 5.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 1861574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, I., and E. C. Gotschlich. 2001. ComE, a competence protein from Neisseria gonorrhoeae with DNA-binding activity. J. Bacteriol. 1833160-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeFlaun, M. F., J. H. Paul, and D. Davis. 1986. Simplified method for dissolved DNA determination in aquatic environments. Appl. Environ. Microbiol. 52654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeFlaun, M. F., J. H. Paul, and W. H. Jeffrey. 1987. Distribution and molecular weight of dissolved DNA in subtropical estuarine and oceanic environments. Mar. Ecol. Prog. Ser. 3865-73. [Google Scholar]
- 9.Dell'Anno, A., and C. Corinaldesi. 2004. Degradation and turnover of extracellular DNA in marine sediments: ecological and methodological considerations. Appl. Environ. Microbiol. 704384-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eagon, R. G. 1962. Pseudomonas natriegens, a marine bacterium with a generation time of less than 10 minutes. J. Bacteriol. 83736-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Focareta, T., and P. A. Manning. 1991. Distinguishing between the extracellular DNases of Vibrio cholerae and development of a transformation system. Mol. Microbiol. 52547-2555. [DOI] [PubMed] [Google Scholar]
- 12.Focareta, T., and P. A. Manning. 1987. Extracellular proteins of Vibrio cholerae: molecular cloning, nucleotide sequence and characterization of the deoxyribonuclease (DNase) together with its periplasmic localization in Escherichia coli K-12. Gene 5331-40. [DOI] [PubMed] [Google Scholar]
- 13.Focareta, T., and P. A. Manning. 1991. Genetic analysis of the export of an extracellular DNase of Vibrio cholerae using DNase-beta-lactamase fusions. Gene 10831-37. [DOI] [PubMed] [Google Scholar]
- 14.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50101-104. [DOI] [PubMed] [Google Scholar]
- 15.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 11869-82. [DOI] [PubMed] [Google Scholar]
- 17.Li, C. L., L. I. Hor, Z. F. Chang, L. C. Tsai, W. Z. Yang, and H. S. Yuan. 2003. DNA binding and cleavage by the periplasmic nuclease Vvn: a novel structure with a known active site. EMBO J. 224014-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang, W., A. Pascual-Montano, A. J. Silva, and J. A. Benitez. 2007. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology 1532964-2975. [DOI] [PubMed] [Google Scholar]
- 19.Lipp, E. K., A. Huq, and R. R. Colwell. 2002. Effects of global climate on infectious disease: the cholera model. Clin. Microbiol. Rev. 15757-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorenz, M. G., D. Gerjets, and W. Wackernagel. 1991. Release of transforming plasmid and chromosomal DNA from two cultured soil bacteria. Arch. Microbiol. 156319-326. [DOI] [PubMed] [Google Scholar]
- 21.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda, M., and N. Taga. 1974. Occurrence and distribution of deoxyribonucleic acid-hydrolyzing bacteria in sea water. J. Exp. Mar. Biol. Ecol. 14157-169. [Google Scholar]
- 23.Meibom, K. L., M. Blokesch, N. A. Dolganov, C.-Y. Wu, and G. K. Schoolnik. 2005. Chitin induces natural competence in Vibrio cholerae. Science 3101824-1827. [DOI] [PubMed] [Google Scholar]
- 24.Meibom, K. L., X. B. Li, A. T. Nielsen, C. Y. Wu, S. Roseman, and G. K. Schoolnik. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. USA 1012524-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 26.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110303-314. [DOI] [PubMed] [Google Scholar]
- 27.Miller, M. C., D. P. Keymer, A. Avelar, A. B. Boehm, and G. K. Schoolnik. 2007. Detection and transformation of genome segments that differ within a coastal population of Vibrio cholerae strains. Appl. Environ. Microbiol. 733695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newland, J. W., B. A. Green, J. Foulds, and R. K. Holmes. 1985. Cloning of extracellular DNase and construction of a DNase-negative strain of Vibrio cholerae. Infect. Immun. 47691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen, A. T., N. A. Dolganov, G. Otto, M. C. Miller, C. Y. Wu, and G. K. Schoolnik. 2006. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2E109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul, J. H., M. F. DeFlaun, and W. H. Jeffrey. 1988. Mechanisms of DNA utilization by estuarine microbial populations. Appl. Environ. Microbiol. 541682-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul, J. H., W. H. Jeffrey, and M. F. DeFlaun. 1987. Dynamics of extracellular DNA in the marine environment. Appl. Environ. Microbiol. 53170-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pukatzki, S., A. T. Ma, D. Sturtevant, B. Krastins, D. Sarracino, W. C. Nelson, J. F. Heidelberg, and J. J. Mekalanos. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 1031528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamplin, M. L., A. L. Gauzens, A. Huq, D. A. Sack, and R. R. Colwell. 1990. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 561977-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21319-346. [DOI] [PubMed] [Google Scholar]
- 35.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53497-515. [DOI] [PubMed] [Google Scholar]
- 37.Yildiz, F. H., and G. K. Schoolnik. 1998. Role of rpoS in stress survival and virulence of Vibrio cholerae. J. Bacteriol. 180773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5647-656. [DOI] [PubMed] [Google Scholar]
- 39.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 993129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]